Abstract
Background
The evolutionary history and ecological associations of Trypanosoma cruzi, the need to identify genetic markers that can distinguish parasite subpopulations, and understanding the parasite’s evolutionary and selective processes have been the subject of a significant number of publications since 1998, the year when the first DNA sequence analysis for the species was published.
Methods
The current analysis systematizes and re-analyzes this original research, focusing on critical methodological and analytical variables and results that have given rise to interpretations of putative patterns of genetic diversity and diversification of T. cruzi lineages, discrete typing units (DTUs), and populations, and their associations with hosts, vectors, and geographical distribution that have been interpreted as evidence for parasite subpopulation specificities.
Results
Few studies use hypothesis-driven or quantitative analysis for T. cruzi phylogeny (16/58 studies) or phylogeography (10/13). Among these, only one phylogenetic and five phylogeographic studies analyzed molecular markers directly from tissues (i.e. not from isolates). Analysis of T. cruzi DTU or lineage niche and its geographical projection demonstrate extensive sympatry among all clades across the continent and no significant niche differences among DTUs. DTU beta-diversity was high, indicating diverse host assemblages across regions, while host dissimilarity was principally due to host species turnover and to a much lesser degree to nestedness. DTU-host order specificities appear related to trophic or microenvironmental interactions.
Conclusions
More rigorous study designs and analyses will be required to discern evolutionary processes and the impact of landscape modification on population dynamics and risk for T. cruzi transmission to humans.
Electronic supplementary material
The online version of this article (doi:10.1186/s13071-016-1918-2) contains supplementary material, which is available to authorized users.
Keywords: Trypanosoma cruzi, Discrete Type Unit, Host specificity, Niche identity, Ecological niche modeling, Chagas disease
Background
Trypanosoma cruzi is the etiological agent of Chagas disease (CD), considered one of the most important parasitic infections in Latin America. Between 25 and 90 million humans are at infection risk via at least one of multiple infection mechanisms [1]. Under natural conditions, the principal transmission modes are transplacental or via one of more than 140 hematophagous triatomine bugs (Reduviidae: Triatominae). Triatomines acquire the parasite from mammal reservoirs due to their obligate blood-feeding (albeit triatomines can also feed on non-reservoir vertebrates such as birds and reptiles) [2–5]. The disease burden for CD in the Latin America and Caribbean region, based on disability-adjusted life-years (DALYs), is at least five times greater than that of malaria, and is approximately one-fifth that of HIV/AIDS [6]. In recent decades, CD has extended to other continents outside natural reservoir or vector distributions due to human migration, with a minimum estimated 10 million individuals infected worldwide [6].
A significant number of studies have been published since 1998 to analyze T. cruzi’s evolutionary history using subpopulation-informative genetic markers (the year when the first DNA sequence analysis for T. cruzi was published). Trypanosoma cruzi is a diploid organism having clonal structure [7–9], strong linkage disequilibrium, and an absence of segregated and recombining genotypes [10]. Only isolated events of recombination or genetic exchange have been documented [8, 11, 12]. Recent technological developments and novel genetic markers are providing new evidence regarding the parasite’s genetic variability [13, 14], prevalence in different hosts, and geographical distribution. Multi-locus enzyme electrophoresis (MLEE), random amplified polymorphic DNA (RAPD), and other methods all demonstrate a broad genetic diversity [15, 16]. Two major lineages of the parasite, T. cruzi I and T. cruzi II, were defined originally using isoenzymes and other molecular markers such as 24S rDNA, and the mini-exon genes [16, 17]. These two clades can be further subdivided into six subclades named discrete typing units (DTU), while subsequent studies using alternative gene sequences have proposed an alternative phylogeny with three principal lineages [11, 17, 18]. The first major lineage proposal [14] designates Lineage I to include only DTUI which has been further subdivided into subgroups a, b, c, d and e based on the microsatellite region of the SL-IR and mini-exon genes [13, 19, 20]. Lineage II from the first scheme includes all other DTUs, originally designated IIa, IIb, IIc, IId and IIe. These are now assigned DTU classification IV, II, III, V and VI, respectively [14]. The newest proposal for three primary lineages is based on a multilocus phylogenetic analysis [18], in which Lineage I continues to include only DTUI, while Lineage II only includes DTUII, and Lineage III includes both DTUs III and IV [18]. The remaining hybrid DTUs, V and VI, are not assigned to any lineage, although they are closest to DTUII.
Understanding T. cruzi population genetics is fundamental to discern parasite’s flow within landscapes, particularly related to fragmentation and land use change, where there are native host community structures and non-native hosts (i.e. livestock, companion animals and humans). Its population dynamics is mediated by the dispersal capacity and interactions of hosts and vector species within landscapes, key components of transmission risk to humans. Since knowledge of parasite population dynamics is fundamental to design effective barriers to prevent human-vector contact and to guide effective patient treatment and clinical care [11, 21], what evidence currently exists regarding the phylogenetic patterns (i.e. speciation events) or phylogeography (i.e. geographical variation of genetic diversity) of T. cruzi? Is there a significant association between lineage, DTU, or subtype, with particular host taxa or vector species, ecotopes (sylvatic, transformed, domestic/peridomestic), landscape change (conserved, matrix, urbanized), biome, or latitudinal gradient? What associations exist between these variables and genetic patterns, or phylogeography of the parasite? Despite previous reviews of evidence regarding genetic diversity and phylogeography of T. cruzi, most have focused on gathering concluding information from publications without systematically analyzing coherence among study aims, design, sampling methods, and criteria used to reach conclusions regarding clonal evolution, presence of DTUs, eco-epidemiology, or clinical associations of parasite populations [9, 11, 13, 22]. Former proposed patterns can be re-evaluated by new evidence or as the result of incomplete experimental designs, or biased field data collections [23, 24]. An additional and more pressing problem for analyzing T.cruzi’s genetic diversity, variation, or structuring from a landscape perspective is the fact that, in general, most data are generated from culture or laboratory animal selected populations (parasite isolates). These difficulties to analyze parasite populations in all hosts have created voids in knowledge of intra-host and metapopulation dynamics, in addition to limitations from study designs and sampling methods, including ex-host selection (in vitro or in vivo) documented previously [25–27].
A complete review of gene sequences used in molecular diagnosis and genotyping summarizes evidence regarding their specificity and sensitivity from certain hosts [28]. MLEE [29], RAPD [30] and kDNA amplification [31], were the first techniques used for T. cruzi molecular identification and characterization. Methods for phylogenetic and phylogeographic studies have used the mini-exon [32], kinetoplast DNA (kDNA), ribosomal DNA (rDNA) [17], GPI [33] and cytb genes [34]. kDNA and satellite DNA (DNAsat) are the principal sequences amplified for human parasite diagnosis, the latter being more specific than kDNA in humans [28, 35]. There are different pairs of primers used for kDNA PCR (s35/36, s34/67, 32f/148r and 121/122), each with different sensitivities, and all produce false positive bands (amplification of homologous host or symbiont DNA). Hence, a combination of both kDNA and DNAsat markers have been considered an optimum solution for patient diagnosis and other aims, if sequencing is not procedural [28]. It is important to note that the sensitivity and specificity of most markers and primers have been analyzed using culture or animal model selected “isolates”, which may indicate a bias for homogeneous populations having non-polymorphic sequences, and may not be specific or sensitive additionally for those populations not tolerant to in vitro methods. Genotyping and classification techniques for T. cruzi have evolved from the use of RAPDs and isoenzymes, to the use of microsatellites and single nucleotide polymorphisms (SNPs). New strategies using both conventional and real time (rt) PCR techniques improve specific amplification for diagnosis in blood by using lower DNA template quantity without the high sequencing costs [28]. However, these methods have not been validated with original samples from a broad range of hosts. Several of these typing methods combined with other markers are also used for DTU classification [36–38].
The term DTU was defined for a group of isolates genetically more similar than each clade to the others, using several molecular markers [39]. Six DTUs (I-VI) were defined as noted above [14, 30], and more recently Tcbat was proposed, and the most frequently used technique to classify DTUs is a multi-primer combined PCR-RFLP analysis that discriminates between the six. Diversity within the various DTUs has required additional sub-classifications, based on additional genetic markers [19, 20, 40]. The multiplex rtPCR is a more recent quantitative method which attempts to standardize parasite DTU typing along with parasite detection, which again will depend on sensitivity to amplify a broad range of haplotypes [11, 28].
From an ecological perspective, host diversity of a parasite represents one of its ecological niche breadth components, since it reflects the diversity of resources used [41]. From an evolutionary perspective, host diversity is not merely a function of how many host species can be exploited, but also which of these are exploited, and how closely-related are they to each other [42]. A parasite’s host spectrum is a result of both location and breadth of its multidimensional ecological niche which can be measured using theoretical methods developed for community ecology [43]. Since all T. cruzi DTUs have been identified across a wide geographical range in the American continent, analyzing host diversity implies understanding host specificity patterns across the continent. A parasite can be highly host specific at local scales, and opportunistic at global scales, or vice versa. In the former case, the parasite uses few hosts that may be substituted across areas (host species turnover), while in the latter case, a parasite may exploit optimally a consecutive subset of host species that are regionally restricted (nested host communities). These two facets of host diversity are additive and contrasting and can be used to evaluate turnover and nestedness of T. cruzi (DTU or lineage) using host niches across regions [44, 45].
The current re-analysis first reviews diagnostic methods and their ability to identify subpopulations from all hosts, their DTUs and lineages, since they are fundamental tools to analyze the evolutionary history and ecological relationships of T. cruzi. It systematizes and weighs original research on T. cruzi population genetics, based on critical methodological and analytical variables. Current genetic and geographical evidence from appropriate studies have then been submitted to a re-analysis of associations with specific host taxa and geographical areas. Particular questions guiding this re-analysis were whether: (i) parasite populations typed as DTUs or lineage schemes including that proposing TcI and TcII (L1) [14], or an alternative lineage scheme proposing TcI, TcII, and TcIII (L2) [18] are broadly distributed and sympatric (or not); (ii) the suggested relationship between marsupials and armadillos and TcI and TcII of lineage scheme L1, respectively, are sustained when contrasted with representative mammal community analyses from other landscapes, and (iii) greater DTU or lineage diversity is found in any particular ecotope or host. The re-analysis evaluates whether in fact these questions could be answered and quantitatively analyzed based on currently published evidence.
Methods
Evidence for T. cruzi population genetics
A systematic search of published evidence was carried out using PubMed and Google Scholar, using the following terms: “molecular diagnosis AND Trypanosoma cruzi”, “discrete typing units AND Trypanosoma cruzi”, “population genetics AND Trypanosoma cruzi”, “genetic diversity AND Trypanosoma cruzi “, “population structure AND Trypanosoma cruzi “,“landscape genetics AND Trypanosoma cruzi”, “phylogeography AND Trypanosoma cruzi” and “phylogenetics AND Trypanosoma cruzi”, “host association AND Trypanosoma cruzi “, and “geographic distance and Trypanosoma cruzi”. An additional search was carried out, using some of the most frequent authors of the first search: “Tibayrenc”, “Barnabé”, “Brisse”, “Miles MA”, “Breniere” “Zingales”, “Oliveira RP”, “Machado”,”Guhl”, “Schijmann”, “Llewellyn”, etc., adding “AND Trypanosoma cruzi”. The criteria for inclusion of data from publications were that (i) they were original research on molecular diagnosis, genotyping, phylogeny, or phylogeography of T. cruzi as related to associations with reservoirs, vectors, geographical range, or disease associations; (ii) that they were published after 1998, the year when reproducible techniques were established for genotyping; (iii) they include DNA sequence analysis, which may also include use of restriction fragment length polymorphisms (RFLP) or the low stringency single specific primer (LSSP); and (iv) that they were published in peer reviewed indexed journals. The only exclusion criterion for studies used in re-analysis was the lack of use of statistical analytical methods for phylogeny or phylogeography. Publications were reviewed based on three principal categories: (i) study approach: aim of the study, formulation of hypotheses, and level of analysis; (ii) criteria for sample selection: origin of the population analyzed (direct from tissue, culture isolates, etc.), taxon of origin of the parasite population, and number of samples per taxon; and (iii) use of one of the following molecular markers or methods: DTU classification, mitochondrial or nuclear gene markers, size of sequences, use and number of microsatellites, and use and type of outgroups. A database was created with the above information along with information from each study results and conclusions.
All studies were classified and categorized according to four main criteria based not only on each study conclusions, but rather based on study aim and methods: molecular analytical methods, evidence generated regarding phylogenetics, evidence generated regarding phylogeography or landscape genetics, and evidence of association between parasite populations and illness. Most studies, both for phylogeny or phylogeography, did not use parasite populations directly from the host, but rather used “isolates” (58/59 for phylogeny and 11/14 for phylogeography). Sample size, geographical scale, and analytical methods were compared separately for studies using isolates vs original parasite populations.
Biotic niches of T. cruzi DTUs and lineages
We assessed biotic (i.e. mammal and triatomine hosts) and abiotic (i.e. bioclimatic/topographic conditions) niche dimensions and divergence among current T. cruzi populations. Biotic niche divergence was evaluated complementarily by testing if each T. cruzi DTU or lineage was singularly more associated to any vector genus or mammal according to a proportional distribution of frequencies (i.e. no singular association), using a Chi-square independence test. Since there are nil or less than 5 samples per DTU or taxa in certain categories, a secondary analysis using confidence intervals to analyze for the null hypothesis that all DTUs had the same probability (17%) to be found in all taxa. We used three different classification schemes to analyze the prevalence of T. cruzi across reservoir orders and vector genera: (i) individual DTUs I, II, III, IV, V and VI; (ii) the L1 [14] major Lineages I and II; and (iii) the L2 [18] lineages I, II and III classifications (see Additional file 1: Table S1). A contingency table was generated with the observed frequencies reported from the literature database for Triatoma, Rhodnius and Panstrongylus, and separately for mammal orders, with no consideration of spatial location.
Given increasing suggestion of sorted host usage by different parasite clades, we analyzed specificity for vector and mammal community assemblages as opposed to single host orders, a measure of host specificity that reflects spatial variation in host species composition or “host spectrum” [45]. This method uses the turnover component of beta-diversity (free from the effect of nestedness) as a new measure of host specificity ("beta-specificity"), since it reflects the "pure" ability of a parasite to shift hosts from one region to another independently of any non-random and/or any nested patterns. Nestedness tends to inflate beta-diversity (and thus beta-specificity) but does not inform whether a parasite is able to shift host composition across scales, only that the parasite population infests subsets of hosts that are nested within the broader host spectrum, in a specific location. A beta-specificity index was calculated to reflect the total beta-diversity (βSOR) based on the Sorensen dissimilarity, by adding the dissimilarity due to "pure" host species turnover (βSIM, a measure of multi-site spatial turnover free from the influence of species richness based on the Simpson dissimilarity index) and βSNE dissimilarity due to nestedness. The mathematical properties of these indices have been tested using different ecological circumstances and have been proven to be highly robust [46–48]. Beta specificity was calculated using the package beta-multi.R [49] for the R software environment (R version 3.0.2). Beta-specificity for each DTU or lineage scheme was analyzed to determine if T. cruzi has consistent host niche patterns (i.e. if lineages/DTUs behave differently in their host spectrum thus reflecting a difference in generalization). Presence-absence matrices were constructed for each DTU or lineage scheme by assigning regions to rows (composed of 50 × 50 km units), and host species that harbored specific lineage in a specific geographic pixel to columns. A matrix reflecting the variation in species composition and host spectrum was constructed for each DTU or lineage, and matrices were grouped by vectors, mammal hosts, or both combined.
Abiotic niche divergence
We constructed bioclimatic niches and assessed abiotic niche divergence by conducting a two-step framework. First, ecological niche models were constructed for each DTU or lineage scheme (L1 and L2), and projected to the American continent for distribution range maps. Subsequently spatial overlap among DTUs and lineages was analyzed using a statistical framework based on null models, to determine if niche was more similar than expected randomly. The same dataset used for biotic niche evaluations (see Additional file 1: Table S1) was used to develop ecological niche models (ENM) for all DTUs, for which collection sites were reported. A total of 234 (out of 270 reported) unique data points were used for T. cruzi DTUI, 8 (out of 14) for DTUII, 27 (out of 38) for DTUIII, 36 (out of 53) for DTUIV, 13 (out of 13) for DTUV, and 49 data-points for DTUVI. These data points were grouped for Lineage I/L1 and L2 (234 data points), Lineage II/L1 (108 data points), Lineage II/L2 (8 data points), or Lineage III/L2 (63 data points). A total of 1997 data points for more than one DTU or lineage or for which georeference resolution was not reported, were not used for model construction. The American continent was divided into 2,632,469 pixels at a resolution of 2.5 min (0.0416667° ≈ 5 km) for latitude and longitude. Nine bioclimatic data layers (BIO1, BIO4, BIO5, BIO6, BIO7, BIO12, BIO13, BIO14 and BIO15) were obtained from Worldclim (www.worldclim.org) at a resolution of 2.5 min [50]. These bioclimatic variables were selected from a total of 19 by choosing the more meaningful variables hypothesized to limit species distributions at coarse-grain scales, after analysis of multi-collinearity in a correlation matrix [51]. The final dataset layer included variables with relatively low correlation (r < 0.75). Additionally, four topographic layers (aspect, slope, topographic index and elevation) were used from the Hydro 1 k data set (http://lta.cr.usgs.gov/GTOPO30). ENM’s based on occurrence data, bioclimatic, and topographic layers were constructed using the Genetic Algorithm for Rule-set Prediction (GARP) [52]. For GARP, we used a convergence criterion of 0.01 and 1000 maximum iterations; a consensus of replicate model was achieved via a 20% relative omission threshold, retaining the central 50% of the distribution of proportional areas predicted as suitable. The software randomly divides occurrence points into calibration data for model building (70%) and evaluation data for model testing (30%) [53, 54]. Each ENM was evaluated using partial ROC (receiver operating curve) using software developed by [55, 56]. The AUC (area under the ROC) was calculated using the values of “sensitivity” in the y-axis and the commission error in the x-axis, measuring the maximum inflection point where both errors are minimized. AUC values are significantly different than random when above 1.0 [51]. The 10 best subsets for each model were projected along with the data points. We used a minimum presence threshold criterion of 95% (E = 5%) in order to generate a binary map (presence/absence) of each projection from the 0–100 range of model output.
To compare similarity between niche models among DTUs or the major lineages (schemes L1 and L2), we used an identity test with the 13 climate and topographic variables for the entire continent. Niche models for identity tests were developed using MaxENT since this tool is not available for GARP [57], with 75% random test, bootstrap replicated runs, 500 maximum iterations, and minimum training presence using ENMtools v.13.2 (http://enmtools.com/). The Hellinger’s and Schoener’s indices for niche identity were calculated for all pairwise combinations of DTUs, or lineages from L1 and L2. The empirical measure of niche similarity between DTUs or lineages was compared to a null distribution for significant difference from that generated for niche models constructed with data points extracted randomly from the distribution range. The hypothesis of niche identity is rejected when the empirically observed value for the two distance indices falls to the left side of the similarity axis, outside of the frequency distribution of the random models. Observed similarities falling inside the frequency distribution or to the right of the axis are considered as insufficient evidence for lack of niche identity [58].
Results
The dataset for re-analysis of T. cruzi DTUs or lineages included 59 studies on T. cruzi phylogeny (Tables 1 and 2) and 14 studies on its phylogeography (Tables 3 and 4). Characteristics of studies excluded from analyses are summarized in Additional file 2: Table S2 and Additional file 3: Table S3. Study design, sample selection, and criteria for molecular marker analysis of T. cruzi phylogeny are summarized in Table 1. Only two studies contained sample size and type, and analytical design based on a hypothesis testing approach (use of original parasite samples and outgroups). The number of samples used in each study was highly variable and almost all, except for four studies, analyzed isolates selected from previous studies (either using culture and/or laboratory animal selection). Two studies analyzed both cultured isolates and original parasite samples, although differential results for both are not presented. Complete or partial classification of DTUs was carried out in 45 of 59 studies, some of these used samples that had been classified in previous studies (27/59). Only a minor proportion of the phylogenetic studies used quantitative methods to validate results (appropriate outgroup, probabilities, and confidence intervals) (17/59) (Table 2).
Table 1.
Classification of phylogenetic studies based on review categories
| Review category | Classification | No. of studies | Reference |
|---|---|---|---|
| Spatial level | Continental | 29 | [18, 33, 40, 88–113] |
| National | 17 | [19, 20, 27, 73, 114–126] | |
| Regional | 13 | [127–139] | |
| Aims | Phylogenetic relationship and/or genetic diversity | 42 | [19, 20, 33, 40, 73, 96, 100–113, 120–125, 132–139] |
| Population structure and/or genetic diversity | 9 | [88–91, 93, 94, 114, 117, 126] | |
| Genetic diversity and/or host association | 3 | [27, 116, 127] | |
| Genetic diversity and genome or proteome association | 5 | [18, 92, 95, 115, 128] | |
| Hypothesis | Study and sample design based on a hypothesis | 2 | [133, 136] |
| Study and sample design not based on a hypothesis | 57 | [18–20, 27, 33, 40, 73, 88–132, 134, 135, 137–139] | |
| Sample source | Humans, other mammals, and triatomines | 43 | [18–20, 33, 40, 73, 88, 90, 92–99, 102–120, 122–124, 126–128, 131, 139] |
| Triatomines and humans | 6 | [27, 89, 100, 121, 130, 134] | |
| Triatomines and other mammals | 2 | [125, 137] | |
| Triatomines | 4 | [129, 133, 135, 136] | |
| Other mammals | 1 | [91] | |
| Humans | 3 | [131, 132, 138] | |
| Parasite population analyzed | Isolates selected using in vitro or in vivo methods | 53 | [18–20, 33, 73, 88–128, 130–132, 134, 135, 137, 139] |
| Original sample | 4 | [27, 129, 133, 136] | |
| Isolates + original | 2 | [40, 138] | |
| Molecular marker used for parasite diagnosis and genotyping | DTU | 9 | [89, 90, 94, 97, 99, 104, 112, 122, 127] |
| MLEE +/or RAPD + DTUs | 9 | [18, 33, 95, 105–109, 111] | |
| Mini-exon + 24S rRNA | 9 | [91, 96, 113, 116, 130–132, 135, 138] | |
| MLEE +/or RAPD | 6 | [100, 103, 114, 118, 137, 139] | |
| Mini-exon + 24S rRNA + DTU | 2 | [73, 110] | |
| Mini-exon + MLEE+/or RAPD | 2 | [88, 102] | |
| GPI | 2 | [117, 128] | |
| Cyt b | 1 | [125] | |
| 24SrRNA | 6 | [92, 93, 98, 115, 127, 133] | |
| Mini-exon | 10 | [19, 20, 40, 119–121, 123, 124, 134, 136] | |
| kDNA (121/122) | 1 | [126] | |
| kDNA (S35/S36) | 2 | [27, 129] | |
| DTU | Classified | 19 | [19, 20, 27, 40, 73, 98, 116, 119–121, 123, 125, 128, 130, 133–137] |
| Previously classified | 26 | [18, 33, 89–91, 94, 95, 97, 100, 102, 104, 106–112, 114, 117, 118, 122, 124, 126, 127, 131] | |
| Not classified | 14 | [16, 88, 92, 93, 96, 99, 103, 105, 113, 115, 129, 132, 138, 139] | |
| Population genetic analyses | Nuclear | 23 | [19, 20, 33, 40, 89, 96–99, 101, 105, 106, 110–113, 117, 120, 127, 133, 135–137] |
| Mitochondrial | 3 | [108, 116, 125] | |
| Nuclear + mitocondrial | 7 | [18, 73, 100, 103, 107, 118, 122] | |
| Nuclear + mitochondrial + microsatelite | 4 | [88, 90, 104, 131] | |
| Nuclear + microsatellite | 1 | [121] | |
| Mitochondrial + microsatellite | 2 | [94, 128] | |
| Microsatelite | 10 | [27, 91–93, 102, 109, 114, 115, 130, 132] | |
| LSSP or RFLP | 9 | [95, 119, 123, 124, 126, 129, 134, 138, 139] | |
| Outgroups | T. c. marinkelli | 9 | [95, 106–108, 116, 125, 127, 135, 136] |
| T. c. marinkelli + T. dionisii | 1 | [73] | |
| T. c. marinkelli + T. rangeli | 2 | [33, 101] | |
| T. c. marinkelli + T. brucei | 1 | [122] | |
| T. c. marinkelli + Leishmania major + L. donovani | 1 | [96] | |
| T. c. marinkelli + T. vespertilionis | 3 | [18, 100, 103] | |
| T. c. marinkellei + T. rangeli + T. brucei | 1 | [90] | |
| T. rangeli + T. dionisii | 1 | [119] | |
| T. rangeli | 5 | [113, 123, 124, 126, 133] | |
| TcII and/ or TcIV | 4 | [20, 117, 120, 128] | |
| Leishmania chagasi | 1 | [134] | |
| B. caudatus + T. borreli + T. rangeli | 1 | [98] | |
| Not used | 29 | [19, 27, 40, 88, 89, 91–94, 97, 99, 102, 104, 105, 109–112, 114, 115, 118, 121, 129–132, 137–139] |
Table 2.
Phylogenetic studies which use quantitative analytical methods
| Article | Hypothesis/Aim | Parasite population | Sample size | Geographical scale | Temporal scale (yrs) | Population genetic analysis | Outgroup | Statistical analytical method |
|---|---|---|---|---|---|---|---|---|
| Flores-Lopez & Machado [18] | Reconstruction of the evolutionary history of Tc | Isolates | 7 | Tc stocks | md | Nucleotide sequences from 32 loci | T. c. marinkellei, T. vespertilionis | Test of selection, divergence time estimates |
| Venegas et al. [27] | Specific host-parasite association in Chilean populations of Tc | Original | 117 | Chile | 2 | Microsatellite loci | none | Phylogram tree (NJ)/Genetic differentiation |
| Barnabe et al. [114] | Subsample analyses of MLMT structuring among reference stocks belonging to known DTUs | Isolates | 94 | Bolivia and Peru | 32 | Microsatellite loci | none | NJ trees/Fixation indices FIS and FST/Genetic diversity Hs/ANOVA |
| Freitas et al. [88] | Dissect the multilocus genotypes into their constituent haploid genome blocks to understand Tc evolutionary history | Isolates | 75 | Brazil | md | Microsatellite loci, 24SrRNA, cox2 sequencing/ RFLP | none | Distance matrices, multidimensional scaling and NJ tree/Haplotype inference and network construction |
| Ienne et al. [89] | Test hybridization hypothesis | Isolates | 9 | Tc stocks | md | 195 SAT | none | Phylogenetic inference (NJ)/Network |
| Lauthier et al. [127] | Stability of multilocus genotypes as the required condition for any molecular epidemiology approach (strain typing) | Isolates | 32 | Argentina | md | Multilocus sequence typing (10 targets) | T. c. marinkellei | Phylogenetic tree/Genotype network |
| Lewis et al. [90] | Origins and evolution of Tc at several overlapping levels | Isolates | 35 | South America | md | GPI, cox2-nad1, microsatellite loci | none | Bayesian Inference, microsatellite analysis |
| Llewellyn et al. [91] | Within-host diversity in TcI | Isolates | 211 | Bolivia, Venezuela and Brazil | 5 | Microsatellite loci | none | Genetic distance (DAS), FIS (FSTAT), AMOVA and index of association |
| Macedo et al. [92] | Usefulness of microsatellite typing in population genetic studies of Tc | Isolates | 53 | Tc stocks | md | Microsatellite loci | none | Wagner network |
| Oliveira et al. [115] | Population structure of the parasite | Isolates | 30 | Brazil and Colombia | md | 8 microsatellites | none | Wagner network |
| Oliveira et al. [93] | Population structure of Tc | Isolates | 54 | md | md | 8 microsatellites | none | Tests for Hardy-Weinberg and linkage disequilibrium/Wagner network |
| Pena et al. [94] | Population structure of TcI | Isolates | 75 | Tc stocks | md | Microsatellite and mitochondrial sequences | none | NJ/ Network |
| Ramirez et al. [116] | Genetic variability within TcI clones and concordance with the established genotypes | Isolates | 70 | Colombia | md | cytb sequencing | T. c. marinkellei | Phylogenetic tree/Genotype network |
| Ramirez et al. [128] | Contemporary cryptic sexuality in Tc | Isolates | 369 | Colombia | 10 | Microsatellite and mitochondrial sequences | DTUII and DTUIV | Genetic diversity/NJ/ML/BEAST |
| Ramirez et al. [117] | Nuclear MLST markers to unravel the genetic structure of TcI in Colombia | Isolates | 50 | Colombia | 11 | Nuclear multilocus sequence typing | DTUII and DTUIV | Genetic diversity and diploid sequence types (DSTs) |
| Telleria et al. [95] | Association between Tc subspecific phylogenetic diversity and levels of protein expression | Isolates | 26 | Tc stocks | md | Proteomics data | T. c. marinkellei | MLEE genetic distances and proteomic Euclidian distances |
| Tomazi et al. [96] | Hybrids are of polyphyletic origin, evolving independently from various hybridization events | Isolates | 26 | South America | md | Sequences of SSU rDNA, EF-1α, actin, DHFR-TS and TR genes | none | Phylogeny inference and network geneologies |
Abbreviations: md missing data, Tc Trypanosoma cruzi
Table 3.
Classification of phylogeographic studies based on review categories
| Review category | Classification | No. of studies | Reference |
|---|---|---|---|
| Spatial scale | Continental | 5 | [67, 68, 140–142] |
| National | 4 | [77, 78, 143, 144] | |
| Regional | 5 | [64, 80, 81, 84, 145] | |
| Aims | Phylogenetic relationships | 4 | [64, 142, 144, 145] |
| Genetic diversity and phylogenetic relationships | 2 | [78, 141] | |
| Genetic diversity and spatial associations | 4 | [67, 68, 81, 143] | |
| Population structure and spatial associations | 2 | [77, 84] | |
| Spatial association and haplotype-host association | 2 | [80, 140] | |
| Hypothesis | Study and sample design based on a hypothesis | 2 | [80, 84] |
| Study and sample design not based on a hypothesis | 12 | [64, 67, 68, 77, 78, 81, 140–145] | |
| Sample source | Humans, other mammals and triatomines | 8 | [64, 68, 80, 84, 140–142, 144] |
| Triatomines and other mammals | 5 | [67, 77, 78, 81, 145] | |
| Triatomines | 1 | [143] | |
| Parasite population analyzed | Isolates selected using in vitro or in vivo methods | 11 | [64, 67, 68, 77, 78, 81, 140–142, 144, 145] |
| Original sample | 3 | [80, 84, 143] | |
| Molecular marker used for diagnosis and genotyping | Mini-exon | 3 | [80, 140, 144] |
| 24S rRNA | 1 | [67] | |
| 24S rRNA + mini-exon | 3 | [64, 68, 142] | |
| S35/S36 kDNA | 2 | [81, 143] | |
| S35/S36 + TcZ1/TcZ2 + mini-exon + 24S rRNA + 18S rRNA | 1 | [145] | |
| MLEE +/or RAPD+ miniexon | 1 | [84] | |
| LSU rDNA + HSP60 + GPI | 1 | [77] | |
| GPI | 2 | [78, 141] | |
| DTU classification | Classified in study | 13 | [64, 67, 68, 77, 78, 80, 81, 84, 141–145] |
| Previously classified | 1 | [140] | |
| Population genetic analysis | Nuclear sequences | 3 | [80, 140, 145] |
| Nuclear + mitochondrial sequences | 2 | [64, 142] | |
| Microsatellites | 3 | [68, 81, 143] | |
| Nuclear sequence and microsatellite | 1 | [67] | |
| Mitochondrial sequence and microsatellite | 3 | [77, 78, 141] | |
| RFLP or LSSP-PCR | 2 | [84, 144] | |
| Outgroups | T. c. marinkelli | 1 | [67] |
| T. c. marinkelli + T. dionisii | 1 | [142] | |
| DTUI | 1 | [77] | |
| DTUII | 2 | [84, 144] | |
| DTUIII + DTUIV | 1 | [141] | |
| DTUIV | 1 | [78] | |
| none | 7 | [64, 68, 80, 81, 140, 143, 145] |
Table 4.
Phylogeographic studies using quantitative analytical methods
| Article | Hypothesis/Aim | Parasite population | Sample size | Geographical scale | Temporal scale (yrs) | Population genetic analysis | Outgroup | Statistical analytical method |
|---|---|---|---|---|---|---|---|---|
| Llewellyn et al. [67] | Population genetics of sylvatic TcIIc from South America (diversity, spatial structure and climatic associations) | Isolates | 53 | Colombia, Brazil, Venezuela, Bolivia and Paraguay | 25 | GPI sequencing/ microsatellite | T. c. marinkellei | NJ/Genetic diversity (Ar/He/Ho/HD)/Mantel test |
| Llewellyn et al. [68] | Population genetics of sylvatic TcI/diversity associated with epidemiological relevance | Isolates | 135 | Americas | 22 | Microsatellite | none | Genetic diversity (Ar/He/Ho/HD)/Mantel test |
| Messenger et al. [77] | Population structure, hybridization and role for humans in parasite dispersal | Isolates | 199 | Bolivia | 6 | Microsatellite and maxicircle | DTUI | Population genetic parameters/NJ/Fst/ Mantel test/ML |
| Lima et al. [78] | Genetic diversity, genetic exchange and impact of ecological disturbance | Isolates | 107 | Brazil | md | Microsatellite and maxicircle | DTUIV | Genetic diversity parameters/ML |
| López-Cancino et al. [80] | Relationships between parasite diversity, host metacommunities, and vectors in a human-disturbed gradient | Original | 81 | Yucatán, México | 5 | Mini-exon sequencing | none | Network analysis/Phylogenetic (ML) |
| Ocaña-Mayorga et al. [81] | Genetic subdivision by transmission cycle, and anthropogenic dispersal between communities and panmixia among strains | Isolates | 81 | Loja Province, Ecuador | md | Microsatellite | none | Genetic diversity (Ar/PA/FIS/FST)/ AMOVA/DAS/Mantel test |
| Rodriguez et al. [84] | Transmission dynamics of Tc genotypes | Original | 121 | Colombia | md | LSSP-PCR, Southern Blot | DTUII | Nei's distance and NJ/AMOVA |
| Herrera et al.[140] | Sequence variability of the SL-IR | Isolates | 244 | 11 Latin American countries | md | Mini-exon sequencing | none | Network/PCA |
| Herrera et al. [145] | Genotype diversity of Tc in Louisiana | Isolates | 15 | New Orleans, Louisiana (USA) | 4 | Miniexon sequencing | none | Phylogenetic (ML) |
| Venegas et al. [143] | Geographical structure and genetic differences among or within lineages in Chilean Tc populations | Original | 64 | Chile | 2 | Microsatellite | none | Fisher's exact test, AMOVA, FST, Mantel test |
| Zumaya-Estrada et al. [141] | Dispersal among domestic transmission cycles of Tcdom in northern South America, sister group of North American strains | Isolates | 72 | Americas | md | Microsatellite | DTUIII, DTUIV | Genetic diversity/Bayesian topology/NJ/MYA geographic calibration point |
Abbreviations: md missing data, Tc Trypanosoma cruzi, Tcdom, Trypanosoma cruzi domestic genotype
Fourteen studies analyzed an association between T. cruzi genetic and geographical distance (Table 3), although only one analyzed genetic distance at the landscape level. Two studies analyzed population structure and spatial associations, and another two studies analyzed spatial and haplotype-host associations. Only two studies had compatible specific aim and sample or analytical design, while only three studies analyzed original samples as opposed to isolates. DTUs were classified in 13 of 14 studies, even though some of these did not complete DTU typing. Most of these studies (11/14) used quantitative analytical methods to validate results (Table 4).
A total of 1402 vector specimen records from 56 species across 6 genera were included in the present analysis, although sufficient data points for quantitative analysis with DTUs or lineages was available only for the three major genera: Triatoma, Rhodnius and Panstrongylus (Table 5). A total of 975 mammal specimens infected with any T. cruzi DTU or lineage were recorded from 95 mammal species, categorized by order (Table 5). The summary of T. cruzi DTUs recorded from mammals and used in this re-analysis is summarized by order, including Carnivora (with and without pets) and Primates without humans, respectively (see Additional file 4: Table S4). Current data do not report any presence of certain DTUs in specific hosts, or there are less than 5 reports per taxa-DTU. An analysis of confidence intervals indicates that the probability of true negative was below 67% for DTUII, DTUIII, DTUIV and DTUV only in Artiodactyla, while in all other taxa and for all DTUs, the probability was greater than 98%.
Table 5.
Trypanosoma cruzi DTUs and lineages reported from vector and mammal species and samples in literature
| No. of species of vectors | Vector samples (n) | No. of species of mammals | Mammal samples (n) | |
|---|---|---|---|---|
| DTUI | 29 | 1204 | 44 | 640 |
| DTUII | 5 | 14 | 10 | 30 |
| DTUIII | 7 | 52 | 10 | 87 |
| DTUIV | 10 | 36 | 21 | 103 |
| DTUV | 2 | 41 | 3 | 41 |
| DTUVI | 3 | 55 | 7 | 74 |
| L1-I | 29 | 1204 | 44 | 640 |
| L1-II | 27 | 198 | 51 | 335 |
| L2-I | 29 | 1204 | 44 | 640 |
| L2-II | 5 | 14 | 10 | 30 |
| L2-III | 17 | 88 | 31 | 190 |
Trypanosoma cruzi DTU associations with specific vectors, hosts and geographical range
DTU frequenciess across reservoir taxa was not independent of DTU class, indicating a significant difference from expected (χ 2 = 1,334.084, df = 40, P < 0.0001). This was particularly the case for Carnivora, Didelphimorphia, Primates (with and without humans), and Rodentia. Only DTUI and VI have been reported in Artiodactyla, with a significant 10-fold increase from expected for DTUVI (P < 0.001) (Fig. 1). All DTUs were reported in Carnivora, although significantly more DTUIV (P < 0.05) and VI (P < 0.001), while significantly less than expected DTUI (P < 0.001) and III (P < 0.05). If pets were excluded from Carnivora, only an increase of DTUIV (P < 0.001) and a decrease of DTUI (P < 0.001) were significant, since DTUVI and DTUIII were only recorded in pets. DTUII was reported significantly more than expected in Chiroptera (P < 0.001); both DTUVI and DTUI were also reported from this latter order, but their frequencies were not significantly different from expected. Statistical inferences for the three previous groups must be considered preliminary, given the low sample size currently genotyped.
Fig 1.
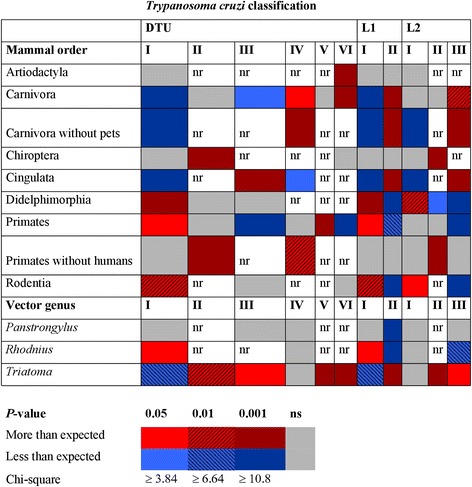
Frequency significance for Trypanosoma cruzi DTUs and lineages in mammal orders and three primary vector genera. Abbreviations: L1, Lineage L1 [14]; L2, Lineage L2 [18]. Abbreviations: nr, not reported; ns, not significant
Only DTUI, III and IV have been reported from the Cingulata, with significantly greater than expected DTUIII (P < 0.001), although significantly less DTUI (P < 0.001) and DTUIV (P < 0.05) in the order. DTUI, II and III have been reported from Didelphimorphia, with significantly more than expected DTUI (P < 0.001) and no significant difference from expected frequencies of the other two. Only DTUs I, III and VI have been reported in Rodentia, although only DTUI was reported significantly more than expected (P < 0.01).
All DTUs were recorded from Primates, although only DTUs I, II and IV have been reported in non-human Primates (Fig. 1). If non-human Primates are analyzed alone, both DTUII (P < 0.001) and IV (P < 0.01) were recorded significantly more than expected; neither was significant if humans were included. Primates including humans had significantly greater frequencies of DTUI (P < 0.05) and DTUV (P < 0.001); the significance is inferred only for Homo sapiens since the frequencies were not significant if humans were not included. DTUIII and VI (along with DTUV) were recorded less than expected (P < 0.001) only if humans were included in the Primate group.
When DTUs were grouped into one of the two major lineage schemes, there were differences in recorded frequencies across reservoir taxa (P < 0.001, Fig. 1). With the exception of Artiodactyla, Chiroptera and non-human Primates, all frequencies for individual mammal orders were significant for Lineage II/L1, similar to DTUI frequencies. The Lineage I/L2 frequency for Primates was no longer significant due to modified DTU frequencies. The Lineage II/L2, similar to DTUII, was more frequently reported from Chiroptera and Primates without humans, although significantly less than expected in Didelphimorphia. Lineage II/L1 was recorded significantly less than expected in Didelphimorphia, all Primates, and Rodentia, while more than expected in Cingulata and Carnivora, with or without pets, similar for Lineage III/L2.
Similar to that observed for reservoir taxa, there were significant differences of DTU frequencies in the three principal vector genera Triatoma, Panstrongylus and Rhodnius (P < 0.001, Fig. 1). Interestingly, there were no significant frequencies for any of the vector genera with DTUIV, and no significant frequencies of any DTU (I, III, or IV) in Panstrongylus, the latter potentially due to few records. DTUs II, V and VI have only been occasionally reported from other than the genus Triatoma in which they are recorded significantly more than expected (P < 0.01, P < 0.001 and P < 0.001, respectively). DTUIII was only recorded significantly in Triatoma (P < 0.05). Lineage II/L1 was recorded significantly less than expected in Rhodnius and Panstrongylus (P < 0.001), while more than expected in Triatoma (P < 0.001). Lineage III/L2 was recorded also significantly less in Rhodnius (P < 0.01). The frequency of DTUI in Rhodnius was significantly more than expected (P < 0.05), while that in Triatoma was significantly less (P < 0.01). The lineage I/L1 frequency was also significantly greater in Rhodnius (P < 0.05), while not significant for Lineage I/L2 in this genus. Lower or nil significance of DTUI in Triatoma was also observed for Lineage I/L1 and Lineage I/L2.
Beta-specificity
Overall beta-specificity (βSOR) approached 1 for all DTUs, reflecting a high rate of species turnover when all hosts (mammals and vectors) were considered (Fig. 2). In general, T. cruzi is opportunistic across geographical scales since beta-specificity free from species turnover (βSIM) was highest and uniform in contrast to nestedness (βSNE), for all DTUs and for all lineages of the L1 and L2 schemes (data shown only for mammals, although similar for vectors) (Fig. 2c, d). Beta specificity free from species turnover was lowest for DTUs II, V and VI, with ranging from 0.71–0.78 in both vectors and mammal hosts (Fig. 2a, b). Beta specificity free from species turnover averaged for L1 and for L2 (Fig. 2c, d), while it was slightly lower for all DTUs. DTUs I and IV had lowest specificity due to nestedness in both mammals and vectors.
Fig. 2.
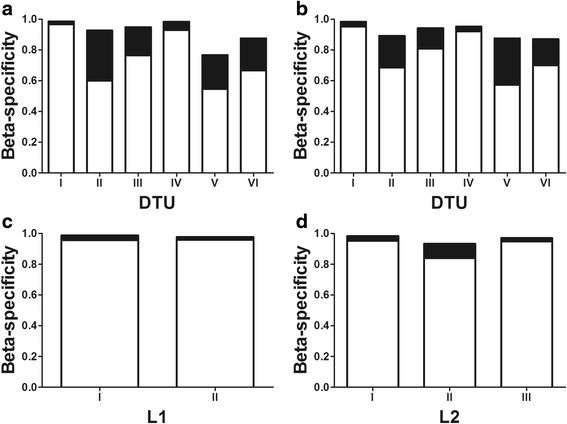
Host beta-specificity for Trypanosoma cruzi DTUs (a) for the three primary vector genera and (b) for mammal orders. Beta specificity for mammals according to major lineage schemes (c) L1 and (d) L2. Bars in white are beta dissimilarity due to host species turnover and in black due to nestedness. The range is between 0 (absence of species interchange across multiple regions) and 1 (complete species interchange across regions)
Ecological niche models of T. cruzi
Ecological niche models were constructed separately for all DTUs (Fig. 3) and both lineage schemes (Fig. 4). All niche models were highly accurate well above random expectation, the AUC values ranging from 1.2–1.9; the lowest AUC ratio was for Lineage II/L1 (see Additional file 5: Figure S1). The DTUIV ENM spans from the Nearctic region of Mexico, across the southeast USA to Argentina (Fig. 3e). DTUIII projects to a more reduced range, from southern Mexico to Argentina, with less coverage on the dryer Pacific coast (Fig. 3c). The ENM of DTUI (and Lineage I) projects throughout the Neotropical region from mid-Mexico to northeastern Argentina (Fig. 3a). The ENM for Lineage III/L2 includes projection similar to that of DTU III and IV, extending from the southeast and southwest USA to southern Argentina (Fig. 4b). In contrast to the geographical range of the former three clades, DTUII (and Lineage II/L2) has far more sparse geographical projection, which is primarily across Central America (CA), and the non-Amazon regions of Venezuela and southern Brazil (Fig. 3b). The ENM of DTUV and VI are more dense, although focused to the non-equatorial Neotropical regions of the continent (Mexico, Central America, southern Brazil, Bolivia) (Fig. 3d, f). The ENM for Lineage II/L1 (DTUs II–VI) includes areas from all individual constituent DTUs (Fig. 4a). The null hypothesis for lack of niche identity was rejected, i.e. niche dissimilarity was only significant for comparison of DTUIV and DTUVI (P <0.01). Distributions for all other comparisons were no different from random (Table 6).
Fig. 3.
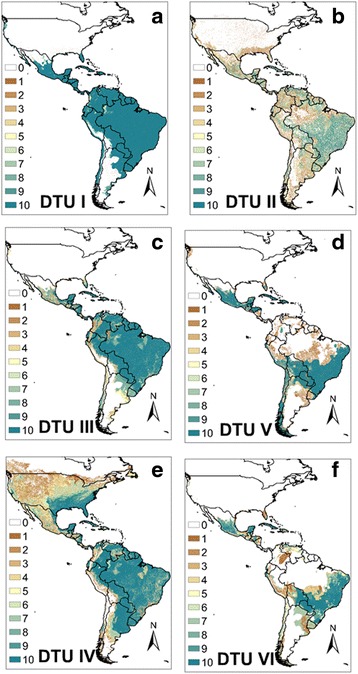
Ecological niche models for all Trypanosoma cruzi DTUs with classification of best (10) to worse (1) subsets. a DTUI. b DTUII. c DTUIII. d DTUV. e DTUIV. f DTUVI
Fig. 4.
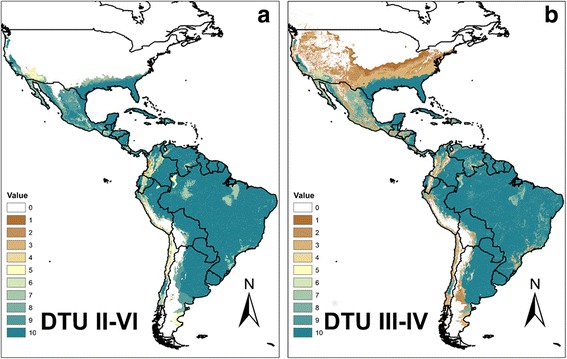
Ecological niche models for lineages from both principal schemes. Ecological niche models for Trypanosoma cruzi lineage I of L1 (a) and lineage III of L2 (b) with classification of best (10) to worse (1) subsets
Table 6.
Hellinger’s Index and Schoener’s D Index and corresponding 95% confidence intervals (CI) for niche dissimilarity calculated for all pairwise combinations of DTUs or lineages from L1 and L2 schemes
| Group | Comparison | n a /n b | Hellinger’s Index | 95% CI | Schoener’sIndex | 95% CI |
|---|---|---|---|---|---|---|
| I vs II | 234/8 | 0.93 | 0.35–0.95 | 0.73 | 0.15–0.95 | |
| I vs III | 234/27 | 0.91 | 0.55–0.90 | 0.70 | 0.35–0.90 | |
| I vs IV | 234/36 | 0.69 | 0.50–0.95 | 0.45 | 0.25–0.90 | |
| I vs V | 234/13 | 0.85 | 0.55–0.95 | 0.64 | 0.30–0.90 | |
| I vs VI | 234/49 | 0.80 | 0.70–0.95 | 0.53 | 0.50–0.90 | |
| II vs III | 8/27 | 0.96 | 0.20–0.85 | 0.79 | 0.10–0.80 | |
| DTU | II vs IV | 8/36 | 0.67 | 0.30–0.80 | 0.42 | 0.10–0.80 |
| II vs V | 8/13 | 0.83 | 0.35–0.90 | 0.67 | 0.25–0.90 | |
| II vs VI | 8/49 | 0.79 | 0.15–0.80 | 0.52 | 0.00–0.80 | |
| III vs IV | 27/49 | 0.71 | 0.55–0.90 | 0.48 | 0.40–0.85 | |
| III vs V | 27/13 | 0.82 | 0.40–0.95 | 0.63 | 0.25–0.90 | |
| III vs VI | 27/49 | 0.77 | 0.45–0.90 | 0.50 | 0.20–0.90 | |
| IV vs V | 36/13 | 0.70 | 0.30–0.80 | 0.43 | 0.20–0.70 | |
| IV vs VI | 36/49 | 0.38* | 0.50–0.85 | 0.18* | 0.30–0.85 | |
| V vs VI | 13/49 | 0.69 | 0.30–0.75 | 0.42 | 0.15–0.70 | |
| Lineage (L1) | I vs II | 234/133 | 0.91 | 0.80–0.95 | 0.73 | 0.65–0.95 |
| I vs II | 234/8 | 0.93 | 0.25–0.90 | 0.73 | 0.00–0.90 | |
| Lineage (L2) | I vs III | 234/63 | 0.86 | 0.70–0.95 | 0.65 | 0.55–0.95 |
| II vs III | 8/63 | 0.84 | 0.25–0.85 | 0.63 | 0.00–0.85 |
Abbreviation: n a /n b, number of samples used for each comparison
*P < 0.01
Discussion
Continuous reports of natural infections with Trypanosoma cruzi in many phylogenetically distant mammal orders provide evidence that it is a generalist parasite. Nonetheless, some attempts have been made to statistically test hypotheses for certain DTU specificities with host genera or orders. While this is a valid question, the broad extension of the T. cruzi clade across the American continent would require an ambitious sampling design and coverage of all mammal orders in study sites, which would require important funding, currently unlikely given reduction in science and technology budgets. The present study analyzes existing data from both single and multisite studies, by following a systematic and critical review, key associations between parasite clades and vector or mammal host taxa [59, 60]. Very few studies have included appropriate host diversity analyses, thus limiting assessment of potential DTU specificity. This would provide evidence for overlapping patterns of all DTU ranges (suggesting true generalization), or of a composite range of individual DTU ranges (indicating some degree of ecological or spatial restrictions that can be interpreted as specialization). To our knowledge, this is the first study that systematically and directly analyzes the published literature that intentionally or not reports the interaction between Trypanosoma cruzi and its hosts, after having been filtered based on methodological criteria for DTU detection, if the specimens in fact harbored the parasite. Two methodological assumptions regarding DTU specificity have been made, the first that the gene markers analyzed and the primers used can identify all DTUs (no DTU primer specificity), and the second is that host tissues used as a source of DTU DNA, can harbor any DTU with equal probability (no DTU tissue specificity). These two assumptions have been rarely assessed and when they have, are generally limited, and hence we encourage efforts to test the validity of these assumptions. Since the data analyzed herein rely on published information, and there is a paucity of reports from certain taxa and certain DTUs, we also caution potential analytical bias.
The majority of T. cruzi phylogenetic studies use additive retrospective collections, analyzing a sum of samples generated from multiple previous studies, without common hypothesis-driven design, thus with limited control of potential confounder factors and bias. These latter studies compile previously generated gene sequences in order to generate evolutionary information from those obtained, but not for the parasite, i.e. they lack external validity. We have used previous data filtered for certain criteria to analyze new questions regarding associations, rather than a meta-analysis approach, which would generate information on the magnitude and variability of effects. Previous individual studies were included based on reliability and certain core conceptual or experimental criteria, and obviously imply heterogeneous geographical coverage. This latter bias cannot be reconciled for this analysis, but should be considered specifically for future studies.
Many technical and methodological problems have affected the study of T. cruzi population genetics, not the least of which is the continued difficulty to sample bug species and reservoir communities in conserved or partially modified habitats, across regions, or the continent. Previous studies provide evidence that hosts do not circulate all parasite populations all the time, in addition to segregation of infra-populations in different tissues and blood [16, 25, 27]. These studies clearly demonstrate that in vitro or in vivo passage of parasite populations further selects for haplotypes, which bias interpretation of genetic diversity [61, 62]. Despite improvement of molecular detection sensitivity and specificity, high polymorphism in primer binding sequences continues to affect parasite detection and hence operative needs for efficient patient and healthcare management. We assume that these gene markers and methods afford equal probability to detect all DTUs when present, but there is no evidence that this may be correct. Few population-based studies have appropriate sampling design and or subpopulation representation, and in order to analyze existing evidence for DTU host specificity and geographical or landscape associations, most studies not analyzing original samples, cannot be used. The present re-analysis uses three robust methods to synthesize and evaluate current evidence related to T. cruzi diversity based on DTU or the two existing lineage schemes: similarity of environmental/geographical associations (abiotic ecological niche), biotic niche structure (beta specificity), and mammal as well as vector host associations (frequency analysis). Results from this re-analysis have been contrasted with conclusions regarding associations from individual studies.
DTUIV (and subclades III and I) and DTUII (and subclades V and VI) are primary phylogenetic branches, both reported frequently in non-human Primates [18]. However, while DTUIV was reported significantly in Carnivora, DTUII is reported significantly more in Chiroptera. Since we assume in this analysis that the methodological probability of identifying all DTUs is similar for all taxa, reservoir, or geographical heterogeneity, and considering the reduced number of samples analyzed, differential DTU distributions may be due to historical access, bioclimatic niche, or host specificity. In terms of vectors, DTUII was frequently reported in Triatoma, while there was no significant association of DTUIV with any vector genera. Current data suggest a high frequency of DTUIV in human oral transmission, although more robust evidence will be required to analyze potential specificity of its transmission via this mechanism [63, 64].
Subclades DTUIII and I are derived from DTUIV, the former recorded from many mammal orders (5), although significant only with Cingulata (i.e. armadillos), grassland and tropical forest insectivores and one of the most important burrow excavating mammal groups in tropical regions [4]. Interestingly, DTUIII was frequently reported only from Triatoma species, which share similar terrestrial habitats. Similar DTU associations would be expected for Panstrongylus species, since they have been assumed to be vectors for T. cruzi transmission to humans currently in Brazil [65]. However, based on current evidence, the present analysis does not support any specific or significant associations of any DTU with this vector genus, and its importance for human infections will require future population genetics analyses which include vectors and all taxa. Despite previous suggestions of DTUIII having broad geographical distribution and spatial structuring specifically in Cingulata across South America, this was not supported by existing evidence across the continent [66, 67].
Although early studies suggested DTUI (lineage LI) specificity for Didelphimorphia [66], this relationship is not exclusive, since it is the most generalist DTU, having been recorded in seven mammal orders, and is found significantly more in small and medium-sized mammals such as Rodentia, Didelphimorphia and Primates (including humans) [66, 68]. Curiously, vector host interaction frequencies are significant for DTUI only with the vector genus Rhodnius, important organisms along with Rodentia, Didelphimorphia and birds, in palm communities [66]. The lack of significant association of the genus Triatoma with DTUI is surprising, but cannot be spurious, especially given its distribution across the continent and number of samples reported. The current analysis of beta specificity does not allow us to ascertain whether spill-over of the DTUI is other than a “permissive” event, given the generalist character of most Triatoma species. In fact, the spill-over could have occurred from Triatoma to Rhodnius, but current evidence justifies less this hypothesis than a direct relationship with the latter genus. This DTU’s specificity with the diverse genus Rhodnius may have been a key mechanism for its dispersal, both north Central America and North America, and south from its principal distribution in the north of South America, and diversification [69, 70]. Overlapped ranges between Rhodnius spp. and the Triatoma dimidiata complex in Colombia, Venezuela, French Guyana, Ecuador and Peru, north through Central America to the Neotropical region of Mexico (at least up to the Tehuantepec Isthmus) in medium forest ecotopes, may have provided novel dispersal and diversification opportunities for DTUI to the north [68]. The Mesoamerican biodiversity corridor and regional indigenous human interchange (Mesoamerican and Andean cultures) have provided ample opportunity for movement of DTUI through the dimidiata complex species, through humans and additional hosts throughout the Neotropical region north of the Amazon basin. North of the Tehuantepec Isthmus (in Mexico), the distribution range of dimidiata complex overlaps with most phyllosoma species complex [71], thus providing a spatial network of ecological connectivity between the vectors as well as with other vector complexes (e.g. protracta, rubida and lecticularia) (Ibarra-Cerdeña, personal communication). This sympatry may have provided additional mechanisms for northward dispersal of DTUI, an important strategy when there is high regional species turnover. Similar mechanisms may have driven southward dispersal where Rhodnius species are sympatric with the Triatoma brasiliensis complex, T. sordida in the infestans complex, and multiple Panstrongylus species [72]. The frequency of DTUI in humans may, however, be related to methodological bias, but given altered reservoir communities in human-modified habitats, its frequency may also be related to the human footprint and mobility in and among modified reservoir communities. A much more specialized analysis of DTUI diversity would need to accompany a more specific sampling effort across regions with or without sympatric Rhodnius to test a relevant hypothesis. Future studies should be designed to generate appropriate evidence for the impact of landscape type and modification on this DTU’s transmission. Recently, a new clade designated Tcbat, genetically more closely related to TcI than to any other DTU, was described [73]. Some authors have recently proposed that Tcbat be recognized as an additional DTU within T. cruzi [74, 75], even though there are too few results from Tcbat to confirm or reject its validity [76]. Association of this clade with specific hosts or vectors will have to await more appropriate gene markers, since it is poorly recognized by existing techniques, and at least in the Neotropical region of Mexico, it has not been identified using the 18SrRNA (Izeta, personal communication).
The DTUII/V/VI group has the highest relative mammal host dissimilarity due to nestedness, indicating that host communities across regions represent non-random subsets of highly diverse successive assemblages. DTU beta-diversity was generally high, also indicating highly diverse host assemblages across regions. Previously proposed geographical restrictions for certain lineages and these DTUs are not supported by re-analysis of current evidence, despite potential host specificities (for DTUIII and DTUVI) [11, 19, 20]. Host dissimilarity was high across the continent (80–100%) principally due to host species turnover, and to a much lesser degree than to nestedness.
Host-DTU specificities coincide closely with parasite phylogeny, and not with aggregated lineage level clades [14, 18]. Significant frequencies for individual DTUs are lost when they are combined as Lineage II/L1. However, significant frequencies of combined DTUs III and IV in Lineage III/L2 maintain significance. Highest DTU diversity is observed in the higher mammal trophic orders (Carnivora, Primates). Orders inhabiting upper canopy and having long distance dispersal such as Chiroptera and non-human Primates, significantly concentrate DTUII, DTUVI and DTUI. DTUIII, although only significant in the insectivorous Cingulata, is recorded in Carnivora and other lower trophic level orders (Rodentia, Didelphimorphia), perhaps due to food chain interactions, or shared nesting habits. DTUI is the dominant subclade in lower trophic level taxa.
In general, there were no significant niche differences among DTUs (except between DTUIV and VI), or lineage schemes, at coarse spatial scales. This is similar to that recently proposed at the landscape level [77, 78], rather than the long-standing hypothesis of host or geographical segregation [11]. The present study has analyzed 2,377 data records from the continent from all T. cruzi hosts, using robust modeling methods, and found that despite greater frequency of DTUI reports, which may be related to detection and isolation methods, its potential distribution is majorly sympatric with other DTUs. Given the high significance of niche models included in this study, the Amazon headwater region and few sparse areas of Central America predict low niche for the DTUII/V/VI, while both regions predict high presence for DTUIV/III/I. The former group may have extended associations with hosts where other DTUs are not present, outside certain latitudinal limits (in northern Mexico, USA and central Argentina). Given that the majority of studies have used selected parasite isolates, it is not possible to discern whether reporting differences are due to methodological bias, or due to real molecular differences in DTU presence or abundance. As an example for the former, the first study using parasite isolates and MLEP in Mexico found greater than 98% Lineage I [79], while recent genetic marker studies using DNA amplification directly from host tissues finds an equal proportion of DTUVI at least in the Neotropical region in Mexico [80].
A key issue that current public health programs must address to limit vector-borne transmission of T. cruzi is the source of parasite populations that are in contact with humans. Human contact with infected vectors not only occurs in domestic areas, but in all fragments, with broad vector gene flow within landscapes [80, 81]. Metapopulation dynamics is assisted by human activity and by its impact on resource availability temporally and spatially, although also by altered reservoir assemblages in different landscape fragments [82, 83]. Vector interactions and their contact with numerous host/reservoir communities also affect parasite metapopulation dynamics in humans and domesticated fauna (livestock and pets). There is insufficient evidence currently, in any landscape to understand microevolutionary processes such as gene flow, gene drift, or parasite selection on a local scale [81, 84]. It is hoped that recognizing the void and the importance of this evidence will motivate more robust analyses. Without this evidence, sustainable transmission barriers will lack an evidence base.
There are many studies using assemblages of T. cruzi samples collected with different purposes in different regions (commonly by different groups of researchers or health system personnel). The samples are often not collected using specific sampling design and a hypothesis testing framework (i.e. without proper replicates or a common sampling design which affects internal and external validity of designs), yet analyzed and interpreted as though they had. Although these contributions are important for certain genetic information, they are affected by statistical noise that masks the nature of the process behind data, leading to flawed conclusions regarding parasite population patterns [62]. It is clear that in order to understand T. cruzi metapopulation dynamics and microevolutionary patterns, more appropriate gene markers and methods are necessary, including those that can be used for sylvatic reservoirs, humans and livestock/pets, as well as all triatomine vectors.
Conclusions
In order to analyze DTU associations with host taxa or geography, a greater number of samples and quantity of DNA is required, which highlights the need for assays with better sensitivity [35, 85, 86]. Marker design and selection, amplification methods, and potential non-specific amplification (including other trypanosome species) need to be tested for broad use and genotyping of parasite clades in order to analyze metapopulation dynamics [87]. Technological advances are urgently needed in this area, since analyses of T. cruzi landscape genetics will continue to be partial and biased unless we can confidently identify and analyze metapopulations from all hosts in assemblages over distribution ranges and according to ecological interactions and temporal dynamics.
Acknowledgements
We are grateful to Carlos Machado, Michael W. Gaunt and Miguel A. Munguía-Rosas for comments and suggestions on early versions of this study and to three anonymous reviewers for their constructive suggestions.
Funding
Conacyt FONSEC161405 and SEP166828 to Janine M. Ramsey, doctoral scholarship to Conacyt to Amaia Izeta (410717).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Authors’ contributions
AIA carried out the systematic search of published evidence and was a major contributor in writing the manuscript. CNIC and JMR analyzed and interpreted the data regarding the re-analysis, T. cruzi associations and ecological niche models. DAML performed the analysis for ecological niche modeling; AIA, JMR and CNIC wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- AUC
area under the ROC
- CA
Central America
- CD
Chagas disease
- CI
confidence interval
- DALYs
disability-adjusted life-years
- DNAsat
satellite DNA
- DTU
discrete typing unit
- ENM
ecological niche models
- kDNA
kinetoplast DNA
- LSSP
low stringency single specific primer
- MLEE
multi-locus enzyme electrophoresis
- NA
North America
- RAPD
random amplified polymorphic DNA
- rDNA
ribosomal DNA
- rtPCR
real time PCR
- RFLP
restriction fragment length polymorphisms
- ROC
receiver operating curve
- SNPs
single nucleotide polymorphisms
- SA
South America
- Tc
Trypanosoma cruzi
Additional files
Data used for re-analysis (some species are not correctly specified in original articles and are georeferenced according to the locality or community). (DOCX 489 kb)
Phylogenetic studies not using analytical methods. (DOCX 30 kb)
Phylogeographic studies not using analytical methods. (DOCX 15 kb)
Number of species and records reported in different orders. (DOCX 16 kb)
Ecological niche significance values. (TIF 40 kb)
Contributor Information
Amaia Izeta-Alberdi, Email: amaiaiz@gmail.com.
Carlos N. Ibarra-Cerdeña, Email: cibarra@cinvestav.mx
David A. Moo-Llanes, Email: davidmooll@comunidad.unam.mx
Janine M. Ramsey, Email: janineramseyw@gmail.com
References
- 1.World Health Organization . Chagas disease: control and elimination. Sixty-third World Health Assembly. Geneva: WHO; 2010. [Google Scholar]
- 2.Noireau F. Wild Triatoma infestans, a potential threat that needs to be monitored. Mem Inst Oswaldo Cruz. 2009;104:60–64. doi: 10.1590/S0074-02762009000900010. [DOI] [PubMed] [Google Scholar]
- 3.Ramsey JM, Gutiérrez-Cabrera AE, Salgado-Ramírez L, Peterson AT, Sánchez–Cordero V, Ibarra-Cerdeña CN. Ecological connectivity of Trypanosoma cruzi reservoirs and Triatoma pallidipennis hosts in an anthropogenic landscape with endemic Chagas disease. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0046013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orozco MM, Enriquez GF, Alvarado-Otegui JA, Cardinal MV, Schijman AG, Kitron U, et al. New sylvatic hosts of Trypanosoma cruzi and their reservoir competence in the humid Chaco of Argentina: a longitudinal study. Am J Trop Med Hyg. 2013;88(5):872–882. doi: 10.4269/ajtmh.12-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocha FL, Roque ALR, Arrais RC, Santos JP, Lima VDS, Xavier SCDC, et al. Trypanosoma cruzi TcI and TcII transmission among wild carnivores, small mammals and dogs in a conservation unit and surrounding areas, Brazil. Parasitology. 2013;140(02):160–170. doi: 10.1017/S0031182012001539. [DOI] [PubMed] [Google Scholar]
- 6.Hotez PJ, Bottazzi ME, Franco-Paredes C, Ault SK, Periago MR. The neglected tropical diseases of Latin America and the Caribbean: a review of disease burden and distribution and a roadmap for control and elimination. PLoS Negl Trop Dis. 2008;2 doi: 10.1371/journal.pntd.0000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyler KM, Engman DM. The life cycle of Trypanosoma cruzi revisited. Int J Parasitol. 2001;31(5–6):472–81. doi: 10.1016/S0020-7519(01)00153-9. [DOI] [PubMed] [Google Scholar]
- 8.Gaunt MW, Yeo M, Frame IA, Stothard JR, Carrasco HJ, Taylor MC, et al. Mechanism of genetic exchange in American trypanosomes. Nature. 2003;421:936–939. doi: 10.1038/nature01438. [DOI] [PubMed] [Google Scholar]
- 9.Miles MA, Llewellyn MS, Lewis MD, Yeo M, Baleela R, Fitzpatrick S, et al. The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: looking back and to the future. Parasitology. 2009;136:1509–1528. doi: 10.1017/S0031182009990977. [DOI] [PubMed] [Google Scholar]
- 10.Tibayrenc M, Ayala FJ. Evolutionary genetics of Trypanosoma and Leishmania. Microbes Infect. 1999;1(6):465–472. doi: 10.1016/S1286-4579(99)80050-1. [DOI] [PubMed] [Google Scholar]
- 11.Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MM, et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12(2):240–253. doi: 10.1016/j.meegid.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Baptista RP, D’Ávila DA, Segatto M, do Valle IF, Franco GR, Valadares HM, et al. Evidence of substantial recombination among Trypanosoma cruzi II strains from Minas Gerais. Infect Genet Evol. 2014;22:183–91. doi: 10.1016/j.meegid.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 13.Macedo AM, Machado CR, Oliveira RP, Pena SDJ. Trypanosoma cruzi: Genetic structure of populations and relevance of genetic variability to the pathogenesis of Chagas disease. Mem Inst Oswaldo Cruz. 2004;99:1–12. doi: 10.1590/S0074-02762004000100001. [DOI] [PubMed] [Google Scholar]
- 14.Zingales B, Andrade SG, Briones MRS, Campbell DA, Chiari E, Fernandes O, et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104(7):1051–1054. doi: 10.1590/S0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 15.Macedo AM, Pena SDJ. Genetic variability of Trypanosoma cruzi: Implications for the pathogenesis of Chagas disease. Parasitol Today. 1998;14:119–124. doi: 10.1016/S0169-4758(97)01179-4. [DOI] [PubMed] [Google Scholar]
- 16.Tibayrenc M. Genetic subdivisions within Trypanosoma cruzi (Discrete Typing Units) and their relevance for molecular epidemiology and experimental evolution. Kinetoplastid Biol Dis. 2003;3:2–12. doi: 10.1186/1475-9292-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol Biochem Parasitology. 1996;83(2):141–152. doi: 10.1016/S0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- 18.Flores-Lopez CA, Machado CA. Analyses of 32 loci clarify phylogenetic relationships among Trypanosoma cruzi lineages and support a single hybridization prior to human contact. PLoS Negl Trop Dis. 2011;5 doi: 10.1371/journal.pntd.0001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrera C, Bargues MD, Fajardo A, Montilla M, Triana O, Vallejo GA, et al. Identifying four Trypanosoma cruzi I isolate haplotypes from different geographic regions in Colombia. Infect Genet Evol. 2007;7(4):535–539. doi: 10.1016/j.meegid.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Falla A, Herrera C, Fajardo A, Montilla M, Vallejo GA, Guhl F. Haplotype identification within Trypanosoma cruzi I in Colombian isolates from several reservoirs, vectors and humans. Acta Trop. 2009;110:15–21. doi: 10.1016/j.actatropica.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 21.D’Ávila DA, Gontijo ED, Lages-Silva E, Meira WSF, Chiari E, Galvão LMC. Random amplified polymorphic DNA profiles of Trypanosoma cruzi isolates from chagasic patients with different clinical forms. Parasitol Res. 2006;98:455–61. doi: 10.1007/s00436-005-0043-5. [DOI] [PubMed] [Google Scholar]
- 22.Guhl F, Ramírez JD. Trypanosoma cruzi I diversity: Towards the need of genetic subdivision? Acta Trop. 2011;119:1–4. doi: 10.1016/j.actatropica.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Abad-Franch F, Valenca-Barbosa C, Sarquis O, Lima MM. All that glisters is not gold: Sampling-process uncertainty in disease-vector surveys with false-negative and false-positive detections. PLoS Negl Trop Dis. 2014;8 doi: 10.1371/journal.pntd.0003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morand S, Krasnov BR, Timothy D, Littlewood J. Parasite diversity and diversification: evolutionary ecology meets phylogenetics. Cambridge: Cambridge University Press; 2015. [Google Scholar]
- 25.Deane MP, Jansen AM, Mangia RHR, Goncalves AM, Morel CM. Are our laboratory “strains” representative samples of Trypanosoma cruzi populations that circulate in nature? Mem Inst Oswaldo Cruz. 1984;79:19–24. doi: 10.1590/S0074-02761984000500006. [DOI] [Google Scholar]
- 26.Devera R, Fernandes O, Coura JR. Should Trypanosoma cruzi be called cruzi complex? A review of the parasite diversity and the potential of selecting population after in vitro culturing and mice infection. Mem Inst Oswaldo Cruz. 2003;98:1–12. doi: 10.1590/S0074-02762003000100001. [DOI] [PubMed] [Google Scholar]
- 27.Venegas J, Díaz F, Rojas T, Miranda S, Jercic MI, González C, et al. Microsatellite loci-based distribution of Trypanosoma cruzi genotypes from Chilean chronic Chagas disease patients and Triatoma infestans is concordant with a specific host-parasite association hypothesis. Acta Parasitol. 2013;58:139–148. doi: 10.2478/s11686-013-0123-0. [DOI] [PubMed] [Google Scholar]
- 28.Schijman AG, Bisio M, Orellana L, Sued M, Duffy T, Jaramillo AMM, et al. International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS Negl Trop Dis. 2011;5(1) doi: 10.1371/journal.pntd.0000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnabé C, Brisse S, Tibayrenc M. Population structure and genetic typing of Trypanosoma cruzi, the agent of Chagas disease: a multilocus enzyme electrophoresis approach. Parasitology. 2000;120(05):513–526. doi: 10.1017/S0031182099005661. [DOI] [PubMed] [Google Scholar]
- 30.Brisse S, Barnabé C, Tibayrenc M. Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int J Parasitol. 2000;30:35–44. doi: 10.1016/S0020-7519(99)00168-X. [DOI] [PubMed] [Google Scholar]
- 31.Sturm NR, Degrave W, Morel C, Simpson L. Sensitive detection and schizodeme classification of Trypanosoma cruzi cells by amplification of kinetoplast minicircle DNA sequences: use in diagnosis of Chagas’ disease. Mol Biochem Parasitol. 1989;33(3):205–214. doi: 10.1016/0166-6851(89)90082-0. [DOI] [PubMed] [Google Scholar]
- 32.Fernandes O, Santos SS, Cupolillo E, Mendonça B, Derre R, Junqueira AC, et al. A mini-exon multiplex polymerase chain reaction to distinguish the major groups of Trypanosoma cruzi and T.rangeli in the Brazilian Amazon. Trans R Soc Trop Med Hyg. 2001;95:97–99. doi: 10.1016/S0035-9203(01)90350-5. [DOI] [PubMed] [Google Scholar]
- 33.Broutin H, Tarrieu F, Tibayrenc M, Oury B, Barnabé C. Phylogenetic analysis of the glucose-6-phosphate isomerase gene in Trypanosoma cruzi. Exp Parasitol. 2006;113(1):1–7. [DOI] [PubMed]
- 34.Brisse S, Henriksson J, Barnabé C, Douzery EJ, Berkvens D, Serrano M, et al. Evidence for genetic exchange and hybridization in Trypanosoma cruzi based on nucleotide sequences and molecular karyotype. Infect Genet Evol. 2003;2(3):173–183. doi: 10.1016/S1567-1348(02)00097-7. [DOI] [PubMed] [Google Scholar]
- 35.Duffy T, Bisio M, Altcheh J, Burgos JM, Diez M, Levin MJ, et al. Accurate real-time PCR strategy for monitoring bloodstream parasitic loads in Chagas disease patients. PLoS Negl Trop Dis. 2009;3(4) doi: 10.1371/journal.pntd.0000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgos JM, Altcheh J, Bisio M, Duffy T, Valadares H, Seidenstein ME, et al. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int J Parasitol. 2007;37(12):1319–1327. doi: 10.1016/j.ijpara.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 37.D’Ávila DA, Macedo AM, Valadares HMS, Gontijo ED, de Castro AM, Machado CR, et al. Probing population dynamics of Trypanosoma cruzi during progression of the chronic phase in chagasic patients. J Clin Microbiol. 2009;47(6):1718–1725. doi: 10.1128/JCM.01658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis MD, Ma J, Yeo M, Carrasco HJ, Llewellyn MS, Miles MA. Genotyping of Trypanosoma cruzi: systematic selection of assays allowing rapid and accurate discrimination of all known lineages. Am J Trop Med Hyg. 2009;81(6):1041–1049. doi: 10.4269/ajtmh.2009.09-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tibayrenc M. Genetic epidemiology of parasitic protozoa and other infectious agents: the need for an integrated approach. Int J Parasitol. 1998;28(1):85–104. doi: 10.1016/S0020-7519(97)00180-X. [DOI] [PubMed] [Google Scholar]
- 40.Cura CI, Mejía-Jaramillo AM, Duffy T, Burgos JM, Rodriguero M, Cardinal MV, et al. Trypanosoma cruzi I genotypes in different geographical regions and transmission cycles based on a microsatellite motif of the intergenic spacer of spliced-leader genes. Int J Parasitol. 2010;40(14):1599–1607. doi: 10.1016/j.ijpara.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Futuyma DJ, Moreno G. The evolution of ecological specialization. Annu RevEcol Syst. 1988;207–233.
- 42.Poulin R, Mouillot D. Parasite specialization from a phylogenetic perspective: a new index of host specificity. Parasitology. 2003;126(05):473–480. doi: 10.1017/S0031182003002993. [DOI] [PubMed] [Google Scholar]
- 43.Poulin R, Krasnov BR, Mouillot D. Host specificity in phylogenetic and geographic space. Trends Parasitol. 2011;27:355–361. doi: 10.1016/j.pt.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Baselga A, Jiménez-Valverde A, Niccolini G. A multiple-site similarity measure independent of richness. Biol Lett. 2007;3:642–645. doi: 10.1098/rsbl.2007.0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krasnov BR, Mouillot D, Shenbrot GI, Khokhlova IS, Poulin R. Beta-specificity: The turnover of host species in space and another way to measure host specificity. Int J Parasitol. 2011;41:33–41. doi: 10.1016/j.ijpara.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Baselga A. The relationship between species replacement, dissimilarity derived from nestedness, and nestedness. Glob Ecol Biogeogr. 2012;21:1223–1232. doi: 10.1111/j.1466-8238.2011.00756.x. [DOI] [Google Scholar]
- 47.Baselga A. Partitioning the turnover and nestedness components of beta diversity. Glob Ecol Biogeogr. 2010;19:134–143. doi: 10.1111/j.1466-8238.2009.00490.x. [DOI] [Google Scholar]
- 48.Baselga A. Disentangling distance decay of similarity from richness gradients: response to Soininen et al. 2007. Ecography. 2007;30(6):838–841. doi: 10.1111/j.2007.0906-7590.05191.x. [DOI] [Google Scholar]
- 49.Baselga A, Orme CDL. Betapart: An R package for the study of beta diversity. Methods Ecol Evol. 2012;3:808–812. doi: 10.1111/j.2041-210X.2012.00224.x. [DOI] [Google Scholar]
- 50.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- 51.Moo-Llanes DA, Ibarra-Cerdeña CN, Rebollar-Téllez EA, Ibáñez-Bernal S, González C, Ramsey JM. Current and future niche of North and Central American sand flies (Diptera: Psychodidae) in climate change scenarios. PLoS Negl Trop Dis. 2013;7:e2421. doi: 10.1371/journal.pntd.0002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stockwell D, Peters D. The GARP modelling systems: Problems and solutions to automated spatial prediction. Int J Geogr Inf Syst. 1999;13:143–158. doi: 10.1080/136588199241391. [DOI] [Google Scholar]
- 53.Anderson R, Lew D, Peterson AT. Evaluating predictive models of species distributions: Criteria for selecting optimal models. Ecol Model. 2003;162:211–232. doi: 10.1016/S0304-3800(02)00349-6. [DOI] [Google Scholar]
- 54.Owens HL, Campbell LP, Dornak LL, Saupe EE, Barve N, Soberón J, et al. Constraints on interpretation of ecological niche models by limited environmental ranges on calibration areas. Ecol Model. 2013;263:10–18. doi: 10.1016/j.ecolmodel.2013.04.011. [DOI] [Google Scholar]
- 55.Barve N. Tool for Partial-ROC version 1. Lawrence: Biodiversity Institute; 2008. [Google Scholar]
- 56.Peterson AT, Papes M, Soberón J. Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecol Model. 2008;213(1):63–72. doi: 10.1016/j.ecolmodel.2007.11.008. [DOI] [Google Scholar]
- 57.Phillips S, Anderson R, Schapire R. Maximum entropy modeling of species geographic distributions. Ecol Model. 2006;190:231–259. doi: 10.1016/j.ecolmodel.2005.03.026. [DOI] [Google Scholar]
- 58.Warren D, Glor R, Turelli M. ENM tools: a toolbox for comparative studies of environmental niche models. Ecography. 2010;33:607–611. doi: 10.1111/j.1600-0587.2009.06041.x. [DOI] [Google Scholar]
- 59.Green S, Higgins JPT. Cochrane handbook for systematic reviews of interventions version 5.1.0. The Cochrane Collaboration; 2011. Available www.handbook.cochrane.org.
- 60.Haddaway NR, Watson MJ. On the benefits of systematic reviews for wildlife parasitology. Int J Parasitol Parasites Wildl. 2016;5(2):184–191. doi: 10.1016/j.ijppaw.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finley RW, Dvorak JA. Trypanosoma cruzi: analysis of the population dynamics of heterogeneous mixtures. J Protozool. 1987;34:409–415. doi: 10.1111/j.1550-7408.1987.tb03202.x. [DOI] [PubMed] [Google Scholar]
- 62.Prugnolle F, De Meeus T. Apparent high recombination rates in clonal parasitic organisms due to inappropriate sampling design. Heredity. 2010;104:135–140. doi: 10.1038/hdy.2009.128. [DOI] [PubMed] [Google Scholar]
- 63.Carrasco HJ, Segovia M, Llewellyn MS, Morocoima A, Urdaneta-Morales S, Martínez C. Geographical distribution of Trypanosoma cruzi genotypes in Venezuela. PLoS Negl Trop Dis. 2012;6(6) doi: 10.1371/journal.pntd.0001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marcili A, Valente V, Valente A, Junqueira ACV, Maia da Silva F, Naiff R, et al. Trypanosoma cruzi in Brazilian Amazonia: lineages TCI and TCIIa in wild primates, Rhodnius spp. and in humans with Chagas disease associated with oral transmission. Int J Parasit. 2009;39(5):615–623. doi: 10.1016/j.ijpara.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 65.Coura JR, Junqueira AC, Fernandes O, Valente SA, Miles MA. Emerging Chagas disease in Amazonian Brazil. Trends Parasitol. 2002;18(4):171–176. doi: 10.1016/S1471-4922(01)02200-0. [DOI] [PubMed] [Google Scholar]
- 66.Yeo M, Acosta N, Llewellyn M, Sánchez H, Adamson S, Miles GA, et al. Origins of Chagas disease: Didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int J Parasit. 2005;35(2):225–233. doi: 10.1016/j.ijpara.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 67.Llewellyn MS, Lewis MD, Acosta N, Yeo M, Carrasco HJ, Segovia M, et al. Trypanosoma cruzi IIc: phylogenetic and phylogeographic insights from sequence and microsatellite analysis and potential impact on emergent Chagas disease. PLoS Negl Trop Dis. 2009;3(9) doi: 10.1371/journal.pntd.0000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Llewellyn MS, Miles MA, Carrasco HJ, Lewis MD, Yeo M, Vargas J, et al. Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog. 2009;5(5) doi: 10.1371/journal.ppat.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abad-Franch F, Palomeque FS, Aguilar HM, Miles MA. Field ecology of sylvatic Rhodnius populations (Heteroptera, Triatominae): risk factors for palm tree infestation in western Ecuador. Trop Med Int Health. 2005;10(12):1258–1266. doi: 10.1111/j.1365-3156.2005.01511.x. [DOI] [PubMed] [Google Scholar]
- 70.Monteiro FA, Barrett TV, Fitzpatrick S, Cordon-Rosales C, Feliciangeli D, Beard C. Molecular phylogeography of the Amazonian Chagas disease vectors Rhodnius prolixus and R. robustus. Mol Ecol. 2003;12(4):997–1006. doi: 10.1046/j.1365-294X.2003.01802.x. [DOI] [PubMed] [Google Scholar]
- 71.Ibarra-Cerdeña CN, Sánchez-Cordero V, Peterson AT, Ramsey JM. Ecology of North American Triatominae. Acta Trop. 2009;110(2):178–186. doi: 10.1016/j.actatropica.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 72.Gurgel-Gonçalves R, Galvao C, Costa J, Peterson AT. Geographic distribution of Chagas disease vectors in Brazil based on ecological niche modelling. J Trop Med. 2012;2012:705326. doi: 10.1155/2012/705326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marcili A, Lima L, Cavazzana M, Junqueira ACV, Veludo HH, Da Silva FM, et al. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology. 2009;136(06):641–655. doi: 10.1017/S0031182009005861. [DOI] [PubMed] [Google Scholar]
- 74.Lima L, Espinosa-Álvarez O, Ortiz PA, Trejo-Varón JA, Carranza JC, Pinto CM, et al. Genetic diversity of Trypanosoma cruzi in bats, and multilocus phylogenetic and phylogeographical analyses supporting Tcbat as an independent DTU (discrete typing unit) Acta Trop. 2015;151:166–177. doi: 10.1016/j.actatropica.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 75.Brenière SF, Walecks E, Barnabé C. Over six thousand Trypanosoma cruzi strains classified into Discrete Typing Units (DTUs): Attempt at an inventory. PLoS Negl Trop Dis. 2016;10(8) doi: 10.1371/journal.pntd.0004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barnabé C, Mobarec HI, Jurado MR, Cortez JA, Brenière SF. Reconsideration of the seven discrete typing units within the species Trypanosoma cruzi, a new proposal of three reliable mitochondrial clades. Infect Genet Evol. 2016;39:176–86. doi: 10.1016/j.meegid.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 77.Messenger LA, Garcia L, Vanhove M, Huaranca C, Bustamante M, Torrico M, et al. Ecological host fitting of Trypanosoma cruzi TcI in Bolivia: mosaic population structure, hybridization and a role for humans in Andean parasite dispersal. Mol Ecol. 2015;24:2406–2422. doi: 10.1111/mec.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lima VS, Jansen AM, Messenger LA, Miles MA, Llewellyn MS. Wild Trypanosoma cruzi I genetic diversity in Brazil suggests admixture and disturbance in parasite populations from the Atlantic Forest region. Parasit Vectors. 2014;7:263–271. doi: 10.1186/1756-3305-7-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bosseno MF, Barnabé C, Gastélum EM, Kasten FL, Ramsey J, Espinoza B, Brenière SF. Predominance of Trypanosoma cruzi lineage I in Mexico. J Clin Microbiol. 2002;40(2):627–632. doi: 10.1128/JCM.40.2.627-632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lopez-Cancino SA, Tun-Ku E, De la Cruz- Felix HK, Ibarra-Cerdeña CN, Izeta-Alberdi A, Pech-May A, et al. Landscape ecology of Trypanosoma cruzi in the southern Yucatan Peninsula. Acta Trop. 2015;151:58–72. doi: 10.1016/j.actatropica.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 81.Ocaña-Mayorga S, Llewellyn MS, Costales JA, Miles MA, Grijalva MJ. Sex, subdivision, and domestic dispersal of Trypanosoma cruzi lineage I in southern Ecuador. PLoS Negl Trop Dis. 2010;4(12) doi: 10.1371/journal.pntd.0000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de la Fuente AL C, Minoli SA, Lopes CM, Noireau F, Lazzari CR, Lorenzo MG. Flight dispersal of the Chagas disease vectors Triatoma brasiliensis and Triatoma pseudomaculata in northeastern Brazil. Acta Trop. 2007;101:115–119. doi: 10.1016/j.actatropica.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 83.Gurtler RE, Cecere MC, Fernandez MP, Vazquez-Prokopec GM, Ceballos LA, Gurevitz JM, et al. Key source habitats and potential dispersal of Triatoma infestans populations in northwestern Argentina: Implications for vector control. PLoS Negl Trop Dis. 2014;8(10) doi: 10.1371/journal.pntd.0003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodriguez IB, Botero A, Mejía-Jaramillo AM, Marquez EJ, Ortiz S, Solari A, et al. Transmission dynamics of Trypanosoma cruzi determined by low-stringency single primer polymerase chain reaction and Southern Blot analyses in four indigenous communities of the Sierra Nevada de Santa Marta, Colombia. Am J Trop Med Hyg. 2009;81(3):396–403. [PubMed] [Google Scholar]
- 85.Duffy T, Cura CI, Ramirez JC, Abate T, Cayo NM, Parrado R, et al. Analytical performance of a multiplex real-time PCR assay using TaqMan probes for quantification of Trypanosoma cruzi satellite DNA in blood samples. PLoS Negl Trop Dis. 2013;7(1) doi: 10.1371/journal.pntd.0002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Higuera SL, Guhl F, Ramírez JD. Identification of Trypanosoma cruzi Discrete Typing Units (DTUs) through the implementation of a High-Resolution Melting (HRM) genotyping assay. Parasit Vectors. 2013;6(1):112. doi: 10.1186/1756-3305-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Charles RA, Kjos S, Ellis AE, Barnes JC, Yabsley MJ. Southern plains woodrats (Neotoma micropus) from southern Texas are important reservoirs of two genotypes of Trypanosoma cruzi and host of a putative novel Trypanosoma species. Vector Borne Zoonotic Dis. 2013;13(1):22–30. doi: 10.1089/vbz.2011.0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Freita JM, Augusto-Pinto L, Pimenta JR, Bastos-Rodrigues L, Gonçalves VF, Teixeira SM, et al. Ancestral genomes, sex, and the population structure of Trypanosoma cruzi. PLoS Pathog. 2006;2(3) doi: 10.1371/journal.ppat.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ienne S, Pedroso A, Carmona EFR, Briones MR, Zingales B. Network genealogy of 195-bp satellite DNA supports the superimposed hybridization hypothesis of Trypanosoma cruzi evolutionary pattern. Infect Genet Evol. 2010;10:601–606. doi: 10.1016/j.meegid.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 90.Lewis MD, Llewellyn MS, Yeo M, Acosta N, Gaunt MW, Miles MA. Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids. PLoS Negl Trop Dis. 2011;5(10) doi: 10.1371/journal.pntd.0001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Llewellyn MS, Rivett-Carnac JB, Fitzpatrick S, Lewis MD, Yeo M, Gaunt MW, et al. Extraordinary Trypanosoma cruzi diversity within single mammalian reservoir hosts implies a mechanism of diversifying selection. Int J Parasitol. 2011;41:609–614. doi: 10.1016/j.ijpara.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Macedo AM, Pimenta JR, De Aguiar RS, Melo AIR, Chiari E, Zingales B, et al. Usefulness of microsatellite typing in population genetic studies of Trypanosoma cruzi. Mem Inst Oswaldo Cruz. 2001;96(3):407–413. doi: 10.1590/S0074-02762001000300023. [DOI] [PubMed] [Google Scholar]
- 93.Oliveira RP, Melo AIR, Macedo AM, Chiari E, Pena SD. The population structure of Trypanosoma cruzi: expanded analysis of 54 strains using eight polymorphic CA-repeat microsatellites. Mem Inst Oswaldo Cruz. 1999;94:65–70. doi: 10.1590/S0074-02761999000700006. [DOI] [PubMed] [Google Scholar]
- 94.Pena SD, Machado CR, Macedo AM. Trypanosoma cruzi: ancestral genomes and population structure. Mem Inst Oswaldo Cruz. 2009;104:108–114. doi: 10.1590/S0074-02762009000900016. [DOI] [PubMed] [Google Scholar]
- 95.Telleria J, Biron DG, Brizard JP, Demettre E, Seveno M, Barnabé C, et al. Phylogenetic character mapping of proteomic diversity shows high correlation with subspecific phylogenetic diversity in Trypanosoma cruzi. Proc Natl Acad Sci USA. 2010;107:20411–20416. doi: 10.1073/pnas.1015496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tomazi L, Kawashita SY, Pereira PM, Zingales B, Briones MRS. Haplotype distribution of five nuclear genes based on network genealogies and Bayesian inference indicates that Trypanosoma cruzi hybrid strains are polyphyletic. Genet Mol Res. 2009;8:458–476. doi: 10.4238/vol8-2gmr591. [DOI] [PubMed] [Google Scholar]
- 97.Bhattacharyya T, Brooks J, Yeo M, Carrasco HJ, Lewis MD, Llewellyn MS, et al. Analysis of molecular diversity of the Trypanosoma cruzi trypomastigote small surface antigen reveals novel epitopes, evidence of positive selection and potential implications for lineage-specific serology. Int J Parasitol. 2010;40:921–928. doi: 10.1016/j.ijpara.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 98.Briones MR, Souto RP, Stolf BS, Zingales B. The evolution of two Trypanosoma cruzi subgroups inferred from rRNA genes can be correlated with the interchange of American mammalian faunas in the Cenozoic and has implications to pathogenicity and host specificity. Mol Biochem Parasitol. 1999;104(2):219–232. doi: 10.1016/S0166-6851(99)00155-3. [DOI] [PubMed] [Google Scholar]
- 99.Cerqueira GC, Bartholomeu DC, Darocha WD, Hou L, Freitas-Silva DM, Machado CR, et al. Sequence diversity and evolution of multigene families in Trypanosoma cruzi. Mol Biochem Parasitol. 2008;157(1):65–72. doi: 10.1016/j.molbiopara.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 100.Iwagami M, Higo H, Miura S, Yanagi T, Tada I, Kano S, et al. Molecular phylogeny of Trypanosoma cruzi from Central America (Guatemala) and a comparison with South American strains. Parasitol Res. 2007;102(1):129–134. doi: 10.1007/s00436-007-0739-9. [DOI] [PubMed] [Google Scholar]
- 101.Kawashita SY, Sanson GF, Fernandes O, Zingales B, Briones MR. Maximum-likelihood divergence date estimates based on rRNA gene sequences suggest two scenarios of Trypanosoma cruzi intraspecific evolution. Mol Biol Evol. 2001;18(12):2250–2259. doi: 10.1093/oxfordjournals.molbev.a003771. [DOI] [PubMed] [Google Scholar]
- 102.Lewis MD, Llewellyn MS, Gaunt MW, Yeo M, Carrasco HJ. Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids. Int J Parasitol. 2009;39(12):1305–17. doi: 10.1016/j.ijpara.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Machado CA, Ayala FJ. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc Natl Acad Sci USA. 2001;98(13):7396–7401. doi: 10.1073/pnas.121187198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Messenger LA, Llewellyn MS, Bhattacharyya T, Franzén O, Lewis MD, Ramirez JD, et al. Multiple mitochondrial introgression events and heteroplasmy in Trypanosoma cruzi revealed by maxicircle MLST and next generation sequencing. PLoS Negl Trop Dis. 2012;6(4) doi: 10.1371/journal.pntd.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O'Connor O, Bosseno MF, Barnabe C, Douzery EJ, Breniere SF. Genetic clustering of Trypanosoma cruzi I lineage evidenced by intergenic miniexon gene sequencing. Infect Genet Evol. 2007;7:587–593. doi: 10.1016/j.meegid.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 106.Rozas M, De Doncker S, Coronado X, Barnabe C, Tibyarenc M, Solari A, et al. Evolutionary history of Trypanosoma cruzi according to antigen genes. Parasitology. 2008;135:1157–1164. doi: 10.1017/S0031182008004794. [DOI] [PubMed] [Google Scholar]
- 107.Subileau M, Barnabe C, Douzery EJ, Diosque P, Tibayrenc M. Trypanosoma cruzi: new insights on ecophylogeny and hybridization by multigene sequencing of three nuclear and one maxicircle genes. Exp Parasitol. 2009;122:328–337. doi: 10.1016/j.exppara.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 108.Telleria J, Lafay B, Virreira M, Barnabe C, Tibayrenc M, Svoboda M. Trypanosoma cruzi: sequence analysis of the variable region of kinetoplast minicircles. Exp Parasitol. 2006;114:279–288. doi: 10.1016/j.exppara.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 109.Venegas J, Conoepan W, Pichuantes S, Miranda S, Jercic MI, Gajardo M, et al. Phylogenetic analysis of microsatellite markers further supports the two hybridization events hypothesis as the origin of the Trypanosoma cruzi lineages. Parasitol Res. 2009;105:191–199. doi: 10.1007/s00436-009-1386-0. [DOI] [PubMed] [Google Scholar]
- 110.Westenberger SJ, Barnabé C, Campbell D, Sturm NR. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics. 2005;171:527–543. doi: 10.1534/genetics.104.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Westenberger SJ, Sturm NR, Campbell DA. Trypanosoma cruzi 5S rRNA arrays define five groups and indicate the geographic origins of an ancestor of the heterozygous hybrids. Int J Parasitol. 2006;36:337–346. doi: 10.1016/j.ijpara.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 112.Yeo M, Mauricio IL, Messenger LA, Lewis MD, Llewellyn MS, Acosta N, et al. Multilocus sequence typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi. PLoS Negl Trop Dis. 2011;5(6) doi: 10.1371/journal.pntd.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zingales B, Stolf BS, Souto RP, Fernandes O, Briones MR. Epidemiology, biochemistry and evolution of Trypanosoma cruzi lineages based on ribosomal RNA sequences. Mem Inst Oswaldo Cruz. 1999;94:159–164. doi: 10.1590/S0074-02761999000700020. [DOI] [PubMed] [Google Scholar]
- 114.Barnabe C, De Meeûs T, Noireau F, Bosseno MF, Monje EM, Renaud F, Brenière SF. Trypanosoma cruzi discrete typing units (DTUs): Microsatellite loci and population genetics of DTUs TcV and TcI in Bolivia and Peru. Infect Genet Evol. 2011;11(7):1752–1760. doi: 10.1016/j.meegid.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 115.Oliveira RP, Broude NE, Macedo AM, Cantor CR, Smith CL, Pena SD. Probing the genetic population structure of Trypanosoma cruzi with polymorphic microsatellites. Proc Natl Acad Sci USA. 1998;95(7):3776–3780. doi: 10.1073/pnas.95.7.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ramirez JD, Duque MC, Guhl F. Phylogenetic reconstruction based on cytochrome b (Cytb) gene sequences reveals distinct genotypes within Colombian Trypanosoma cruzi I populations. Acta Trop. 2011;119(1):61–65. doi: 10.1016/j.actatropica.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 117.Ramirez JD, Tapia-Calle G, Guhl F. Genetic structure of Trypanosoma cruzi in Colombia revealed by a high-throughput nuclear multilocus sequence typing (nMLST) approach. BMC Genet. 2013;14:96. doi: 10.1186/1471-2156-14-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Barnabe C, Breniere SF. Scarce events of mitochondrial introgression inTrypanosoma cruzi: new case with a Bolivian strain. Infect Genet Evol. 2012;12:1879–1883. doi: 10.1016/j.meegid.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 119.Cuervo P, Cupolillo E, Seura I, Saravia N, Fernandes O. Genetic diversity of Colombian sylvatic Trypanosoma cruzi isolates revealed by the ribosomal DNA. Mem Inst Oswaldo Cruz. 2002;97(6):877–880. doi: 10.1590/S0074-02762002000600023. [DOI] [PubMed] [Google Scholar]
- 120.Herrera C, Guhl F, Falla A, Fajardo A, Montilla M, Vallejo GA, et al. Genetic variability and phylogenetic relationships within Trypanosoma cruzi I isolated in Colombia based on miniexon gene sequences. J. Parasitol Res. 2009; article ID 897364, doi:10.1155/2009/897364. [DOI] [PMC free article] [PubMed]
- 121.Martínez I, Nogueda B, Martínez-Hernández F, Espinoza B. Microsatellite and mini-exon analysis of mexican human DTU I Trypanosoma cruzi strains and their susceptibility to nifurtimox and benznidazole. Vector Borne Zoonotic Dis. 2013;13(3):181–187. doi: 10.1089/vbz.2012.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roellig DM, Savage MY, Fujita AW, Barnabé C, Tibayrenc M, Steurer FJ, et al. Genetic variation and exchange in Trypanosoma cruzi isolates from the United States. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Salazar A, Schijman AG, Triana O. High variability of Colombian Trypanosoma cruzi lineage I stocks as revealed by low-stringency single primer-PCR minicircle signatures. Acta Trop. 2006;100(1):110–118. doi: 10.1016/j.actatropica.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 124.Santos SS, Cupolillo E, Junqueira A, Coura JR, Jansen A, Sturm NR, et al. The genetic diversity of Brazilian Trypanosoma cruzi isolates and the phylogenetic positioning of zymodeme 3, based on the internal transcribed spacer of the ribosomal gene. Ann Trop Med Parasitol. 2002;96(8):755–764. doi: 10.1179/000349802125002301. [DOI] [PubMed] [Google Scholar]
- 125.Spotorno AE, Córdova L, Solari A. Differentiation of Trypanosoma cruzi I subgroups through characterization of cytochrome b gene sequences. Infect Genet Evol. 2008;8:898–900. doi: 10.1016/j.meegid.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 126.Triana O, Ortiz S, Dujardin JC, Solari A. Trypanosoma cruzi: Variability of stocks from Colombia determined by molecular karyotype and minicircle Southern blot analysis. Exp Parasitol. 2006;113(1):62–66. doi: 10.1016/j.exppara.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 127.Lauthier JJ, Tomasini N, Barnabé C, Rumi MMM, D'Amato AMA, Ragone PG, et al. Candidate targets for Multilocus Sequence Typing of Trypanosoma cruzi: validation using parasite stocks from the Chaco Region and a set of reference strains. Infect Genet Evol. 2012;12(2):350–358. doi: 10.1016/j.meegid.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 128.Ramirez JD, Guhl F, Messenger LA, Lewis MD, Montilla M, Cucunuba Z, et al. Contemporary cryptic sexuality in Trypanosoma cruzi. Mol Ecol. 2012;21:4216–4226. doi: 10.1111/j.1365-294X.2012.05699.x. [DOI] [PubMed] [Google Scholar]
- 129.Brito CMM, Lima MM, Sarquis O, Pires MQ, Coutinho CF, Duarte R, et al. Genetic polymorphism in Trypanosoma cruzi I isolated from Brazilian northeast triatomines revealed by low-stringency single specific primer polymerase chain reaction. Parasitol Res. 2008;103(5):1111–1117. doi: 10.1007/s00436-008-1102-5. [DOI] [PubMed] [Google Scholar]
- 130.Câmara ACJ, Varela-Freire AA, Valadares HMS, Macedo AM, D'Ávila DA, Machado CR, et al. Genetic analyses of Trypanosoma cruzi isolates from naturally infected triatomines and humans in northeastern Brazil. Acta Trop. 2010;115:205–211. doi: 10.1016/j.actatropica.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 131.Carranza JC, Valadares HM, D'Avila DA, Baptista RP, Moreno M, Galvão LM, et al. Trypanosoma cruzi maxicircle heterogeneity in Chagas disease patients from Brazil. Int J Parasitol. 2009;39(9):963–973. doi: 10.1016/j.ijpara.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 132.D'Avila DA, Macedo AM, Valadares HM, Gontijo ED, De Castro AM, Machado CR, et al. Probing population dynamics of Trypanosoma cruzi during progression of the chronic phase in chagasic patients. J Clin Microbiol. 2009;47(6):1718–25. doi: 10.1128/JCM.01658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hwang WS, Zhang G, Maslov D, Weirauch C. Infection rates of Triatoma protracta (Uhler) with Trypanosoma cruzi in Southern California and molecular identification of trypanosomes. Am J Trop Med Hyg. 2010;83(5):1020. doi: 10.4269/ajtmh.2010.10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Luna-Marin KP, Jaramillo CL, Hernández G, Gutiérrez R, Vallejo GA, Angulo VM. ITS–RFLP-and RAPD-based genetic variability of Trypanosoma cruzi I, human and vector strains in Santander (Colombia) Parasitol Res. 2009;105(2):519–528. doi: 10.1007/s00436-009-1422-0. [DOI] [PubMed] [Google Scholar]
- 135.Martins LPA, Marcili A, Castanho REP, Therezo ALS, Oliveira JCP, Suzuki RB, et al. Rural Triatoma rubrovaria from southern Brazil harbors Trypanosoma cruzi of lineage IIc. Am J Trop Med Hyg. 2008;79:427–434. [PubMed] [Google Scholar]
- 136.Perez E, Monje M, Chang B, Buitrago R, Parrado R, Barnabé C. Predominance of hybrid discrete typing units of Trypanosoma cruzi in domestic Triatoma infestans from the Bolivian Gran Chaco region. Infect Genet Evol. 2013;13:116–123. doi: 10.1016/j.meegid.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 137.Tomasini N, Lauthier JJ, Rumi MMM, Ragone PG, D’Amato AAA, Brandan CP, et al. Interest and limitations of spliced leader intergenic region sequences for analyzing Trypanosoma cruzi I phylogenetic diversity in the Argentinean Chaco. Inf Genet Evol. 2011;11(2):300–307. doi: 10.1016/j.meegid.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 138.Zafra G, Mantilla JC, Jácome J, Macedo AM, González CI. Direct analysis of genetic variability in Trypanosoma cruzi populations from tissues of Colombian chagasic patients. Hum Pathol. 2011;42(8):1159–1168. doi: 10.1016/j.humpath.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 139.Zalloum L, Gomes ML, Kinoshita AT, Toledo MJO, Prioli AJ, Araújo SM. Trypanosoma cruzi: two genetic groups in Paraná State, Southern Brazil. Exp Parasitol. 2005;111(1):55–58. doi: 10.1016/j.exppara.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 140.Herrera CP, Barnabé C, Brenière SF. Complex evolutionary pathways of the intergenic region of the mini-exon gene in Trypanosoma cruzi TcI: a possible ancient origin in the Gran Chaco and lack of strict genetic structuration. Infect Genet Evol. 2013;16:27–37. doi: 10.1016/j.meegid.2012.12.028. [DOI] [PubMed] [Google Scholar]
- 141.Zumaya-Estrada F, Messenger LA, Lopez-Ordonez T, Lewis MD, Flores-Lopez CA, Martínez-Ibarra AJ, et al. North American import? Charting the origins of an enigmatic Trypanosoma cruzi domestic genotype. Parasit Vectors. 2012;5(1):226. doi: 10.1186/1756-3305-5-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Marcili A, Lima L, Valente VC, Valente SA, Batista JS, Junqueira ACV, et al. Comparative phylogeography of Trypanosoma cruzi TCIIc: new host, association with terrestrial ecotopes, and spacial clustering. Inf Genet Evol. 2009;9(6):1265–1274. doi: 10.1016/j.meegid.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 143.Venegas J, Rojas T, Díaz F, Miranda S, Jercic MI, González C, et al. Geographical structuring of Trypanosoma cruzi populations from Chilean Triatoma infestans triatomines and their genetic relationship with other Latino American counterparts. Ann Trop Med Parasitol. 2011;105(8):625–646. doi: 10.1179/2047773211Y.0000000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mejía-Jaramillo AM, Arboleda-Sánchez S, Rodríguez IB, Cura C, Salazar A, Del Mazo J, et al. Geographical clustering of Trypanosoma cruzi I groups from Colombia revealed by low-stringency single specific primer-PCR of the intergenic regions of spliced-leader genes. Parasitol Res. 2009;104(2):399–410. doi: 10.1007/s00436-008-1212-0. [DOI] [PubMed] [Google Scholar]
- 145.Herrera CP, Licon MH, Nation CS, Jameson SB, Wesson DM. Genotype diversity of Trypanosoma cruzi in small rodents and Triatoma sanguisuga from a rural area in New Orleans, Louisiana. Parasit Vectors. 2015;8:123. doi: 10.1186/s13071-015-0730-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.


