(The American Journal of Human Genetics 99, 1206–1216; November 3, 2016)
In the original version of this article that appeared online, Figure 1 did not entirely correspond to the figure legend. Panel (D) was missing data of a breakpoint analysis performed for the deletion of the PIEZO exon and an exon in affected individual D-II. The agarose gel in (D) did not include the presence of the neighboring PIEZO exons. In (E), the graphical representation of PIEZO exons did not depict all exons. Reproduced here are the original and corrected panels (D) and (E). The corrected figure appears in print and online. The authors apologize for this error.
Figure 1.
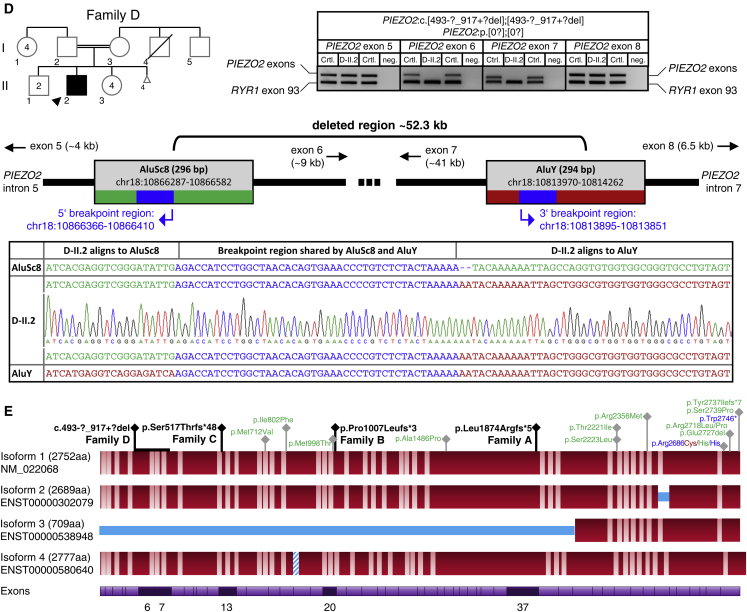
Identification of Homozygous Frameshift Variants in PIEZO2 in Ten Affected Individuals of Four Independent Families (corrected)
(A–D) Pedigrees and Sanger sequencing results of four independent families with homozygous frameshift mutations (A–C) or homozygous exon deletions (D) in PIEZO2. DNA was not available for individuals with gray outline. The unequivocal individual identifier is assembled of family, generation, and individual (e.g., A-III.1). Deletion of a genomic region comprising the PIEZO2 exons 6 and 7 in individual D-II.2 (D) was confirmed by PCR analysis and by identification of the breakpoint region. PIEZO2 exons 5, 6, 7, and 8 were amplified in multiplex with RYR1 exon 93 as PCR-positive control. Breakpoint regions were identified by Sanger sequencing of a long-range PCR product amplified from the genomic region between exon 5 and exon 8.
(E) PIEZO2 protein structure and mutations identified: homozygous loss-of-function mutations (black) and dominant gain-of-function mutations for DA3 (blue), DA5 (green), and MWKS (red) are given. Protein isoforms refer to UniProt entry Q9H5I5. Bright regions indicate putative transmembrane domains (approximated scale); blue regions indicate missing or inserted regions in different isoforms. The bottom line shows exons in which homozygous frameshift mutations were identified.
Figure 1.
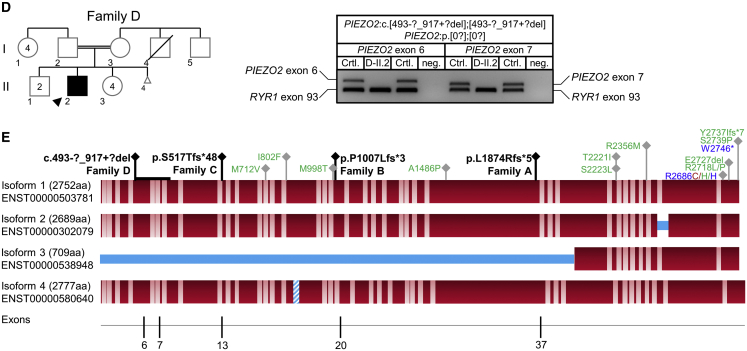
Identification of Homozygous Frameshift Variants in PIEZO2 in Ten Affected Individuals of Four Independent Families (original)
(A–D) Pedigrees and Sanger sequencing results of four independent families with homozygous frameshift mutations (A–C) or homozygous exon deletions (D) in PIEZO2. DNA was not available for individuals with gray outline. The unequivocal individual identifier is assembled of family, generation, and individual (e.g., A-III.1). Deletion of a genomic region comprising the PIEZO2 exons 6 and 7 in individual D-II.2 (D) was confirmed by PCR analysis and by identification of the breakpoint region. PIEZO2 exons 5, 6, 7, and 8 were amplified in multiplex with RYR1 exon 93 as PCR-positive control. Breakpoint regions were identified by Sanger sequencing of a long-range PCR product amplified from the genomic region between exon 5 and exon 8.
(E) PIEZO2 protein structure and mutations identified: homozygous loss-of-function mutations (black) and dominant gain-of-function mutations for DA3 (blue), DA5 (green), and MWKS (red) are given. Protein isoforms refer to UniProt entry Q9H5I5. Bright regions indicate putative transmembrane domains (approximated scale); blue regions indicate missing or inserted regions in different isoforms. The bottom line shows exons in which homozygous frameshift mutations were identified.