Abstract
Human MITF is, by convention, called the “microphthalmia-associated transcription factor” because of previously published seminal mouse genetic studies; however, mutations in MITF have never been associated with microphthalmia in humans. Here, we describe a syndrome that we term COMMAD, characterized by coloboma, osteopetrosis, microphthalmia, macrocephaly, albinism, and deafness. COMMAD is associated with biallelic MITF mutant alleles and hence suggests a role for MITF in regulating processes such as optic-fissure closure and bone development or homeostasis, which go beyond what is usually seen in individuals carrying monoallelic MITF mutations.
Main Text
Mouse Mitf encodes a basic helix-loop-helix zipper protein critical for the development of neural-crest-derived melanocytes, neuroectoderm-derived retinal pigment epithelium (RPE) cells, and hematopoietic-tissue-derived osteoclasts and mast cells. Autosomal-dominant MITF mutations are associated with two highly overlapping deafness and pigmentation disorders: Waardenburg syndrome type 2A (WS2A [MIM: 193510])1 and Tietz syndrome (MIM: 103500).2 Congenital pigmentation defects and sensorineural deafness are attributed to the role of MITF in differentiation and survival of melanocytes in skin and stria vascularis of the cochlea, respectively.3 Autosomal-recessive or compound-heterozygous inheritance of MITF has not been reported previously in humans. Here, we describe two unrelated individuals with compound-heterozygous MITF mutations resulting in a complex phenotype that we term COMMAD (coloboma, osteopetrosis, microphthalmia, macrocephaly, albinism, and deafness) and investigate the underlying molecular mechanisms. Biochemical and functional data for one of the probands demonstrate that mutations do not affect dimerization of MITF with other MiT family transcription factors but rather alter nuclear migration and DNA binding of homo- and heterodimers and thus allow the mutant alleles to act as dominant negative. These observations are in agreement with those of previous studies on the Mitfmi/mi mouse model, where homozygosity of the dominant-negative mi allele causes a similar phenotype.4, 5
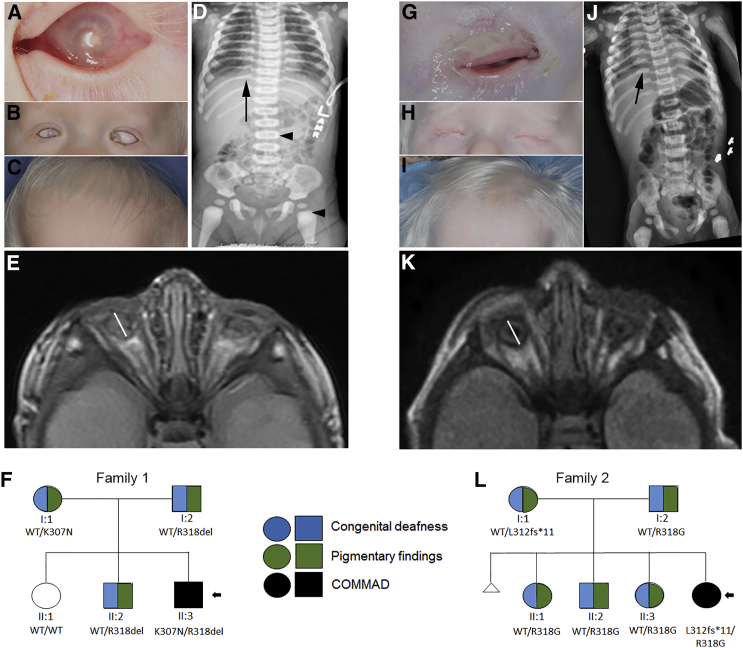
At last examination, proband I was a 5-year, 3-month-old male with colobomatous microphthalmia and microcornea with pannus, dense bilateral cataracts, translucent irides, profound congenital sensorineural hearing loss, and a lack of visible pigment in the hair, skin, and eyes (Figures 1A–1C). Microphthalmia was first detected on prenatal ultrasound. Head circumference was 56.0 cm (>3 SDs for age), consistent with macrocephaly, and weight (17.3 kg [−0.5 SD]) and height (110.0 cm [0.0 SD]) were normal for his age. He had facial dysmorphisms including frontal bossing, shallow orbits, preauricular pits, and posteriorly rotated ears. Skeletal features included a prominent frontal bone, diffuse expansion of the anterior ends of the ribs (Figure 1D, arrow), and bilateral fifth-finger clinodactyly (data not shown). A radiographic skeletal survey performed at 13 months of age showed osteopetrosis (Figure 1D, arrowheads depict areas of increased bone density). Axial magnetic resonance imaging (MRI) of the brain showed small eyes (∼7–8 mm, line on Figure 1E), optic nerves, and chiasm with mild prominence of ventricles, but no other structural abnormalities (Figure 1E). He was delivered at term after an uneventful pregnancy to non-consanguineous parents, both of whom have congenital sensorineural hearing loss, blue irides, fair skin, and premature graying of the hair and are in their third or fourth decade. One male sibling was affected similarly to his parents, and one sister was unaffected (Figure 1F).
Figure 1.
Clinical Features of COMMAD Syndrome
(A–K) COMMAD-affected probands I (A–E) and II (G–K) had microphthalmia and shallow orbits (A and G) with frontal bossing (B and H) and platinum hair (C and I). Additionally, osteopetrosis was noted, prominently in the anterior ribs (arrows) and femoral head (arrowheads) (D and J). Microphthalmia (8 mm line across optic globe) with associated optic-nerve and chiasm hypoplasia was confirmed on brain MRI (E and K), but other structures were normal.
(F and L) Pedigrees of family 1 (F) and family 2 (L).
At last exam, proband II was a 9-month-old female born with severe microphthalmia, profound congenital sensorineural hearing loss, and a lack of pigment in the hair, skin, and eyes (Figures 1G–1I). She had relative macrocephaly (43.0 cm [0 SD for age]), short stature (65.0 cm [−2 SDs]), and low weight (5.2 kg [<−3 SDs]). She had skeletal findings (Figure 1J) and craniofacial dysmorphisms similar to those of proband I, with the addition of micrognathia and wide palatine ridges. She had mild hypotonia throughout. Brain MRI revealed severe microphthalmia (globes 8 mm bilaterally, line on Figure 1K) and small optic nerves, as well as a cavum septum pellucidum et vergae variant and otherwise normal brain structures (Figure 1K). She was also born to non-consanguineous parents, both of whom have congenital sensorineural hearing loss, premature graying, blue irides, and fair skin. She has one male and two female siblings affected similarly to her parents (Figure 1L). Renal ultrasound was normal for both probands.
Interestingly, the phenotypes of probands I and II share homology with the phenotype of mice homozygous for the Mitf mi allele (Mitfmi/mi)4, 5 and are unlike that of any previously reported individuals with heterozygous MITF mutations.6, 7 Clinical diagnosis of Waardenburg syndrome type 2 in the parents and affected siblings prompted sequencing of MITF. Both families were previously unreported. DNA samples were collected after approval by the NIH institutional review board (protocol 06-EI-0169). Informed consent or assent for DNA storage, mutation analysis, and publication of images was obtained from all subjects. Sanger sequencing of the entire MITF coding sequence, including intron-exon boundaries, performed in a laboratory certified by the Clinical Laboratory Improvement Amendments, revealed two mutations in proband I: paternally inherited MITF mutation c.952_954delAGA (p.Arg318del in the isoform MITF-A [GenBank: NM_198159.2], corresponding to p.Arg217del in the isoform MITF-M [GenBank: NM_000248.1]), known to cause WS2A and Tietz syndrome;8 and a maternally inherited, previously unreported MITF mutation, c.921G>C (p.Lys307Asn in the isoform MITF-A). This latter mutation is not reported in 1000 Genomes, dbSNP, NHLBI Exome Sequencing Project (ESP) Exome Variant Server, or the Exome Aggregation Consortium (ExAC) Browser and was predicted to be deleterious with a CADD score of 26. Alteration of Lys307 to Gln, another polar amine-group amino acid, is associated with WS2A.9 A male sibling diagnosed with WS2A was heterozygous for the paternal allele (Figure 1F). The unaffected sister had neither mutation. Proband II was also observed to be compound heterozygous for the paternally inherited c.952A>G (p.Arg318Gly) missense mutation and maternally inherited c.938−1G>A, a canonical splice-site mutation resulting in a truncated protein (p.Leu312fs∗11). None of the affected individuals from both families displayed dystopia canthorum, consistent with a WS2A diagnosis, and none had osteopetrosis, macrocephaly, microphthalmia, or colobomata.
Heteroallelic combinations in Mitf mutant mice have been studied extensively, and only compound heterozygosity of dominant-negative alleles causes the most severe COMMAD-like phenotype. Because MITF p.Arg318del is a known dominant-negative variant, we next studied the functional impact of the c.921G>C (p.Lys307Asn) MITF mutation observed in proband I and his mother. The open reading frame of human MITF (catalog no. FXC03436, Promega; GenBank: NM_198159) was obtained, and isoforms MITF-A and MITF-D were amplified from it (for primer and probe sequences, see Table S1). All experiments were performed in the context of MITF-A (1,563 bp; GenBank: NP_937802.1), the most widely expressed isoform, as opposed to MITF-M (GenBank: NP_000239.1), on which most of the previous studies were based.10, 11 Experiments were repeated with MITF-D (1,407 bp), an isoform expressed only in RPE. For detailed methodology, refer to the Supplemental Data.
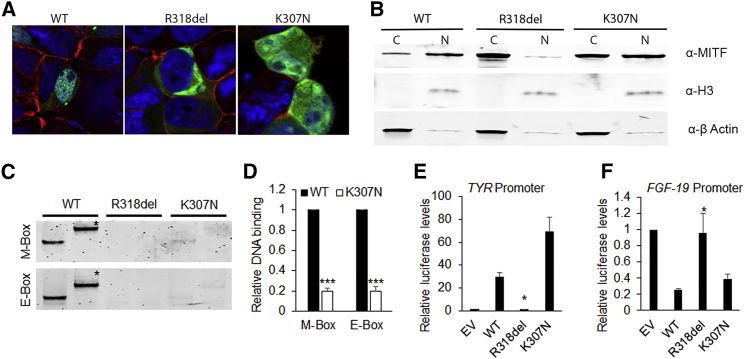
In our study, GFP-tagged wild-type (WT) MITF demonstrated predominantly nuclear localization (Figures 2A and 2B; Figure S1) and strong M-box (TCATGTG) and E-box (CACGTG) binding (Figures 2C and 2D; Figure S2). GFP-tagged MITF-A p.Arg318del mutant isoforms (Figures 2A and 2B; Figure S1) did not migrate to the nucleus in transfected HEK293 cells or bind consensus M-box or E-box DNA sequences in vitro (Figures 2C and 2D; Figure S2). This is most likely due to the fact that the positively charged arginine residue is a part of nuclear-localizing signal (ERRRRF) and is directly involved in interactions with the negatively charged DNA phosphate backbone (Figures S3A–S3C). This allele neither activated the tyrosinase (TYR [MIM: 606933]) promoter nor repressed the FGF19 (MIM: 603891) promoter in dual luciferase reporter assays (Figures 2E and 2F; Figures S4A and S4B). Previously, WS2A-associated p.Arg318del and p.Arg318Gly proteins were demonstrated to lack DNA binding or regulate target-gene promoters, despite reported nuclear localization of the corresponding human MITF-M.12 In comparison, the MITF-A p.Lys307Asn mutant distributed equally between the nucleus and the cytoplasm (Figures 2A and 2B; Figure S1), indicating a partial requirement for the charged p.Lys307 residue for nuclear translocation of the human MITF-A and MITF-D isoforms. Furthermore, compared with WT MITF, p.Lys307Asn MITF-A or MITF-D had only ∼20% DNA-binding ability in vitro, correlating with the loss of a positively charged amino acid (Figures 2C and 2D; Figures S2E and S2F). The p.Lys307Asn protein, nevertheless, had significant transcriptional regulatory potential on TYR and FGF19 promoters (Figures 2E and 2F; Figures S4A and S4B).
Figure 2.
Nuclear Migration, DNA Binding, and Promoter Regulatory Potential of WT and Mutant MITF-A Isoforms
HEK293 cells were cultured in DMEM with 10% FBS and 1% penicillin-streptomycin for 24 hr and then transiently transfected with fluorescent-protein-tagged wild-type (WT) or mutant MITF constructs with X-tremeGENE HP (Roche) according to the manufacturer’s instructions. Cells were fixed with 4% paraformaldehyde 24 hr after transfection, stained with Phalloidin (red) and DAPI, and photographed at 60× magnification with a confocal microscope.
(A) GFP-tagged WT MITF-A isoform was observed predominantly in the nucleus of cells. The p.Arg318del mutant was observed only in the cytoplasm, whereas p.Lys307Asn localized in both the cytoplasm and nucleus, suggesting some disruption of the nuclear migration ability (also see Movie S1).
(B) Subcellular localization of the mutants was further confirmed by western blot analysis of the cytoplasmic (C) and nuclear (N) fractions of cells transfected by WT and mutant MITF-A, in agreement with the fluorescence microscopy data. For protein blotting, cytoplasmic and nuclear fractions were separated with the Nuclear Extract Kit (Active Motif) according to the manufacturer’s instructions. 20 μg of protein (BCA Protein Assay Kit, Life Technologies) was separated by SDS-PAGE on polyacrylamide gels (Bio-Rad). Proteins were transferred onto polyvinylidene fluoride membranes (Bio-Rad) and probed with mouse monoclonal antibody to MITF (clone D5, Thermo Scientific). Rabbit polyclonal antibody to histone H3 and β-actin (Abcam) were used as loading controls. Membranes were then incubated with secondary antibodies (1:10,000; LI-COR) and then scanned with the Odyssey infrared scanner and analyzed with Image Studio Lite v.4.0 (LI-COR).
(C) Electrophoretic mobility shift assay (EMSA) was performed with DNA oligonucleotides harboring M-box and E-box consensus sequences (Table S2) labeled with IR700 dye (Integrated DNA Technologies) with the Odyssey Infrared EMSA Kit (LI-COR) as per the manufacturer’s instructions. WT and mutant MITF isoforms were in vitro translated with the SP6 TnT Quick Coupled Transcription/Translation System (Promega) as per the manufacturer’s instructions. 50 femtoM of duplex oligonucleotide was mixed with binding buffer (100 cmM Tris, 500 mM KCl, and 10 cmM DTT [pH 7.5]), poly(deoxyinosinic-deoxycytidylic), 1 M KCl, and the WT and/or mutant protein in vitro translation mix, incubated for 30 min at room temperature, and then separated on 5% TBE polyacrylamide gels (Bio-Rad) at 100V. The gels were scanned with an Odyssey scanner and analyzed with Image Studio Lite v.4.0 (LI-COR). M-box and E-box sequences showed strong DNA binding by WT MITF-A, no DNA binding by the p.Arg318del mutant, and significantly reduced binding by the p.Lys307Asn mutant. A supershift (∗), produced by MITF-specific antibody, confirmed the specificity of MITF and DNA binding (also see Figure S2).
(D) Quantification of the DNA binding by WT and p.Lys307Asn MITF-A, as calculated from three different trials.
(E and F) The Dual-Luciferase Reporter Assay System (Promega) was used to study the promoter regulatory potential of WT and mutant MITF-A isoforms. With X-tremeGENE HP (Roche), HEK239T cells (1 × 105 seeding density) were co-transfected 24 hr after seeding with 100 ng of MITF-A or -D isoforms, 200 ng of luciferase-promoter reporter construct, 1:1,000 Renilla luciferase vector (pRL Null Renilla), and empty vector to equalize the amount of transfected DNA. All transfections were performed in duplicate. Transfected cells were lysed 24 hr after transfection, and aliquots were used for determining firefly and Renilla luciferase activities in triplicate. Data were normalized to the activity of Renilla luciferase. All experiments were repeated three times. The WT and p.Lys307Asn mutant MITF-A activated the TYR promoter and repressed the FGF19 promoter. The p.Arg318del mutant showed no promoter regulatory potential. All comparisons are in reference to the WT MITF; error bars represent standard errors p values were calculated with the two-sided Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005.
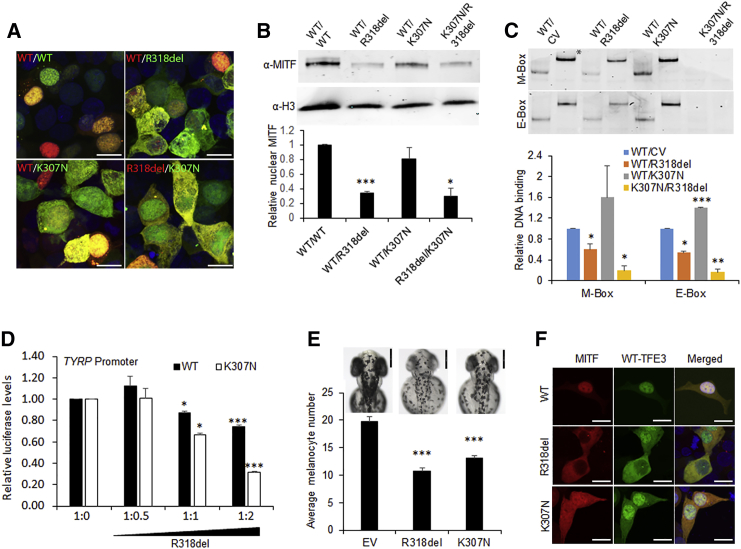
The mouse Mitf mi allele (similar to p.Arg318del of proband I) acts in a dominant-negative manner on MiT family transcription factors by dimerizing with them and interfering with their nuclear localization.11 Dominant-negative mutations in MITF typically affect either the DNA-binding domain or the transcriptional-activation domain, leaving the dimerization domains unaffected.10 Consistent with this observation, homozygosity of the Mitf or (oak ridge) allele (ERR[R/K]RF, proximal to ERRR[R/G]F of proband II) or Mitf wh allele (Ile212Asn) in mice results in some of the salient features of COMMAD, such as a white coat, osteopetrosis, and ocular malformations including microphthalmia and anophthalmia.5 To account for the clinical phenotype of the probands, we studied the interactions between p.Arg318del and p.Lys307Asn proteins, as well as their interaction with WT MITF. Our results showed that co-expression of p.Arg318del and p.Lys307Asn proteins in HEK293 cells resulted in migration of only 30% of MITF into the nucleus, whereas co-expression of the p.Arg318del or p.Lys307Asn mutant and the WT protein resulted in 36% or 81% migration, respectively (Figures 3A and 3B and Movie S1). Electrophoretic mobility shift assay (EMSA) showed that DNA binding of co-in-vitro-translated mutant proteins (p.Arg318del and p.Lys307Asn in equal amounts) was <20% of that of WT MITF for consensus E-box and M-box elements. A WT-p.Arg318del combination resulted in ∼50% reduction of DNA binding, whereas WT-p.Lys307Asn bound both consensus elements better than a WT-control-vector combination (Figure 3C). These results were further supported by in silico DNA-binding simulations (Figure S3D). Furthermore, transcriptional activation of the TYRP1 (MIM: 115501) promoter was reduced more dramatically when p.Lys307Asn was co-expressed with increasing amounts of p.Arg318del (Figure 3D, open bars) than when the WT protein was co-expressed with increasing amounts of p.Arg318del (Figure 3D, solid bars). Transcriptional repression of the FGF19 promoter by MITF was similarly impaired when p.Lys307Asn was co-expressed with increasing amounts of p.Arg318del (Figure S4C, open bars). The subcellular-localization data together with DNA-binding and promoter regulatory studies suggest that the p.Arg318del-p.Lys307Asn combination would significantly diminish MITF activity more than either variant present in combination with the WT protein.
Figure 3.
The Nuclear Migration, DNA Binding, and Promoter Regulatory Potential of WT MITF in the Presence of Mutant MITF Isoforms
(A) Nuclear-cytoplasmic distribution of MITF in the presence of the mutants (scale bars represent 10 μm) was studied by confocal microscopy. The WT and mutant MITF were differentially tagged by GFP or RFP.
(B) Amounts of nuclear MITF were calculated by western blotting from three different experiments and are represented as a graph.
(C) EMSA gels representative of three experiments show DNA-binding ability of the WT-control-vector (CV), WT-p.Arg318del, WT-p.Lys307Asn, and p.Arg318del-p.Lys307Asn combinations (∗supershifted band) to the M-box and E-box DNA sequences.
(D) The dominant effect of p.Arg318del on WT and p.Lys307Asn proteins was studied by a dual luciferase reporter assay with the TYRP promoter. In increasing concentrations (0, 50, 100, and 200 ng), p.Arg318del was co-transfected with a constant amount of WT or p.Lys307Asn (100 ng), along with 200 ng of a luciferase-promoter construct (TYRP or FGF19), 1:1,000 Renilla luciferase vector, and empty vector to equalize the amounts of transfected DNA. The p.Arg318del mutant in increasing amounts had a dominant-negative effect on WT MITF (solid bars) and p.Lys307Asn (open bars).
(E) Effect of mutant zebrafish mitfa overexpression on the average melanocyte numbers of 36 hpf zebrafish embryos. The WT ABTL strain of zebrafish was maintained under standard conditions of fish husbandry according to NIH Animal Use and Care Committee guidelines. WT mitfa and the isoforms harboring the two human mutations, cloned in pCS2+, were linearized and injected into the cells of freshly fertilized zebrafish eggs at the single-cell stage. Linearized pCS2+ vector (EV) was also injected in equal amounts as an experimental control. The volume of injection solution per embryo was ∼1 nL. The melanocytes on a specified dorsal-trunk region of 34–36 hpf zebrafish embryos were photographed and then counted for 30 different embryos pooled from three different trials. Representative images of 48 hpf embryos are shown (scale bars represent 0.1 mm).
(F) Nuclear localization of TFE3 in the presence of the two MITF mutants (scale bars represent 10 μm) was studied by confocal microscopy. All comparisons are in reference to the WT MITF or EV; error bars represent standard error p values were calculated with the two-sided Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005.
Heterozygous MITF mutations in humans cause sensorineural deafness and pigmentation defects distinct from complete albinism. Fully penetrant albinism in the individuals with COMMAD syndrome suggests that MITF function is below the critical threshold required for melanocyte migration, differentiation, and survival. To investigate the effects of these mutants on pigmentation in vivo, we introduced the two analogous human mutations into zebrafish mitfa cDNA (homologous to human MITF-M) and injected them as expression constructs into 1-cell-stage zebrafish embryos. Melanocytes in a specified dorsal-trunk region were quantified at 34–36 hr post-fertilization (hpf). As expected, injection of p.Arg318del mutant MITF resulted in a significantly lower number of melanocytes than did injection of similar amounts of empty vectors. Injection of p.Lys307Asn also led to significantly reduced melanocyte numbers (Figure 3E), consistent with its effects observed in vitro.
Mitfmi/mi mice exhibit colobomatous microphthalmia, which has never been reported for human MITF mutations. The severe eye phenotype observed in COMMAD syndrome could be attributed to the deregulation of genes involved in the development of RPE and the neural retina.13, 14 For instance, FGF15 in mice (homologous to human FGF19) is a direct target of MITF and plays an important role in retinal differentiation. We therefore performed a reporter assay by using the RPE-specific Best1 promoter15 and observed that the p.Arg318del-p.Lys307Asn combination showed significantly less transactivation of the Best1 promoter than the WT-WT, WT-p.Arg318del, and WT-p.Lys307Asn combinations (Figure S4D). These data fit well with the observation that only biallelic MITF mutations in mice and humans result in severe eye phenotypes. Our study provides evidence that MITF mutations can cause colobomatous microphthalmia in humans.
MITF acts as a heterodimer with Mi-T family transcription factors TFE3, TFEB, and TFEC.10 Osteopetrosis in Mitf mutant mice has been observed only with homozygosity of semi-dominant mutant alleles, e.g., in Mitfmi/mi mice. In these mice, MITF interferes with the activity of binding partner TFE3, which is important for osteoclast function. Consistent with this, osteoclasts are normal in Mitf- or Tfe3-null mice, whereas combined loss of both proteins results in severe osteopetrosis.16 In HEK293 cells, we observed cytoplasmic retention of GFP-tagged TFE3 when it was co-expressed with RFP-tagged p.Arg318del or p.Lys307Asn, but not when it was co-expressed with WT MITF (Figure 3F). In contrast to COMMAD syndrome, none of the reported cases of human MITF mutations exhibit osteopetrosis, suggesting that the two mutant alleles together exert a stronger dominant-negative effect on the entire Mi-T family than a single dominant-negative allele.
Developmentally, mouse Mitf expression is tightly regulated and is first detected throughout the optic vesicle of 22-somite-stage embryos, followed by expression in presumptive RPE of the optic cup.17 Shortly after its detection in the eye, MITF expression is also observed in neural-crest-derived melanoblasts, which are melanocyte precursors.18 Quite interestingly, early eye development is normal in individuals with heterozygous MITF mutations, but the spectrum of pigmentation and sensorineural defects varies greatly.19 Also, retinal-pigmentation defects in humans are observed only with the dominant-negative allele.19 In adult mice, Mitf expression is observed in osteoclasts,20 mast cells,21 heart, and skeletal muscle,22 in addition to melanocytes and RPE. The Mitfmi/mi mouse displays defects in all of these cell and tissue types. Although heterozygous human MITF mutations in Waardenburg and Tietz syndromes are not associated with these defects, apart from pigmentation and related sensorineural defects,8 all of the COMMAD syndrome phenotypes associate well with abnormalities observed in the Mitfmi/mi mouse.
To our knowledge, no published reports have shown a complete phenotypic overlap with the clinical features of COMMAD syndrome. Mutations in MAF (associated with Ayme-Gripp syndrome [MIM: 601088]) cause coloboma, congenital cataracts, sensorineural deafness, intellectual disability, and dysmorphic facies, among others.23 Some other genes known to cause syndromic or non-syndromic uveal coloboma—such as PAX6 (MIM: 607108), SHH (MIM: 600725), GDF3 (MIM: 606522), RBP (MIM: 180250), CHX10 (MIM: 142993), SOX2 (MIM: 184429), OTX2 (MIM: 600037), and RAX (MIM: 601881)—result in a range of ocular phenotypes, including microphthalmia, cataracts, and microcornea. None of the reported incidences of syndromic or isolated cases of coloboma have been associated with albinism or osteopetrosis. Moreover, these abnormalities have not been reported previously for human MITF disorders. Description of this syndrome is perhaps particularly important, given that intermarriage within the deaf community is relatively common, and neither set of parents in this study recognized they had WS2A prior to having their child with COMMAD. We would also stress the need for genotyping individuals suspected to have Waardenburg syndrome at the molecular level and providing appropriate genetic counselling.
In summary, we report an MITF allele (c.921G>C [p.Lys307Asn]) that causes WS2A and, when present with the dominant-negative allele (c.952_954delAGA [p.Arg318del]), results in a syndrome that we have termed COMMAD. This condition was seen again in another person with a probably dominant-negative mutation (p.Arg318Gly) and a canonical splice-site mutation (c.938−1G>A). We propose that an allelic combination involving at least one dominant-negative mutation, inherited in a recessive manner, represents the underlying molecular mechanism leading to COMMAD syndrome, thus delineating important roles for MITF in ocular morphogenesis and bone homeostasis in humans.
Acknowledgments
We are grateful to the probands and their families. We thank Dr. Heinz Arnheiter for critical comments on the manuscript, Dr. James Lister (Virginia Commonwealth University, Richmond, VA) for the zebrafish mitfa construct, Drs. Noriko Esumi (Wilmer Eye Institute, Johns Hopkins University, Baltimore, MD) and Sridhar Mani (Albert Einstein College of Medicine, Bronx, NY) for luciferase promoter constructs, Drs. Chun Gao and Robert Farris (Biological Imaging Core, National Eye Institute [NEI]) for expert assistance with confocal laser-scanning microscopy experiments, Ramakrishna Alur for technical assistance, Delphine Blain for genetic counseling, the NEI Clinical Photography Section for outstanding technical assistance, and Anna Larson and Tyler Fayard for zebrafish breeding and maintenance. This work was supported by the intramural program of the NEI.
Published: November 23, 2016
Footnotes
Supplemental Data include four figures, one table, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.11.004.
Web Resources
1000 Genomes, http://www.1000genomes.org/
Combined Annotation Dependent Depletion (CADD), http://cadd.gs.washington.edu/
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/
OMIM, http://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
UCSF Chimera, http://www.cgl.ucsf.edu/chimera/
UniProt: MITF, http://www.uniprot.org/uniprot/O75030
YASARA, http://www.yasara.org/
Supplemental Data
Top: nuclear migration of GFP- and RFP-tagged WT MITF (left) and nuclear migration of RFP-tagged WT MITF in the presence of GFP-tagged p.Arg318del mutant (right). Bottom: nuclear migration of RFP-tagged WT MITF in the presence of GFP-tagged p.Lys307Asn mutant (left) and nuclear migration of mutant MITF in the presence of GFP-tagged p.Lys307Asn mutant and RFP-tagged p.Arg318del mutant (right).
References
- 1.Waardenburg P.J. A new syndrome combining developmental anomalies of the eyelids, eyebrows and nose root with pigmentary defects of the iris and head hair and with congenital deafness. Am. J. Hum. Genet. 1951;3:195–253. [PMC free article] [PubMed] [Google Scholar]
- 2.Tietz W. A syndrome of deaf-mutism associated with albinism showing dominant autosomal inheritance. Am. J. Hum. Genet. 1963;15:259–264. [PMC free article] [PubMed] [Google Scholar]
- 3.Price E.R., Fisher D.E. Sensorineural deafness and pigmentation genes: melanocytes and the Mitf transcriptional network. Neuron. 2001;30:15–18. doi: 10.1016/s0896-6273(01)00259-8. [DOI] [PubMed] [Google Scholar]
- 4.Hero I. The optic fissure in the normal and microphthalmic mouse. Exp. Eye Res. 1989;49:229–239. doi: 10.1016/0014-4835(89)90093-6. [DOI] [PubMed] [Google Scholar]
- 5.Steingrímsson E., Moore K.J., Lamoreux M.L., Ferré-D’Amaré A.R., Burley S.K., Zimring D.C., Skow L.C., Hodgkinson C.A., Arnheiter H., Copeland N.G. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat. Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- 6.Hughes A.E., Newton V.E., Liu X.Z., Read A.P. A gene for Waardenburg syndrome type 2 maps close to the human homologue of the microphthalmia gene at chromosome 3p12-p14.1. Nat. Genet. 1994;7:509–512. doi: 10.1038/ng0894-509. [DOI] [PubMed] [Google Scholar]
- 7.Tassabehji M., Newton V.E., Read A.P. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat. Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 8.Tassabehji M., Newton V.E., Liu X.Z., Brady A., Donnai D., Krajewska-Walasek M., Murday V., Norman A., Obersztyn E., Reardon W. The mutational spectrum in Waardenburg syndrome. Hum. Mol. Genet. 1995;4:2131–2137. doi: 10.1093/hmg/4.11.2131. [DOI] [PubMed] [Google Scholar]
- 9.Léger S., Balguerie X., Goldenberg A., Drouin-Garraud V., Cabot A., Amstutz-Montadert I., Young P., Joly P., Bodereau V., Holder-Espinasse M. Novel and recurrent non-truncating mutations of the MITF basic domain: genotypic and phenotypic variations in Waardenburg and Tietz syndromes. Eur. J. Hum. Genet. 2012;20:584–587. doi: 10.1038/ejhg.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steingrímsson E., Copeland N.G., Jenkins N.A. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 11.Takebayashi K., Chida K., Tsukamoto I., Morii E., Munakata H., Arnheiter H., Kuroki T., Kitamura Y., Nomura S. The recessive phenotype displayed by a dominant negative microphthalmia-associated transcription factor mutant is a result of impaired nucleation potential. Mol. Cell. Biol. 1996;16:1203–1211. doi: 10.1128/mcb.16.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grill C., Bergsteinsdóttir K., Ogmundsdóttir M.H., Pogenberg V., Schepsky A., Wilmanns M., Pingault V., Steingrímsson E. MITF mutations associated with pigment deficiency syndromes and melanoma have different effects on protein function. Hum. Mol. Genet. 2013;22:4357–4367. doi: 10.1093/hmg/ddt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bharti K., Gasper M., Ou J., Brucato M., Clore-Gronenborn K., Pickel J., Arnheiter H. A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genet. 2012;8:e1002757. doi: 10.1371/journal.pgen.1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raviv S., Bharti K., Rencus-Lazar S., Cohen-Tayar Y., Schyr R., Evantal N., Meshorer E., Zilberberg A., Idelson M., Reubinoff B. PAX6 regulates melanogenesis in the retinal pigmented epithelium through feed-forward regulatory interactions with MITF. PLoS Genet. 2014;10:e1004360. doi: 10.1371/journal.pgen.1004360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esumi N., Kachi S., Campochiaro P.A., Zack D.J. VMD2 promoter requires two proximal E-box sites for its activity in vivo and is regulated by the MITF-TFE family. J. Biol. Chem. 2007;282:1838–1850. doi: 10.1074/jbc.M609517200. [DOI] [PubMed] [Google Scholar]
- 16.Steingrimsson E., Tessarollo L., Pathak B., Hou L., Arnheiter H., Copeland N.G., Jenkins N.A. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc. Natl. Acad. Sci. USA. 2002;99:4477–4482. doi: 10.1073/pnas.072071099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen M., Arnheiter H. Signaling and transcriptional regulation in early mammalian eye development: a link between FGF and MITF. Development. 2000;127:3581–3591. doi: 10.1242/dev.127.16.3581. [DOI] [PubMed] [Google Scholar]
- 18.Nakayama A., Nguyen M.T., Chen C.C., Opdecamp K., Hodgkinson C.A., Arnheiter H. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech. Dev. 1998;70:155–166. doi: 10.1016/s0925-4773(97)00188-3. [DOI] [PubMed] [Google Scholar]
- 19.Izumi K., Kohta T., Kimura Y., Ishida S., Takahashi T., Ishiko A., Kosaki K. Tietz syndrome: unique phenotype specific to mutations of MITF nuclear localization signal. Clin. Genet. 2008;74:93–95. doi: 10.1111/j.1399-0004.2008.01010.x. [DOI] [PubMed] [Google Scholar]
- 20.Weilbaecher K.N., Hershey C.L., Takemoto C.M., Horstmann M.A., Hemesath T.J., Tashjian A.H., Fisher D.E. Age-resolving osteopetrosis: a rat model implicating microphthalmia and the related transcription factor TFE3. J. Exp. Med. 1998;187:775–785. doi: 10.1084/jem.187.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King R., Peterson A.C., Peterson K.C., Mihm M.C., Jr., Fisher D.E., Googe P.B. Microphthalmia transcription factor expression in cutaneous mast cell disease. Am. J. Dermatopathol. 2002;24:282–284. doi: 10.1097/00000372-200206000-00017. [DOI] [PubMed] [Google Scholar]
- 22.Hodgkinson C.A., Moore K.J., Nakayama A., Steingrímsson E., Copeland N.G., Jenkins N.A., Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 23.Niceta M., Stellacci E., Gripp K.W., Zampino G., Kousi M., Anselmi M., Traversa A., Ciolfi A., Stabley D., Bruselles A. Mutations Impairing GSK3-Mediated MAF Phosphorylation Cause Cataract, Deafness, Intellectual Disability, Seizures, and a Down Syndrome-like Facies. Am. J. Hum. Genet. 2015;96:816–825. doi: 10.1016/j.ajhg.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top: nuclear migration of GFP- and RFP-tagged WT MITF (left) and nuclear migration of RFP-tagged WT MITF in the presence of GFP-tagged p.Arg318del mutant (right). Bottom: nuclear migration of RFP-tagged WT MITF in the presence of GFP-tagged p.Lys307Asn mutant (left) and nuclear migration of mutant MITF in the presence of GFP-tagged p.Lys307Asn mutant and RFP-tagged p.Arg318del mutant (right).