Abstract
Epileptic encephalopathies are a catastrophic group of epilepsies characterized by refractory seizures and cognitive arrest, often resulting from abnormal brain development. Here, we have identified an epileptic encephalopathy additionally featuring cerebral calcifications and coarse facial features caused by recessive loss-of-function mutations in DENND5A. DENND5A contains a DENN domain, an evolutionarily ancient enzymatic module conferring guanine nucleotide exchange factor (GEF) activity to multiple proteins serving as GEFs for Rabs, which are key regulators of membrane trafficking. DENND5A is detected predominantly in neuronal tissues, and its highest levels occur during development. Knockdown of DENND5A leads to striking alterations in neuronal development. Mechanistically, these changes appear to result from upregulation of neurotrophin receptors, leading to enhanced downstream signaling. Thus, we have identified a link between a DENN domain protein and neuronal development, dysfunction of which is responsible for a form of epileptic encephalopathy.
Main Text
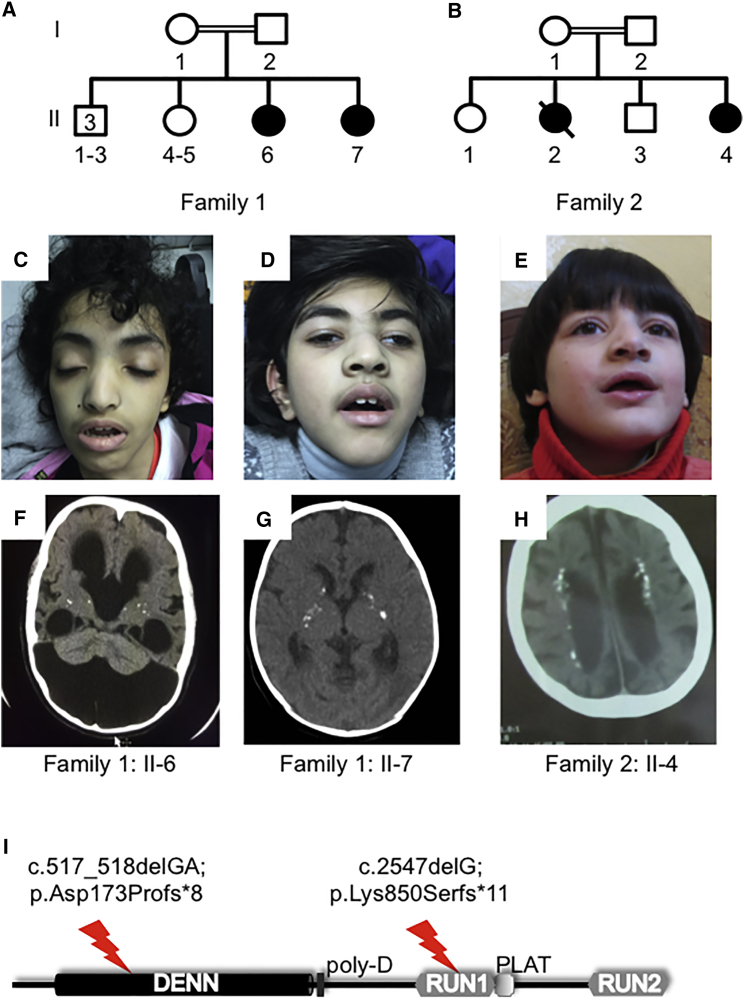
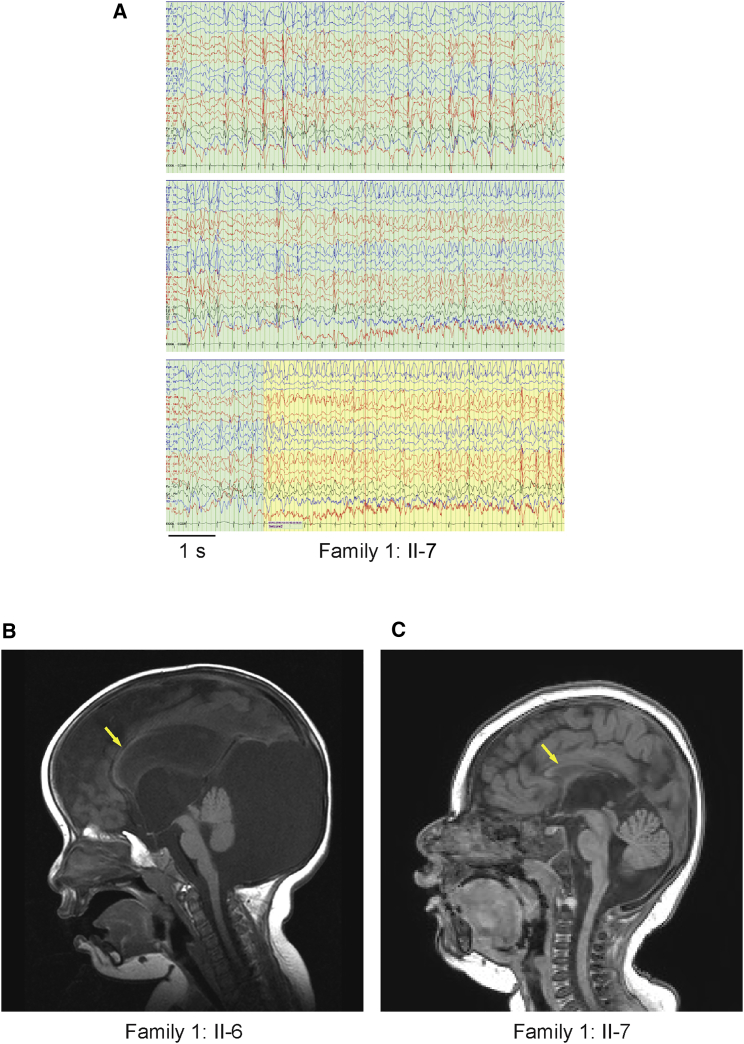
With a lifetime incidence of 3%, epilepsy is a common neurological disorder. Epileptic encephalopathies are a rare but devastating subform of epilepsies that often result from gross alterations in brain development.1 Epileptic encephalopathy is frequently of unknown etiology, but newly identified genetic forms of the syndrome provide insight into disease mechanisms. A consanguineous family from Saudi Arabia (family 1) presented with two affected sisters with epileptic encephalopathy (Figures 1A, 1C, and 1D). The proband (individual II-6 in Figure 1) is 14 years old and had a normal term delivery after an uncomplicated pregnancy. In the first day of life, she was noticed to have facial twitching, multifocal limb jerking, and clonic seizures. Presently, she has global developmental delay and spastic tetraplegia and is non-verbal and dependent on support for all activities of daily living. She has multiple daily seizures, namely tonic, myoclonic, and generalized tonic-clonic. Her younger sister is similarly affected but has additional exaggerated startle and startle-induced seizures. Electroencephalographs (EEGs) in both individuals reveal slow background and frequent frontally dominant lateralized, bifrontal, or generalized spike-wave discharges interictally or in association with myoclonus (shown for individual II-7 in Figure 2A). Ictal activity consisted of rhythmic runs of the same, culminating in generalized convulsion (Figure 2A). Both sisters have microcephaly. At age 7 years, the occipitofrontal circumference (OFC) for individual II-7 was 40 cm, below the second percentile. That of II-6 at age 10 years was 54 cm, corresponding to the 50th percentile, but it should be noted that this child had hydrocephalus in the neonatal period and has a large posterior fossa arachnoid cyst (Figures 1F and 2B). The OFCs of the parents were not obtained, but neither appeared microcephalic. Magnetic resonance imaging (MRI) revealed dysgenesis of the corpus callosum and ventriculomegaly (Figures 2B and 2C). Individual II-6 additionally demonstrates Dandy-Walker malformation, hypoplastic vermis, and diffuse hydrocephalus (Figure 2C). Computed tomography (CT) images revealed multiple small foci of calcification in the basal ganglia for both individuals (Figures 1F and 1G). Routine cerebrospinal fluid analysis on these individuals, including chemistry, was normal.
Figure 1.
Two Families Affected by Epileptic Encephalopathy Featuring Cerebral Calcification and Coarse Facial Features
(A and B) Pedigrees of family 1 (A) and family 2 (B).
(C–E) Photographs of affected individuals II-6 (C) and II-7 (D) of family 1 and II-4 (E) of family 2. Note the common coarse facial features, including nasal prominence and frontal bossing.
(F–H) CT scans of the same individuals as above. Note the generalized atrophy and calcification (white spots) of the basal ganglia (F, II-6; G, II-7) and periventricular cortex (H).
(I) Schematic overview of the structure of DENND5A, which contains DENN, RUN, and PLAT domains. The positions of the identified truncating variants are indicated.
Figure 2.
EEG and MRI Analysis of Individuals from Family 1
(A) Wakeful EEG of individual II-7 from family 1. Interictal activity (green background) is characterized by near-continuous independent or synchronous bilateral or generalized bifrontally dominant spike-wave discharges. Ictus (seizure; yellow background) is associated with continuous generalized bifrontally dominant rhythmic activity.
(B and C) MRI demonstrating dysgenesis of the corpus callosum (arrows) and ventriculomegaly in affected individuals II-6 (B) and II-7 (C), as well as Dandy-Walker malformation, a posterior fossa cyst, hypoplastic vermis, and diffuse hydrocephalus in individual II-6.
A second consanguineous family (family 2) from Jordan also presented with two affected sisters (Figures 1B and 1E) after uneventful pregnancies and deliveries. Both children developed seizures in the neonatal period. Individual II-4 was examined at age 7 years and was noted to have severe global developmental delay. She has generalized tonic-clonic seizures, speaks only a few words, and exhibits major anxiety and hyperactivity. Only interictal EEG was available and revealed slow background with bifrontal epileptiform discharges. Her growth parameters are in the low range, and she is microcephalic (OFC 41 cm < the 1st percentile), whereas the OFC of her father and unaffected sibling II-3 are 54 and 51 cm, respectively (around the 50th percentile). The mother’s OFC was not measured, but she did not appear microcephalic. CT showed multiple small foci of calcification in the white matter along the lateral ventricles (Figure 1H). MRI scans were not performed. Individual II-2 of family 2 died at age 10 years and was not clinically assessed, but her parents reported that she had the same symptoms as her sister.
All four affected individuals have (or had) similar coarse facial features (Figures 1C–1E). Specifically, they present with an open mouth with a slightly tented full upper lip and thick everted lower lip, short philtrum, and large nostrils of a prominent nose. They also have thick curved eyebrows, long eyelashes, and large ears (Figures 1C–1E). Individual II-4 of family 2 was also noted to have a high palate. In summary, the affected individuals have a severe neonatal-onset neurological disorder including severe epilepsy. To what extent the latter influences their neurodevelopment is unclear. The syndrome also appears to be characterized by relatively uniform facial features.
This project was approved by the ethics committees of the Hospital for Sick Children and University of Toronto (research ethics board file no. 1000033784), Johns Hopkins Aramco Healthcare (institutional review board no. 42), the University of Tübingen (protocol no. 594/2015BO1), and Philadelphia University Jordan (protocol no. Feb 25,2015/1). Signed informed consent for collecting biological materials and publishing the results was obtained from legal guardians. All studies were performed in compliance with the Declaration of Helsinki.
To identify the genetic mutations responsible for the phenotypes, we performed whole-exome sequencing (WES) on individuals from both families 1 and 2. WES for the affected siblings of family 1 was performed on the Ion Proton system after exonic amplification with the Ion AmpliSeq Exome Kit (Life Technologies). Data analysis was performed with Ion Torrent Suite version 4.2.1.0. The resulting variant calls were annotated with a custom pipeline incorporating ANNOVAR.2 For individuals II-6 and II-7 in family 1, the average read depth was 82× and 130×, respectively, and more than 88% and 93% of target bases, respectively, were covered at >20×. A total of 53,996 (individual II-6) and 56,008 (individual II-7) variants (prior to application of any quality filtering) were detected. WES for individual II-4 of family 2 was performed on a NextSeq 500 platform (Illumina) after enrichment with SureSelectXT Human All Exon V5 (Agilent). Data analysis was performed through an in-house pipeline including BWA-MEM3 for alignment. Variant calling utilized freebayes.4 Variant annotation was performed with SnpEff, a program for annotating and predicting the effects of SNPs.5 We achieved an average read depth of 131.1×, and 86% of the target region was covered at least 20×. A total of 50,785 variants were identified with this approach.
To identify potentially pathogenic variants in both families, data analysis focused on rare variants (minor allele frequency of <1%) with a likely effect on protein function (e.g., nonsense, frameshift, splice-site, and missense variants) in both public databases (1000 Genomes Project,6 the Exome Aggregation Consortium [ExAC] Browser,7 and the NHLBI GO Exome Sequencing Project Exome Variant Server) and our in-house databases. Because the pattern of inheritance was expected to be autosomal recessive as a result of consanguinity in the parents, homozygous variants were assessed first. In family 1, the parents are second cousins (a coefficient of relationship of at least 3.13%). In family 2, the parents are first cousins (a coefficient of relationship of at least 12.5%). Exome data indicated a level of homozygosity of approximately 6.5% for family 1 and approximately 10% for family 2. We consider these results to be reasonable given that these families are likely to have had many more consanguineous marriages, probably for many generations. In family 1, we found that the two affected siblings share ten high-quality homozygous variants, one of which is a c.517_518delGA (p.Asp173Profs∗8) mutation leading to a frameshift in DENND5A (GenBank: NM_015213.3) (Figure 1I). From the exome data, the homozygous regions spanning the variant in individuals II-6 and II-7 are approximately chr11: 8,014,479–19,901,632 and chr11: 8,959,545–19,256,250, respectively. In family 2, we identified a homozygous c.2547delG (p.Lys850Serfs∗11) mutation leading to a frameshift in DENND5A (Figure 1I), and the homozygous region is at least chr11: 7,110,751–2,063,940. Neither of these variants had been previously reported in public databases, and both were absent from the ExAC Browser. DENND5A is very conserved in the general population and is highly intolerant of loss-of-function variants. The pLI score is 0.99,7 and no loss-of-function variant was found to be homozygous in the ExAC Browser. We screened retrospectively for all rare variants in previously reported genetic neurological disorders, including intellectual disability and epileptic encephalopathy, and no other potentially pathogenic variants were detected in either family.
The DENND5A variants were confirmed by Sanger sequencing, heterozygous in the parents, and absent or heterozygous in unaffected siblings (Figure S1). By virtue of the affected exons, both mutations are expected to lead to nonsense-mediated mRNA decay. DENND5A (also called Rab6-interacting protein 1) is composed of an N-terminal DENN (differentially expressed in normal and neoplastic cells) domain followed by two RUN (RPIP8 [RaP2-interacting protein 8], UNC-14, and NESCA [new molecule containing SH3 at the carboxyl terminus]) domains flanking a PLAT (polycystin-1, lipoxygenase, alpha-toxin) domain (Figure 1I). In the rare case that no nonsense-mediated mRNA decay should occur, affected individuals in family 1 would have a truncated DENN domain that would almost certainly lead to misfolding of this module, and individual II-4 of family 2 would have a truncated N-terminal RUN domain (RUN1) with a similar consequence (Figure 1I). It is highly likely that either situation would lead to clearance of the misfolded protein. Thus, loss-of-function mutations in DENND5A cause a form of epileptic encephalopathy. We therefore sought to examine the role of DENND5A in neuronal development.
Through RUN1, DENND5A binds Rab6, a protein that regulates membrane trafficking and localizes to the trans-Golgi network (TGN).8, 9 Through this interaction, Rab6 targets DENND5A to the TGN.9 Through the C-terminal RUN domain (RUN2), DENND5A interacts with sorting nexin 1, a component of endosome-derived vesicles that target to the TGN, and thus DENND5A is thought to function in the trafficking of cargo from endosomes to the TGN.10, 11 The DENN domain is an evolutionarily conserved module that functions globally as a GEF for Rabs,12, 13, 14, 15 which are critical regulators of intracellular membrane trafficking,16 and DENND5A has been reported to function as a GEF for Rab39.14 Two distinct gene products encode highly related isoforms of Rab39: Rab39A, which is associated with late endosomes and lysosomes,17 and Rab39B, which is concentrated at the Golgi complex, where it regulates trafficking to synaptic terminals.18 Thus, DENND5A functions in membrane trafficking at a crossroads between the Golgi and the endosomal system, but the physiological outputs of DENND5A function remain unknown.
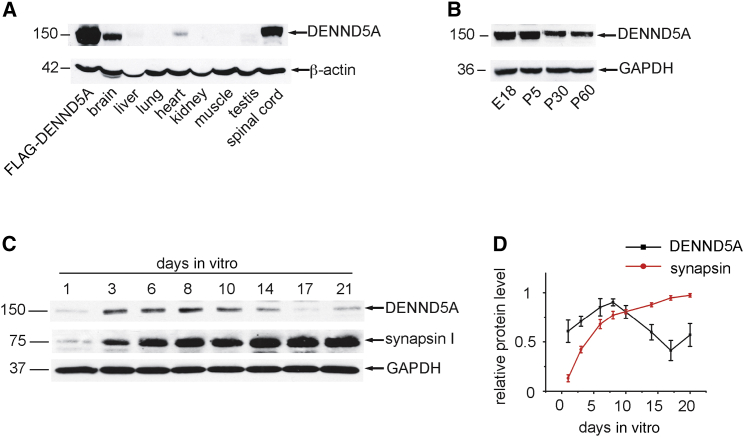
The observation that loss-of-function mutations in DENND5A cause epileptic encephalopathy suggests that DENND5A controls membrane trafficking pathways critical for normal neuronal development. We thus sought to examine the potential function of DENND5A in this process. Importantly, western blot detected DENND5A predominantly in neuronal tissues (Figure 3A), and in a panel of cell lines, it was detectable only in PC12 cells, a neuronal-like line (Figure S2A). However, we cannot rule out that more sensitive tests such as RT-PCR could detect low levels of expression in non-neuronal lines and tissues. Knockdown of DENND5A in PC12 cells confirmed that the protein band detected by the antibody is endogenous DENND5A (Figures S2B and S2C). In rat brain, DENND5A was at its highest levels at embryonic day 18 and continually decreased through to postnatal day 60 (Figure 3B). In rat cortical neurons in culture, the amount of DENND5A increased from 1 day in vitro (DIV) through to 8 DIV and then gradually declined up to 21 DIV (Figures 3C and 3D). In contrast, synapsin, a marker of mature synapses, increased throughout culture development (Figures 3C and 3D).19 Thus, DENND5A is present predominantly in developing neuronal tissue.
Figure 3.
DENND5A Is Detected Predominantly in Developing Neurons
(A) Equal protein aliquots of lysates from various tissues were blotted with antibodies recognizing DENND5A (polyclonal antibody, Abcam) and actin (monoclonal antibody C4, Millipore) as indicated. Lysates from HEK293T cells expressing FLAG-tagged DENND5A (FLAG-DENND5A) were included as a control.
(B) Crude lysates were prepared from brains dissected from embryonic day 18 (E18) or postnatal day 5, 30, or 60 (P5, P30, or P60) rats. Equal protein aliquots were processed for western blot with antibodies recognizing DENND5A and GAPDH (monoclonal antibody, Santa Cruz) as indicated.
(C) Primary rat cortical neurons were seeded in culture dishes, and lysates were prepared from the neurons at the indicated days in vitro. Equal protein aliquots were processed for western blots with antibodies recognizing DENND5A, GAPDH, and synapsin.19
(D) Quantification of the relative amounts of DENND5A and synapsin from blots as in (C). The highest-intensity values were set as 1.0. Data represent the mean ± SEM from eight independent experiments.
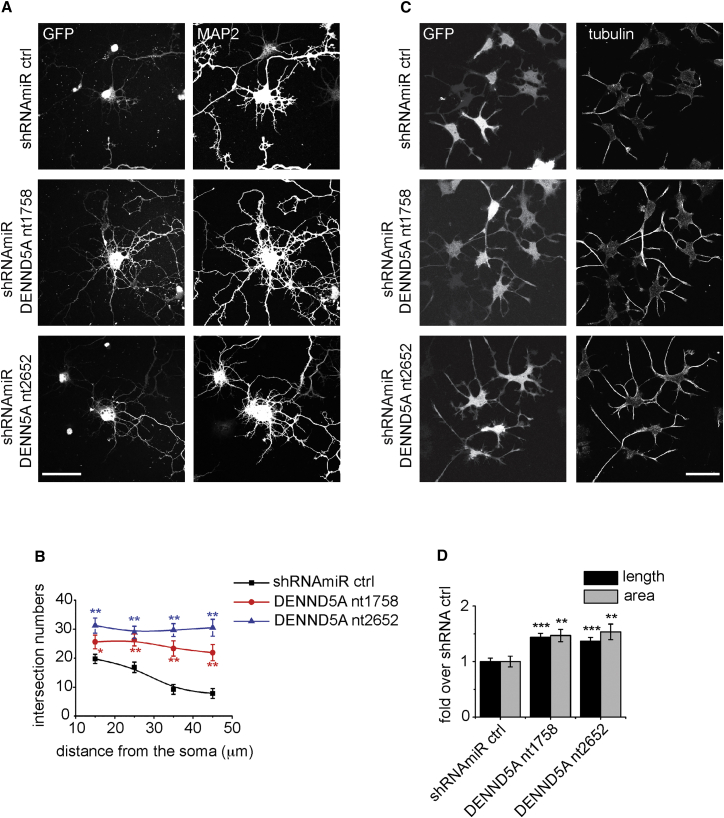
We next sought to examine whether loss of function of DENND5A influences neuronal development. For these experiments, we chose hippocampal cultures, a well-characterized system for examining neuronal development.20 The cultures were transduced with two distinct lentiviruses encoding shRNAmiRs targeting DENND5A. The shRNAmiR system involves small hairpin RNAs (shRNAs) that are expressed in the structural context of a microRNA21 and was adapted by our laboratory as previously described.13 Targeting DENND5A led to enhanced outgrowth of dendrites at 6 DIV, as marked by MAP2, in comparison to transduction with a lentivirus encoding a control shRNAmiR (Figure 4A). To quantify these differences, we performed a Sholl analysis, which involves drawing concentric rings around neurons and counting the number of times that a dendrite intersects a ring at each distance from the middle. At all distances from the soma, the knockdown cells had more intersections, indicating that they have longer dendrites with greater branching complexity (Figure 4B).
Figure 4.
DENND5A Knockdown Enhances Process Outgrowth in Neurons and PC12 Cells
(A) Primary cultured hippocampal neurons at 1 day in vitro (DIV)22 were transduced with lentivirus expressing shRNAmiRs targeting rat DENND5A mRNA (GenBank: NM_001107546.2). The 21 nt inhibitory sequences match DENND5A starting at nucleotide 1,758 (shRNAmir DENND5A nt1758: 5′-ACTCAGGATTTACCAGCTAAA-3′) and nucleotide 2,652 (shRNAmiR DENND5A nt2652: 5′-GAGCCACGGGCTACAAGTAAA-3′). Non-targeting shRNAmiR virus was used as a control (shRNAmiR ctrl).23 At 6 DIV, transduced neurons were fixed and processed for GFP fluorescence and for indirect immunofluorescence with antibody recognizing MAP2 (chicken polyclonal antibody, EnCor Biotechnology) to reveal the somatodendritic region of the neurons. GFP was expressed as part of the viral expression cassette to verify transduction. Images were captured on a laser-scanning confocal microscope (LSM 710, Carl Zeiss) equipped with a Plan Apochromat 40× oil objective (numerical aperture 1.3; Carl Zeiss). Acquisition was performed with ZEN 11.0 software. The scale bar represents 50 μm.
(B) Sholl analysis for dendritic complexity of transduced hippocampal neurons from representative images as in (A) was performed with ImageJ with the ShollAnalysis plugin. Data represent the mean ± SEM from two independent experiments measuring a minimum of 18 neurons per condition per experiment; repeated-measure one-way ANOVA followed by a Dunnett’s post-test revealed a significant difference between control and knockdown neurons. ∗p < 0.05, ∗∗p < 0.01.
(C) PC12 cells were transduced with control lentivirus (shRNAmiR ctrl) or lentivirus to knockdown rat DENND5A (shRNAmiR DENND5A nt1758 and shRNAmiR DENND5A nt2652). Transduced PC12 cells were serum starved for 24 hr and then treated with 50 ng/mL NGF (2.5S NGF, Cederlane) in serum-free medium. After 24 hr treatment, cells were fixed and processed for GFP fluorescence and for indirect immunofluorescence with antibody against α-tubulin (polyclonal antibody, ICN Biomedicals). GFP was expressed as part of the viral expression cassette to verify transduction. The scale bar represents 50 μm.
(D) Length of the longest neurite (length) and total area of neurites (area) for each cell from representative images as in (C) were analyzed with ImageJ. Data show the mean ± SEM from two independent experiments measuring a minimum of 30 cells per condition per experiment; repeated-measure one-way ANOVA followed by a Dunnett’s post-test revealed a significant difference between control and knockdown cells. ∗∗p < 0.01, ∗∗∗p < 0.001.
DENND5A is readily detectable in PC12 cells (Figure S2A). PC12 cells have been extensively used as a model system for neuronal development because neurite outgrowth can be initiated at defined times by the addition of nerve growth factor (NGF), and the signaling pathways controlling this process are well defined.24 Knockdown of DENND5A in PC12 cells (Figures S2B and S2C) significantly enhanced NGF-induced differentiation at 24 hr, as indicated by both increased neurite length and increased total surface area of the cells (Figures 4C and 4D). Knockdown of DENND5A also led to a significant increase in PC12 cell differentiation, even in the absence of NGF treatment (Figures S3A and S3B). Thus, it appears that DENND5A plays an inhibitory role in dendrite outgrowth.
During nervous system development, multiple factors, including neurotrophins and activity, drive the development of dendrites and dendritic arborization. Often, these activities are balanced in a “push-pull” fashion.25 For example, neurotrophins can both enhance and limit dendritic growth.25 Disruption of either class of signal leads to improper synaptic connectivity. Naturally occurring cell death is a normal aspect of neuronal development,26 and appropriate synaptic connectivity limits cell death by providing trophic factors and coordinated electrical activity.27 Thus, although nervous system development is vastly complex and difficult to model in culture systems, it is reasonable to consider that disruption of DENND5A during neuronal development in affected individuals could lead to improper synaptic connectivity and result in enhanced neuronal death contributing to epilepsy.
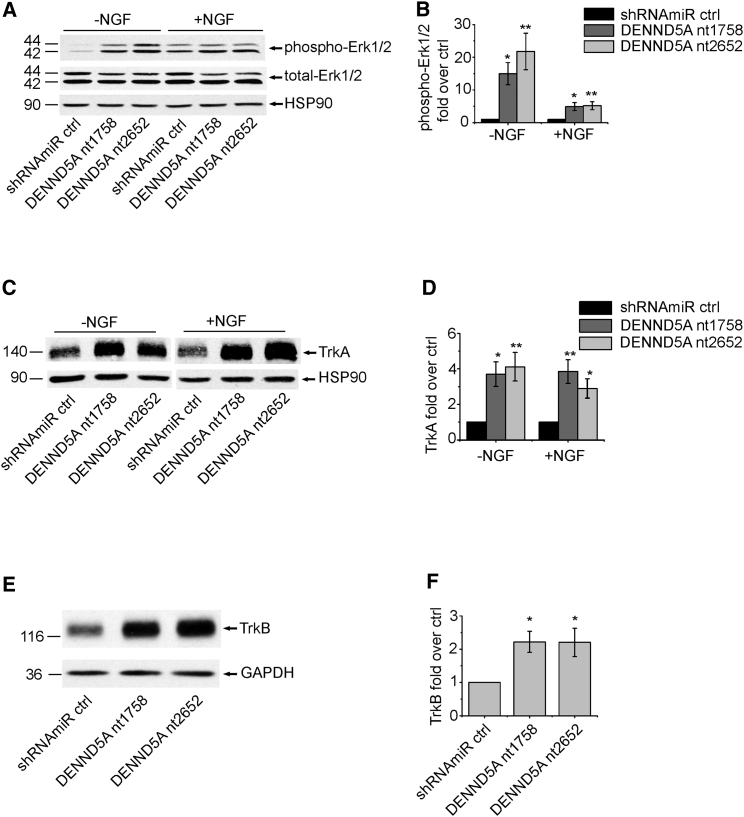
We next sought to explore the mechanisms by which DENND5A contributes to neurite and dendrite development. In PC12 cells, NGF stimulates neurite outgrowth by binding to its receptor, TrkA, leading to sustained activation of Erk.24 We thus tested for possible changes in Erk activation after knockdown of DENND5A. As expected, treating PC12 cells with NGF for 24 hr led to sustained Erk activation under control conditions (compare lanes 1 and 4 in the phospho-Erk1/2 blot in Figure 5A). Knockdown of DENND5A strongly potentiated NGF-induced Erk activation (Figures 5A and 5B). Erk potentiation following knockdown of DENND5A was seen even in the absence of NGF stimulation (Figures 5A and 5B). This is consistent with the observation that knockdown of DENND5A also stimulated neurite outgrowth in the absence of NGF (Figure S3). Thus, loss of DENND5A function leads to overactivation of Erk, and this most likely contributes to disruptions in normal neuronal development.
Figure 5.
Knockdown of DENND5A Leads to Enhanced Erk Activation and Increased Amounts of Neurotrophin Receptors
(A) PC12 cells were transduced with control lentivirus (shRNAmiR ctrl) or lentivirus to knockdown DENND5A (DENND5A nt1758 and DENND5A nt2652). After 24 hr serum starvation, transduced PC12 cells were harvested (−NGF) or treated for an additional 24 hr with NGF (50 ng/mL) (+NGF). Equal protein aliquots of lysates prepared from these cells were processed by western blot with antibodies recognizing phospho-Erk (monoclonal antibody, Cell Signaling Technology) and total Erk (monoclonal antibody, Cell Signaling Technology). HSP90 (monoclonal antibody, Assay Designs Stressgen) was used as a loading control.
(B) Quantification of five experiments performed as in (A). The graphs show the average relative change in phospho Erk1/2 levels normalized to total Erk1/2 levels between cells with knockdown DENND5A and control cells. Error bars represent the SEM, and statistical analysis employed a one-way ANOVA followed by a Bonferroni post-test. ∗p < 0.05, ∗∗p < 0.01.
(C) After 24 hr serum starvation, transduced PC12 cells were harvested (−NGF) or treated for 24 hr with NGF (50 ng/mL) (+NGF). Equal protein aliquots of lysates prepared from these cells were processed by western blot with antibodies recognizing TrkA (polyclonal antibody for pan Trk [TrkA and TrkB] [C17F1], Cell Signaling Technology) and HSP90.
(D) Quantification of TrkA amounts from experiments as in (C) normalized to HSP90 and expressed in relation to those in control shRNAmiR cells. Data show the mean ± SEM from eight independent experiments, and statistical analysis employed a one-way ANOVA followed by a Bonferroni post-test. ∗p < 0.05, ∗∗p < 0.01.
(E) Rat cortical neurons were transduced with control lentivirus (shRNAmiR ctrl) or lentivirus to knockdown DENND5A (DENND5A nt1758 and DENND5A nt2652) at 1 DIV. Cell lysates were prepared at 8 DIV, and equal protein aliquots were processed by western blot with antibodies recognizing the indicated proteins.
(F) Quantification of TrkB levels from experiments as in (E) normalized to GAPDH and expressed in relation to those of control shRNAmiR cells. Data show the mean ± SEM from six independent experiments, and statistical analysis employed a repeated-measure one-way ANOVA followed by a Dunett’s post-test. ∗p < 0.05.
Given that Erk is activated downstream of TrkA in PC12 cells,24 we sought to examine whether there were alterations in the levels of TrkA after DENND5A knockdown. Knockdown of DENND5A led to upregulation of TrkA (Figures 5C and 5D). In contrast to PC12 cells, primary cortical and hippocampal neurons express TrkB, a neurotrophin receptor related to TrkA, and the development of these neurons is strongly influenced by brain-derived neurotrophic factor (BDNF), the ligand for TrkB.24 Knockdown of DENND5A led to upregulation of TrkB in cultured cortical neurons (Figures 5E and 5F). Thus, an increase in the levels of Trk receptors could drive enhanced Erk activation, leading to the altered neurite outgrowth phenotypes observed.
The mechanisms whereby loss of DENND5A leads to enhanced amounts of neurotrophin receptor remain unknown. DENND5A functions as a GEF for Rab39,14 and disruption of both Rab39A and Rab39B causes alterations in neuronal development.17, 18 DENND5A functions at an interface between endosomes and the TGN, as does Rab39. Alterations in these pathways could lead to enhanced stability of neurotrophin receptors by preventing their targeting to lysosomes for degradation. At least in neuroblastoma cells, activation of TrkA in the absence of NGF, which occurs upon TrkA overexpression, has been shown to lead to apoptosis.28 Hence, upregulation of Trk receptors due to DENND5A deficiency could lead to increased apoptosis in the developing brain. Mutations in NTRK1 (MIM: 256800; encoding TrkA) and NTRK2 (MIM: 613886; encoding TrkB) have been reported in individuals with intellectual disability.29, 30
The severe alterations in neuronal development in the affected individuals described here most likely result from multiple factors. For example, all affected individuals have brain calcifications. Three prior established genetic causes of brain calcifications are mutations in PDGFRB (MIM: 615007), PDGFB (MIM: 615483), and SLC20A2 (MIM: 213600), the latter of which are by far the most common.31 SLC20A2 is a phosphate transporter, and PDGFRB and PDGFB have been implicated in modulating levels of SLC20A1, another phosphate transporter.32 Alterations in the levels or localization of these transporters after DENND5A loss of function could contribute to the calcification facet of the pathology seen in the individuals included in this study.
Recently, mutations in DNM1 (MIM: 602377), encoding the GTPase dynamin, which functions in the scission of clathrin-coated vesicles, were found to cause epileptic encephalopathy.33, 34 Truncating alterations in the GEF for ARF6, encoded by IQSEC2 (MIM: 309530), have been reported in individuals with X-linked intellectual disability with epilepsy and microcephaly.35 ARF6 functions in endosomal membrane trafficking. Many other factors playing a role in vesicular trafficking, e.g., RAB3GAP1 (MIM: 600118),36 RAB3GAP2 (MIM: 614225),37 and members of the adaptor protein complexes,38, 39, 40, 41 have been reported in various forms of syndromic and non-syndromic intellectual disability, as well as related disorders. Perhaps of most relevance, mutations in RAB39B (MIM: 300271) cause intellectual disability with epilepsy.18 All together, this evidence appears to indicate that alterations in membrane trafficking are emerging as a central locus in intellectual disability and epileptic encephalopathy, and our study further supports this concept.
While our paper was in review, another two consanguineous families, each with one child affected by biallelic truncating or highly damaging DENND5A mutations, were reported.42 These children are young (21 and 26 months old), but their phenotypes to date are highly similar to those of the individuals described here. These four families together establish this genetic disease and will allow specific diagnosis and genetic counselling in affected families.
Acknowledgments
We thank Dr. Pietro De Camilli for the gift of the synapsin antibody and M. Wilke for help with the analysis of computed tomography scans. This study was supported by a grant from the Canadian Institutes for Health Research (MOP-62684) to P.S.M., by a grant from the German Academic Exchange Service as part of the German-Arab Transformation Program Line4 (project ID 57166498) to O.R. and T.F., and by funding from the Ontario Brain Institute and Genome Canada to B.A.M. C.H. is supported by a fellowship from Fonds de Recherché du Quebec – Santé (Dossier-30199 and Dossier-33963). B.A.M. holds the University of Toronto Michael Bahen Chair in Epilepsy Research. P.S.M. is a James McGill Professor and a Fellow of the Royal Society of Canada.
Published: November 17, 2016
Footnotes
Supplemental Data include three figures and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.10.006.
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
NHLBI GO Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org
Supplemental Data
References
- 1.Carvill G.L., Heavin S.B., Yendle S.C., McMahon J.M., O’Roak B.J., Cook J., Khan A., Dorschner M.O., Weaver M., Calvert S. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 2013;45:825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, arXiv:1303.3997v1 [q-bio.GN], https://arxiv.org/abs/1303.3997.
- 4.Garrison, E., and Marth, G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv, arXiv:1207.3907 [q-bio.GN], https://arxiv.org/abs/1207.3907.
- 5.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goud B., Zahraoui A., Tavitian A., Saraste J. Small GTP-binding protein associated with Golgi cisternae. Nature. 1990;345:553–556. doi: 10.1038/345553a0. [DOI] [PubMed] [Google Scholar]
- 9.Recacha R., Boulet A., Jollivet F., Monier S., Houdusse A., Goud B., Khan A.R. Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure. 2009;17:21–30. doi: 10.1016/j.str.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Wassmer T., Attar N., Harterink M., van Weering J.R., Traer C.J., Oakley J., Goud B., Stephens D.J., Verkade P., Korswagen H.C., Cullen P.J. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev. Cell. 2009;17:110–122. doi: 10.1016/j.devcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandes H., Franklin E., Jollivet F., Bliedtner K., Khan A.R. Mapping the interactions between a RUN domain from DENND5/Rab6IP1 and sorting nexin 1. PLoS ONE. 2012;7:e35637. doi: 10.1371/journal.pone.0035637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato M., Sato K., Liou W., Pant S., Harada A., Grant B.D. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 2008;27:1183–1196. doi: 10.1038/emboj.2008.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allaire P.D., Marat A.L., Dall’Armi C., Di Paolo G., McPherson P.S., Ritter B. The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell. 2010;37:370–382. doi: 10.1016/j.molcel.2009.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshimura S., Gerondopoulos A., Linford A., Rigden D.J., Barr F.A. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 2010;191:367–381. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marat A.L., Dokainish H., McPherson P.S. DENN domain proteins: regulators of Rab GTPases. J. Biol. Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 17.Mori Y., Matsui T., Omote D., Fukuda M. Small GTPase Rab39A interacts with UACA and regulates the retinoic acid-induced neurite morphology of Neuro2A cells. Biochem. Biophys. Res. Commun. 2013;435:113–119. doi: 10.1016/j.bbrc.2013.04.051. [DOI] [PubMed] [Google Scholar]
- 18.Giannandrea M., Bianchi V., Mignogna M.L., Sirri A., Carrabino S., D’Elia E., Vecellio M., Russo S., Cogliati F., Larizza L. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am. J. Hum. Genet. 2010;86:185–195. doi: 10.1016/j.ajhg.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Camilli P., Cameron R., Greengard P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. I. Its general distribution in synapses of the central and peripheral nervous system demonstrated by immunofluorescence in frozen and plastic sections. J. Cell Biol. 1983;96:1337–1354. doi: 10.1083/jcb.96.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verderio C., Coco S., Pravettoni E., Bacci A., Matteoli M. Synaptogenesis in hippocampal cultures. Cell. Mol. Life Sci. 1999;55:1448–1462. doi: 10.1007/s000180050384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stegmeier F., Hu G., Rickles R.J., Hannon G.J., Elledge S.J. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burman J.L., Wasiak S., Ritter B., de Heuvel E., McPherson P.S. Aftiphilin is a component of the clathrin machinery in neurons. FEBS Lett. 2005;579:2177–2184. doi: 10.1016/j.febslet.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Ritter B., Murphy S., Dokainish H., Girard M., Gudheti M.V., Kozlov G., Halin M., Philie J., Jorgensen E.M., Gehring K., McPherson P.S. NECAP 1 regulates AP-2 interactions to control vesicle size, number, and cargo during clathrin-mediated endocytosis. PLoS Biol. 2013;11:e1001670. doi: 10.1371/journal.pbio.1001670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaudry D., Stork P.J., Lazarovici P., Eiden L.E. Signaling pathways for PC12 cell differentiation: making the right connections. Science. 2002;296:1648–1649. doi: 10.1126/science.1071552. [DOI] [PubMed] [Google Scholar]
- 25.McAllister A.K. Cellular and molecular mechanisms of dendrite growth. Cereb. Cortex. 2000;10:963–973. doi: 10.1093/cercor/10.10.963. [DOI] [PubMed] [Google Scholar]
- 26.Oppenheim R.W. Cell death during development of the nervous system. Annu. Rev. Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 27.Aminoff M.J., Daroff R.B. Elsevier Academic Press; 2014. Encyclopedia of the neurological sciences. [Google Scholar]
- 28.Ruggeri P., Cappabianca L., Farina A.R., Gneo L., Mackay A.R. NGF FLIPs TrkA onto the death TRAIL in neuroblastoma cells. Cell Death Dis. 2016;7:e2139. doi: 10.1038/cddis.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Indo Y., Tsuruta M., Hayashida Y., Karim M.A., Ohta K., Kawano T., Mitsubuchi H., Tonoki H., Awaya Y., Matsuda I. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat. Genet. 1996;13:485–488. doi: 10.1038/ng0896-485. [DOI] [PubMed] [Google Scholar]
- 30.Yeo G.S.H., Connie Hung C.C., Rochford J., Keogh J., Gray J., Sivaramakrishnan S., O’Rahilly S., Farooqi I.S. A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nat. Neurosci. 2004;7:1187–1189. doi: 10.1038/nn1336. [DOI] [PubMed] [Google Scholar]
- 31.Lemos R.R., Ramos E.M., Legati A., Nicolas G., Jenkinson E.M., Livingston J.H., Crow Y.J., Campion D., Coppola G., Oliveira J.R. Update and Mutational Analysis of SLC20A2: A Major Cause of Primary Familial Brain Calcification. Hum. Mutat. 2015;36:489–495. doi: 10.1002/humu.22778. [DOI] [PubMed] [Google Scholar]
- 32.Villa-Bellosta R., Levi M., Sorribas V. Vascular smooth muscle cell calcification and SLC20 inorganic phosphate transporters: effects of PDGF, TNF-alpha, and Pi. Pflugers Arch. 2009;458:1151–1161. doi: 10.1007/s00424-009-0688-5. [DOI] [PubMed] [Google Scholar]
- 33.Dhindsa R.S., Bradrick S.S., Yao X., Heinzen E.L., Petrovski S., Krueger B.J., Johnson M.R., Frankel W.N., Petrou S., Boumil R.M., Goldstein D.B. Epileptic encephalopathy-causing mutations in DNM1 impair synaptic vesicle endocytosis. Neurol. Genet. 2015;1:e4. doi: 10.1212/01.NXG.0000464295.65736.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.EuroEPINOMICS-RES Consortium. Epilepsy Phenome/Genome Project. Epi4K Consortium De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoubridge C., Tarpey P.S., Abidi F., Ramsden S.L., Rujirabanjerd S., Murphy J.A., Boyle J., Shaw M., Gardner A., Proos A. Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nat. Genet. 2010;42:486–488. doi: 10.1038/ng.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aligianis I.A., Johnson C.A., Gissen P., Chen D., Hampshire D., Hoffmann K., Maina E.N., Morgan N.V., Tee L., Morton J. Mutations of the catalytic subunit of RAB3GAP cause Warburg Micro syndrome. Nat. Genet. 2005;37:221–223. doi: 10.1038/ng1517. [DOI] [PubMed] [Google Scholar]
- 37.Aligianis I.A., Morgan N.V., Mione M., Johnson C.A., Rosser E., Hennekam R.C., Adams G., Trembath R.C., Pilz D.T., Stoodley N. Mutation in Rab3 GTPase-activating protein (RAB3GAP) noncatalytic subunit in a kindred with Martsolf syndrome. Am. J. Hum. Genet. 2006;78:702–707. doi: 10.1086/502681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montpetit A., Côté S., Brustein E., Drouin C.A., Lapointe L., Boudreau M., Meloche C., Drouin R., Hudson T.J., Drapeau P., Cossette P. Disruption of AP1S1, causing a novel neurocutaneous syndrome, perturbs development of the skin and spinal cord. PLoS Genet. 2008;4:e1000296. doi: 10.1371/journal.pgen.1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abou Jamra R., Philippe O., Raas-Rothschild A., Eck S.H., Graf E., Buchert R., Borck G., Ekici A., Brockschmidt F.F., Nöthen M.M. Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am. J. Hum. Genet. 2011;88:788–795. doi: 10.1016/j.ajhg.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno-De-Luca A., Helmers S.L., Mao H., Burns T.G., Melton A.M., Schmidt K.R., Fernhoff P.M., Ledbetter D.H., Martin C.L. Adaptor protein complex-4 (AP-4) deficiency causes a novel autosomal recessive cerebral palsy syndrome with microcephaly and intellectual disability. J. Med. Genet. 2011;48:141–144. doi: 10.1136/jmg.2010.082263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verkerk A.J., Schot R., Dumee B., Schellekens K., Swagemakers S., Bertoli-Avella A.M., Lequin M.H., Dudink J., Govaert P., van Zwol A.L. Mutation in the AP4M1 gene provides a model for neuroaxonal injury in cerebral palsy. Am. J. Hum. Genet. 2009;85:40–52. doi: 10.1016/j.ajhg.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anazi S., Maddirevula S., Faqeih E., Alsedairy H., Alzahrani F., Shamseldin H.E., Patel N., Hashem M., Ibrahim N., Abdulwahab F. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol. Psychiatry. 2016 doi: 10.1038/mp.2016.113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.