This investigation explores the transcriptional control and subcellular location of proliferator-activated receptor-γ coactivator 1-α (PGC-1α) following exercise in a hot environment. It provides new evidence for decreased transcription of PGC-1α following an acute exercise bout in a hot environment, coinciding with decreased binding of cAMP response element-binding protein (CREB), myocyte enhancer factor 2 (MEF2), and forkhead box class-O1 (FoxO1) to the PGC-1α promoter region. However, translocation of PGC-1α protein from the cytosol into the nucleus following exercise appears to be unaffected by differences in environmental temperature.
Keywords: mitochondrial biogenesis, skeletal muscle, heat stress
Abstract
The purpose of this study was to determine mitochondrial biogenesis-related mRNA expression, binding of transcription factors to the peroxisome proliferator-activated receptor-γ coactivator 1-α (PGC-1α) promoter, and subcellular location of PGC-1α protein in human skeletal muscle following exercise in a hot environment compared with a room temperature environment. Recreationally trained males (n = 11) completed two trials in a temperature- and humidity-controlled environmental chamber. Each trial consisted of cycling in either a hot (H) or room temperature (C) environment (33 and 20°C, respectively) for 1 h at 60% of maximum wattage (Wmax) followed by 3 h of supine recovery at room temperature. Muscle biopsies were taken from the vastus lateralis pre-, post-, and 3 h postexercise. PGC-1α mRNA increased post (P = 0.039)- and 3 h postexercise in C (P = 0.002). PGC-1α, estrogen-related receptor-α (ERRα), and nuclear respiratory factor 1 (NRF-1) mRNA was all lower in H than C post (P = 0.038, P < 0.001, and P = 0.030, respectively)- and 3 h postexercise (P = 0.035, P = 0.007, and P < 0.001, respectively). Binding of cAMP response element-binding protein (CREB) (P = 0.005), myocyte enhancer factor 2 (MEF2) (P = 0.047), and FoxO forkhead box class-O1 (FoxO1) (P = 0.010) to the promoter region of the PGC-1α gene was lower in H than C. Nuclear PGC-1α protein increased postexercise in both H and C (P = 0.029) but was not different between trials (P = 0.602). These data indicate that acute exercise in a hot environment blunts expression of mitochondrial biogenesis-related mRNA, due to decreased binding of CREB, MEF2, and FoxO1 to the PGC-1α promoter.
NEW & NOTEWORTHY
This investigation explores the transcriptional control and subcellular location of proliferator-activated receptor-γ coactivator 1-α (PGC-1α) following exercise in a hot environment. It provides new evidence for decreased transcription of PGC-1α following an acute exercise bout in a hot environment, coinciding with decreased binding of cAMP response element-binding protein (CREB), myocyte enhancer factor 2 (MEF2), and forkhead box class-O1 (FoxO1) to the PGC-1α promoter region. However, translocation of PGC-1α protein from the cytosol into the nucleus following exercise appears to be unaffected by differences in environmental temperature.
mitochondrial adaptation is important for maintaining optimal health as well as improving athletic performance. Interventions to stimulate mitochondrial development are of interest as the dysfunction of mitochondria have been implicated in many conditions including diabetes (10, 19, 45), peripheral arterial disease (32), and aging (40, 46). The growing number of individuals with these mitochondrial-related pathologies demonstrates the importance of understanding human mitochondrial biogenesis and possible interventions to stimulate mitochondrial biogenesis. Exercise training is one known intervention that leads to mitochondrial biogenesis in humans (18). While exercise alone is a valuable therapy to stimulate mitochondrial development, reduced capacity to perform adequate exercise in a sedentary or diseased population exemplifies the need to further enhance the effectiveness of a given exercise bout to stimulate mitochondrial development. Environmental temperature may play an important role in regulation of mitochondrial biogenesis. Mild heat stress has been shown to induce mitochondrial biogenesis in C2C12 myotubes (26). Alternatively, acute heat stress in yeast models has demonstrated a downregulation in mitochondrial function (37), while chronic heat stress has been shown to inhibit mitochondrial development in chickens (3). In humans, early markers of mitochondrial biogenesis appear to be blunted following exercise in a hot environment (41). It is not known if this blunting effect is due to increased temperature during an exercise bout or during the recovery period, as in previous work participants both exercised and recovered in a hot environment (41), which has limited practical applications. Additional insight into this mechanism is necessary to better understand the impact of acute heat stress during exercise on mitochondrial biogenesis in a human model.
Normal function of the mitochondria is largely regulated by peroxisome proliferator-activated receptor-γ coactivator 1-α (PGC-1α), a transcriptional coactivator known as a master regulator of metabolism and mitochondrial biogenesis (48). PGC-1α binds to and coactivates several transcription factors, including nuclear respiratory factor 1 (NRF-1), myocyte enhancer factor 2 (MEF2), estrogen-related receptor-α (ERRα), GA binding protein transcription factor α-subunit 60 kDa (GABPA, also known as NRF-2), and mitochondrial transcription factor A (TFAM) (6, 9, 15, 22). Activation of these downstream transcription factors is important for regulation of mitochondrial biogenesis (22). Exercise is a potent stimulator of PGC-1α mRNA expression in skeletal muscle (28, 33). PGC-1α transcription is modulated by the binding of four different transcription factors to the PGC-1α promoter region, cAMP response element-binding protein (CREB), activating transcription factor 2 (ATF2), MEF2, and forkhead box class-O (FoxO1) (8). Binding of these transcription factors is controlled through several different mechanisms, many of which are stimulated in response to acute exercise (1, 15, 17, 24, 42).
It had been long assumed that increases in mitochondrial biogenesis observed following exercise were attributed to an increase in PGC-1α mRNA (33). However, more recent evidence suggests that exercise-induced mitochondrial biogenesis begins before the exercise-induced increase in PGC-1α mRNA expression (47). Activation of existing PGC-1α protein may be responsible for acute exercise-induced mitochondrial biogenesis, while the observed increase in mRNA expression is a compensatory mechanism to prepare for adaptations to future exercise bouts (25, 36). To exert its coactivating effect on mitochondrial biogenesis-related transcription, PGC-1α must be translocated into the nucleus. At rest, the majority of PGC-1α protein is found in the cytosol (25). However, following endurance exercise, nuclear PGC-1α content increases without a change in total PGC-1α, suggesting a translocation from the cytosol to the nucleus (25). Once inside the nucleus, the deacetylase sirtuin-1 (SIRT1) acts to deacetylate PGC-1α, activating it and allowing it to bind to downstream transcription factors. This PGC-1α-transcription factor complex then binds to DNA and initiates transcription of genes related to mitochondrial biogenesis (13, 14, 36).
Expression of PGC-1α mRNA following exercise has been shown to be temperature sensitive in humans (41). PGC-1α mRNA response to exercise is blunted following exercise and recovery in a hot environment compared with room temperature. However, several other genes associated with mitochondrial biogenesis do not appear to be affected by temperature (41). Currently, the posttranslational mechanisms of PGC-1α protein after exercise in different environmental temperatures are not known. Understanding the stimulus that temperature and exercise has on the regulation of mitochondrial biogenesis is important to optimize training programs to be more effective at increasing performance, maintaining optimal function, and treating individuals with mitochondrial dysfunction. Therefore, the purpose of this study is to determine the acute response of PGC-1α, both transcriptionally and posttranslationally, to exercise in a hot temperature (33°C), compared with a room temperature (20°C), environment. More specifically, this study aims to determine how the normally observed translocation of PGC-1α protein into the nucleus, as well as binding of CREB, ATF2, MEF2, and FOXO1 to the PGC-1α-promoter region of chromosomal DNA, is altered following exercise in a hot environment compared with a room temperature control environment.
METHODS
Participant screening.
Recreationally active males (n = 11) between the ages of 19 and 45 were recruited as participants in this study. Participants self-reported taking part in aerobic exercise 3–7 days per week; their descriptive data can be found in results. After review and approval of all protocols by the University of Nebraska at Omaha Institutional Review Board (IRB), participants were informed of the general methods and procedures, risks, and benefits associated with participating in the study, the measures taken to minimize risk, and a complete description of their rights as a volunteer. Participants signed an IRB-approved informed consent form. All potential participants were stratified for risk according to the American College of Sports Medicine guidelines. Only participants stratified as “low risk” for participation in physical activity were enrolled in the study.
Aerobic capacity tests.
Participants performed a graded exercise test to measure aerobic capacity and to establish cycling intensity for the trial sessions. Participants cycled on a magnetically braked Velotron cycle ergometer (Racermate, Seattle, WA) beginning at a workload of 95 watts (W), and the workload was increased by 35 W every 3 min until volitional fatigue. Expired gases were analyzed using a flow and gas concentration calibrated TrueOne 2400 metabolic cart (ParvoMedics, Sandy, UT). V̇o2 peak was defined as the highest 15-s average V̇o2 during the protocol. Maximum wattage (Wmax) was calculated by taking the time completed in the last stage divided by the total stage duration (3 min) multiplied by 35 W and added to the W of the last completed stage.
Body composition assessments.
For descriptive purposes, participants had their body composition determined by hydrostatic weighing using an electronic load cell based system (Exertech, Dresbach, MN) corrected for estimated residual lung volume (35). Body density from hydrostatic weighing was converted to percent body fat using the Siri equation (39).
Experimental trials.
Each participant completed two trials, using a randomized, counterbalanced cross-over design. Trials consisted of 60 min of cycling at 60% of Wmax in either a room temperature (C) environment (20°C, 60% humidity) or hot (H) environment (33°C, 60% humidity). This exercise protocol was developed to control for total workload, while also ensuring that recreationally trained participants would be able to complete the exercise protocol in the hot condition. Following exercise, participants recovered in a supine position for 3 h at room temperature. Both trials were performed in a temperature and humidity controlled environmental chamber (Darwin, St Louis, MO), separated by at least 5, and no more than 20, days. On the trial day, participants were instructed to arrive at the laboratory after an 8-h fast, having replicated an identical 24-h diet that included abstaining from caffeine, alcohol, and tobacco before both trials. Additionally, participants were instructed to log and repeat any physical activity in the 48 h before each trail and asked to abstain from exercise in the 24 h preceding each trail. Participants ingested a Jonah Core Body Temperature Capsule with 125 ml of water and a fiber bar (29 g carbohydrate, 9 g fiber, 4 g fat, and 2 g protein, Fiber One; General Mills, Minneaplis, MN) to aid in the capsule passing through the stomach and into the small intestine. One hour after ingestion of the core temperature capsule, participants began the exercise protocol in the environmental chamber at the appropriate temperature. During the 60-min cycling bout, participants consumed 125 ml of water every 15 min. The 3-h recovery period began immediately upon completion of the cycling bout. During the recovery period, participants were allowed to drink water ad libitum but remained fasted until the final the conclusion of the protocol.
Muscle biopsies.
Muscle biopsies were taken pre-, post-, and 3 h postexercise in each trial. The final biopsy was performed 3 h postexercise to allow for peak expression of the genes of interest in this investigation (25, 26, 41), as well as to make accurate comparisons to previous work from our laboratory (41). Biopsies were taken from the vastus lateralis muscle using a 5-mm Bergstrom percutaneous muscle biopsy needle with the aid of suction. Each subsequent biopsy during a trial was obtained from the same leg using a separate incision 2 cm proximal to the previous biopsy. After excess blood, connective tissue, and fat were quickly removed, tissue samples were divided into two parts. One was immersed in RNAlater (Qiagen, Valencia, CA) for mRNA analysis and stored at 4°C overnight and then at −80°C until further analysis. The remaining tissue was flash frozen in liquid nitrogen and stored at −80°C for later analysis.
Body core temperature, skin temperature, and heart rate.
Body core temperature was measured by a Jonah Core Body Temperature Capsule ingestible thermistor and transmitted telemetrically to an EQ02 LifeMonitor Sensor Electronics Module (SEM; Hidalgo, Cambridge, UK) every 15 s. The EQ02 LifeMonitor SEM was also used to measure skin temperature (using an infrared thermistor) and heart rate, both of which were also logged every 15 s. Data were averaged for the entire 60 min to represent the cycling bout.
Rating of perceived exertion.
Rating of perceived exertion was assessed at 15, 30, 45, and 60 min of the cycling protocol using the 6–20 Borg Scale (4). An average of all four assessments was used to represent the entire cycling bout.
Oxygen consumption.
Expired gases were collected during exercise for determination of exercise intensity using a flow and gas concentration calibrated TrueOne 2400 metabolic cart (ParvoMedics, Sandy, UT). Gas was collected for 5 min at 10, 25, 40, and 55 min of the cycling protocol. The last 3 min of each segment were averaged to represent the collection period, and all four segments were averaged to represent the entire cycling bout.
mRNA extraction.
A 26.0 ± 10.8 mg piece of skeletal muscle was homogenized in 800 μl of Trizol (Invitrogen, Carlsbad CA) using an electric tissue disruptor (Tissue Tearor; Biosped Products, Bartlesville, OK). Samples were then incubated at room temperature for 5 min, after which 160 μl of chloroform were added. Tubes were shaken vigorously by hand for 15 s. After an additional incubation at room temperature for 2 to 3 min, the samples were centrifuged at 12,000 g for 15 min and the aqueous phase was transferred to a fresh 1.5-ml tube. mRNA was precipitated by adding 400 μl of isopropyl alcohol and incubated overnight at −20°C. The RNA was purified using an RNeasy mini kit (cat. no. 74104; Qiagen) according to the manufacturer's protocol using the additional DNase digestion step (RNase-free DNase set; Qiagen). RNA was then quantified using a nano-spectrophotometer (nano-drop ND-1000, Wilmington DE). RNA integrity was assessed using an Agilent RNA 6000 Kit (Agilent Technologies, Santa Clara, CA) according to manufacturer instructions, and read on a 2100 Bioanalyzer (Agilent Technologies). These quality control measures confirmed pure, intact RNA (260:280 ratio = 2.02 ± 0.13; 260:230 ratio = 1.56 ± 0.33; RIN = 8.0 ± 0.8).
cDNA synthesis.
First-strand cDNA synthesis was achieved using Superscript-first-strand synthesis system for RT-PCR kit (Invitrogen) according to the manufacturer's protocol. The resulting cDNA was then diluted with the appropriate amount of RNase free water to achieve a final cDNA concentration of 0.5 μg/μl in the PCR reaction.
qRT-PCR.
Each 20 μl qRT-PCR reaction contained 500 nM primers, 250 nM probe (PimeTime qPCR assay; Integrated DNA Technologies, Coralville, IA), 10 μl of Brilliant III Ultra-Fast QPCR Master Mix (Agilent Technologies), and 2.5 μl of sample cDNA. PCR was run using a Stratagene mx3005p PCR system (Aligent Technologies) using a two-step Roche protocol (1 cycle at 95°C for 3 min, followed by 40 cycles of 95°C for 5 s followed by 60°C for 20 s). Quantification of mRNA for genes of interest was calculated on post- and 3 h postexercise muscle samples relative to preexercise and stable reference genes using the 2−ΔΔCT method (27, 38). The geometric mean of the most stable reference genes for each participant, as determined by qbase+ geNorm software (Biogazelle, Zwijnaarde, Belgium) was used to calculate fold-changes of the genes of interest according to the 2−ΔΔCT method (44). Fold-change values were log transformed for statistical analysis to yield a normal distribution, as fold-change data are heavily skewed in a linear scale (44).
Subcellular fractionation.
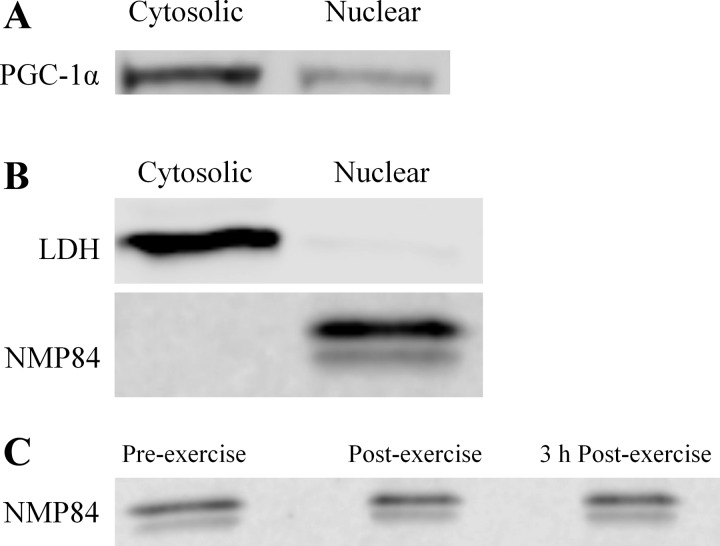
Subcellular fractionation was performed using an NE-PER Nuclear and Cytosolic Extraction Kit (Pierce Biotechnologies, Rockford, IL) according to manufacturer instructions, with additional wash steps to purify the nuclear extract. Briefly, 22.9 ± 1.3 mg of muscle were washed with 500 μl of phosphate-buffered saline (PBS) and centrifuged at 500 g for 5 min at 4°C. Samples were then homogenized in 200 μl of cytoplasmic extraction reagent I supplemented with HALT protease inhibitors (Pierce Biotechnologies) using an electronic tissue disruptor (Tissue Tearor; Biosped Products), vortexed for 15 s and incubated on ice for 10 min. Next, 11 μl of ice cold cytoplasmic extraction reagent II were added and tubes were vortexed and spun in a centrifuge at 16,000 g for 5 min at 4°C to elute the cytoplasmic fraction. The supernatant (the cytosolic fraction) was immediately transferred to a prechilled 1.5-ml tube, and stored at −80°C for later analysis. The pellet containing the nuclear fraction was washed four times with PBS to eliminate any cross contamination from the cytosolic extract. The pellet was then suspended in 100 μl of nuclear extraction regent supplemented with HALT protease inhibitors, and incubated on ice for 40 min, vortexing every 10 min. The nuclear extract was eluted by centrifuging at 16,000 g for 10 min at 4°C, transferred to a prechilled 1.5-ml tube and stored at −80°C for later analysis. This protocol resulted in <2% cross contamination between fractions, as evidenced by a high abundance of lactate dehydrogenase (LDH) and lack of nuclear matrix protein 84 (NMP84) in cytosolic fractions and an absence of LDH and presence of NMP84 in nuclear fractions.
Protein quantification.
Protein quantity of each fraction was measured using a commercially available BCA Protein Assay Kit (Pierce Biotechnologies, Rockford, IL) according to manufacturer instructions. Absorbance of samples was read at 562 nm using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA). Protein concentrations were then calculated using a standard curve according to kit instructions. Sample concentrations were 2,802 ± 380 μg/μl.
Gel electrophoresis and Western blotting.
Proteins were separated by SDS-PAGE. Twenty μg of protein were added to each well of precast 4–20% polyacrylamide gels (NuSep; Homebush, NSW, Australia), along with 7.5 μl of 4× loading buffer containing β-mercaptoethanol [125 mM Tris·HCl, ph 6.8, 50% glycerol, 4% SDS, 0.2% (wt/vol) Orange G, and 143 M β-mercaptoethanol; Li-Cor, Lincoln, NE] and an appropriate volume of homogenization buffer so that each well contained 30 μl. Gels were run at 150 V for 1 h in a Mini-PROTEAN Tetra cell (Bio-Rad, Hercules, CA) in 0.025 M Tris, 0.192 M glycine, and 0.1% SDS running buffer. Gels were then transferred to a nitrocellulose membrane using a Trans-Blot Turbo transfer system according to manufacturer's recommended protocol (Bio-Rad). The membrane was then blocked for 1 h at room temperature with Li-Cor blocking buffer (Li-Cor, Lincoln, NE), and incubated overnight (∼16 h) at 4°C in rabbit anti-human primary antibody (1:200) in Li-Cor blocking buffer for either PGC-1α, LDH, or NMP84 (all primary antibodies from Santa Cruz Biotechnology, Santa Cruz, CA) in a tube rotator. The next day, the membrane was washed with Tris-buffered saline plus Tween 20 (TBST; 50 mM Tris, 150 mM NaCl, 0.05% Tween 20) three times for 5 min each and then incubated in Li-Cor goat anti-rabbit 800 CW secondary antibody (1:5,000 in blocking buffer; Li-Cor) for 1 h. The membrane was again washed with TBST three times for 5 min each and imaged by infrared fluorescence using an Odyssey Fc imaging system (Li-Cor). Bands were quantified using Image Studio 5.2 software (Li-Cor).
Chromatin immunoprecipitation.
Binding of CREB, ATF2, MEF2, and FoxO1 to the PGC-1α promoter region was assessed by chromatin immunoprecipitation (ChIP). There was sufficient sample from nine participants for ChIP analysis (n = 9). ChIP was performed using a commercially available EpiQuick Tissue Chromatin Immunoprecipitation Kit (Epigentek, Farmingdale, NY) according to manufacturer's instructions. Briefly, protein-DNA interactions were fixed by cross linking 24.7 ± 4.9 mg of skeletal muscle with 1% formaldehyde. Samples were then homogenized with an electronic tissue disruptor, and DNA was sheared by sonication. Shearing was accomplished by sonicating samples with four pulses of 15 to 20 s using a Q55 sonicator (Qsonica, Newtown, CT) set to an amplitude of 40, with at least 1 min on ice between pulses. This protocol resulted in average DNA fragment sizes of 400 to 1,200 base pairs. Fragment sizes were checked using an Agilent DNA 7500 Kit (Agilent Technologies) according to manufacturer instructions, and read on a 2100 Bioanalyzer (Agilent Technologies). Protein-bound DNA was immunoprecipitated using Normal Mouse IgG (negative control), anti-RNA polymerase II (positive control), or previously validated (43, 49) anti-CREB (cat. no. sc-377154X), ATF2 (cat. no. sc-187X), MEF2 (cat. no. sc-313X), or FOXO1 (cat. no. sc-11350X) antibodies (all antibodies from Santa Cruz Biotechnology). Cross linking was reversed using proteinase K, and DNA was purified using spin columns and 90% EtOH. Purified immunoprecipitated DNA was quantified using qRT-PCR as outlined above. Custom probe/primer pairs were designed for putative binding sites of each transcription factor within the PGC-1α promoter region (PimeTime qPCR assay; Integrated DNA Technologies), as well as GAPDH to quantify input DNA. Raw PCR data was normalized using the percent input method (16), in which recovered immunoprecipitated DNA is expressed as a percentage of total DNA input into the reaction.
Statistical analyses.
Differences in exercise V̇o2, HR, core temperature, skin temperature, and rating of perceived exertion (RPE) were analyzed using paired t-tests. Differences in gene expression, protein content, and chromatin binding throughout the three time points (pre-, post-, and 3 h postexercise) and between the two trials (control and hot) were analyzed using 3 × 2 repeated measures ANOVAs (time × trial). In the event of a significant F-ratio, Fishers protected least significant difference method was used to detect where differences occurred. A probability of type I error of <5% was considered significant (P < 0.05). Statistical analysis was performed using SPSS Version 22.0 (IBM, Armonk, NY). All data are reported as mean ± SD.
RESULTS
Participant and exercise descriptive data.
Recreationally trained males (n = 11, age: 24 ± 3 yr, height: 178 ± 5 cm, weight: 80.3 ± 8.0 kg, percent body fat: 14.6 ± 3.3%, V̇o2 peak: 4.34 ± 0.84 l/min, Wmax: 275 ± 40 W) completed this study. During the trials, V̇o2 (P = 0.003), percent V̇o2 peak (P = 0.010), heart rate (P < 0.001), skin temperature (P < 0.001), and RPE (P < 0.001) were all higher in H than C, while core body temperature was not different between trials (P = 0.172). Total work was kept constant between trials. Exercise descriptive data are presented in Table 1.
Table 1.
Exercise descriptive data
| Trial | Control | Hot |
|---|---|---|
| Total work, kJ | 617.34 ± 85.38 | 617.34 ± 85.38 |
| V̇o2, l/min | 2.84 ± 0.38 | 3.05 ± 0.46* |
| %V̇o2peak | 64.5 ± 6.0 | 69.7 ± 5.1* |
| Heart rate, beats/min | 156 ± 10 | 166 ± 9* |
| Skin temperature, °C | 32.3 ± 1.1 | 36.6 ± 0.7* |
| Core temperature, °C | 37.8 ± 0.3 | 38.0 ± 0.4 |
| Rating of perceived exertion | 12 ± 1 | 15 ± 1* |
Values are means ± SE; n = 11.
P < 0.05 from control.
Mitochondrial biogenesis-related and transcription factor mRNA expression.
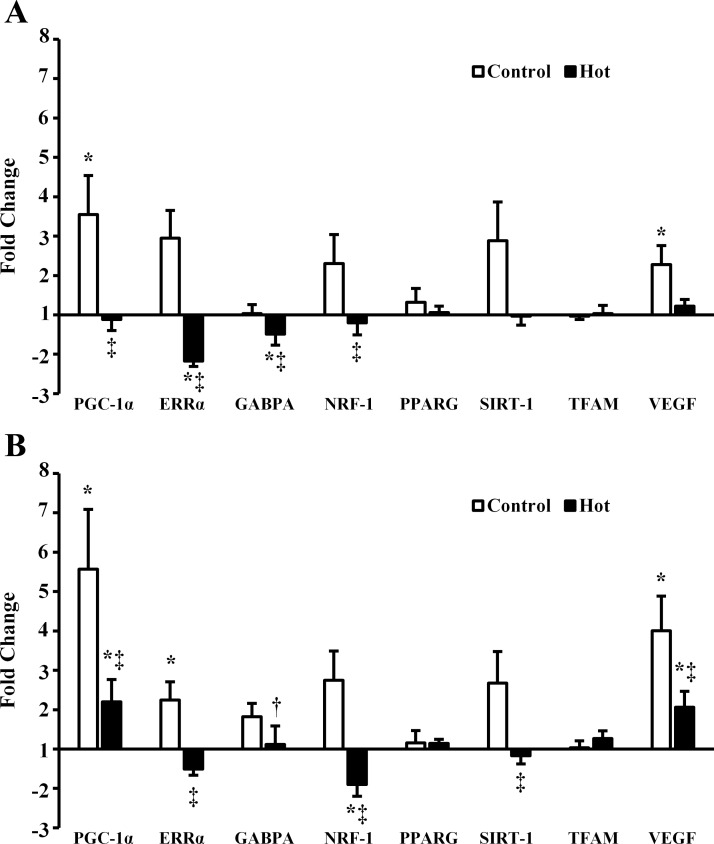
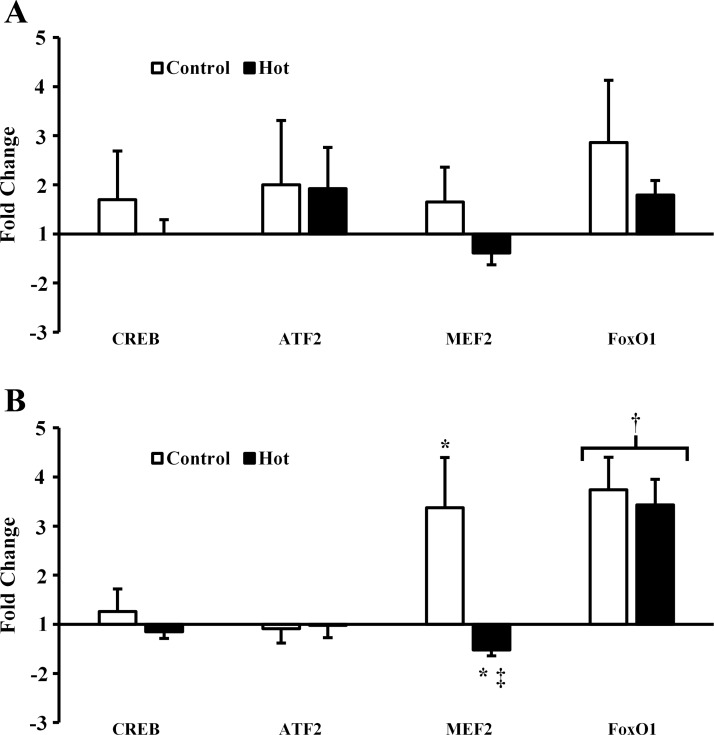
PGC-1α and VEGF mRNA increased postexercise (P = 0.039 and P = 0.014, respectively) and 3 h postexercise (P = 0.002 and P < 0.001, respectively) in C. PGC-1α and VEGF mRNA also increased 3 h postexercise in H (P = 0.037 and P = 0.008, respectively) but to a lesser degree than in C (P = 0.035 and P = 0.009, respectively). PGC-1α was lower immediately postexercise in H than C (P = 0.038). ERRα mRNA decreased postexercise in H (P = 0.009) and was lower in H than C postexercise (P < 0.001) and 3 h postexercise (P = 0.007). By 3 h postexercise ERRα had increased in C (P = 0.011). GABPA decreased postexercise in H (P = 0.019), although by less than onefold and was lower postexercise in H than C (P = 0.033). GABPA was higher 3 h postexercise than postexercise in H (P = 0.043) and not different between H and C 3 h postexercise (P = 0.122). NRF-1 was unchanged post- and 3 h postexercise in C (P = 0.268 and P = 0.066, respectively) but was downregulated 3 h postexercise in H (P = 0.002). NRF-1 mRNA was lower in H than C postexercise (P = 0.030) and 3 h postexercise (P < 0.001). SIRT-1 mRNA was not different from pre-, post (P = 0.064)-, or 3 h postexercise (P = 0.092) in C but was lower in H than C 3 h postexercise (P = 0.021). Peroxisome proliferator-activated receptor-γ (PPARG) and TFAM mRNA were unaffected by exercise in either H or C (P = 0.363 and P = 0.589, respectively). Mitochondrial biogenesis-related gene expression data are presented in Fig. 1. CREB and ATF2 mRNA were unaffected by exercise in H and C (P = 0.191 and P = 0.250, respectively). MEF2 was increased 3 h postexercise in C (P = 0.024) and decreased by less than onefold 3 h postexercise in H (P = 0.037). MEF2 was also lower in H than C 3 h postexercise (P = 0.008). FoxO1 was not different between trials (P = 0.841) but was increased 3 h postexercise (P < 0.001). PGC-1α promoting transcription factor gene expression data is presented in Fig. 2.
Fig. 1.
mRNA expression of mitochondrial biogenesis related genes A: postexercise and B: 3 h postexercise. PGC-1α, proliferator-activated receptor-γ coactivator 1-α; ERRα, estrogen-related receptor-α; GABPA, GA binding protein transcription factor α-subunit 60 kDa; NRF-1, nuclear respiratory factor 1; PPARG, peroxisome proliferator-activated receptor-γ; SIRT-1, sirtuin-1; TFAM, mitochondrial transcription factor A; VEGF, vascular endothelial growth factor. *P < 0.05 from preexercise; †P < 0.05 from postexercise; ‡P < 0.05 from control.
Fig. 2.
mRNA expression of PGC-1α promoting transcription factors. A: postexercise. B: 3 h postexercise. CREB, cAMP response element-binding protein; ATF2, ; MEF2, myocyte enhancer factor 2; FoxO1, FoxO forkhead box class-O1. *P < 0.05 from preexercise; †P < 0.05 from pre- and postexercise; ‡P < 0.05 from control.
CREB, ATF2, MEF2, and FoxO1 DNA binding.
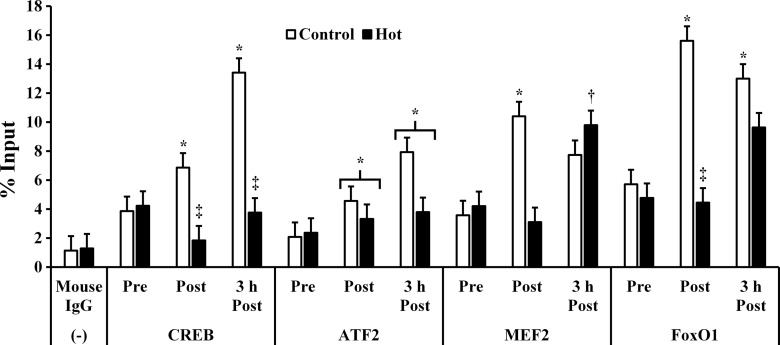
Binding of CREB to the PGC-1α promoter increased post (P = 0.026)- and 3 h postexercise (P = 0.011) in C but did not change in H. CREB binding was lower post (P = 0.005)- and 3 h postexercise (P = 0.004) in H compared with C. ATF2 binding to the PGC-1α promoter was not different between trials (P = 0.124) but was increased post (P = 0.017)- and 3 h postexercise (0.014). MEF2 binding to the PGC-1α promoter increased postexercise in C (P = 0.045) but was not significantly different from preexercise at 3 h postexercise in C (P = 0.105). Conversely, MEF2 binding did not increase from pre- to postexercise in H (P = 0.688) but was higher 3 h postexercise than both pre- and postexercise (P = 0.045 and P = 0.03, respectively). FoxO1 binding to the PGC-1α promoter was increased post (P = 0.035)- and 3 h postexercise (P = 0.036) in C but did not change in H (P = 0.235). FoxO1 binding was higher in C than H postexercise (P = 0.006; Fig. 3). PGC-1α promoting transcription factor DNA binding data is presented in Fig. 3.
Fig. 3.
DNA binding of transcription factors to the PGC-1α promoter. Mouse IgG was used as a negative control, and GAPDH was used as a positive control. *P < 0.05 from preexercise. †P < 0.05 from pre- and postexercise. ‡P < 0.05 from control.
Subcellular location of PGC-1α protein.
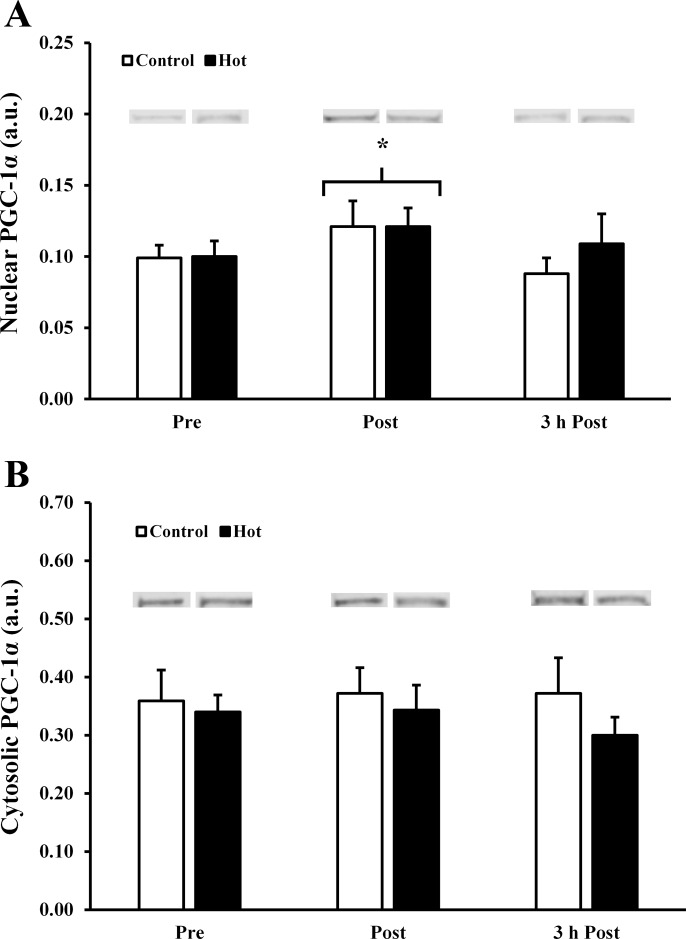
At rest, 77 ± 5% of PGC-1α was present in the cytosol (Fig. 4A). Efficiency of subcellular fractionation was demonstrated by a high abundance of lactate LDH and lack of NMP84 in cytosolic fractions, and an absence of LDH and presence of NMP84 in nuclear fractions (Fig. 4B). Nuclear protein yield was not impacted by exercise, as demonstrated by equal amounts of NMP84 in pre-, post-, and 3 h postexercise samples (Fig. 4C). Postexercise, there was a ∼20% increase in nuclear PGC-1α (P = 0.029, main effect of time; Fig. 5A), indicating a translocation of cytosolic PGC-1α into the nucleus. There were no differences between trials in nuclear (P = 0.602) or cytosolic (P = 0.333) or PGC-1α (Fig. 5, A and B).
Fig. 4.
A: subcellular location of PGC-1α at rest. B: purity of cytosolic and nuclear fractions demonstrated by presence of lactate dehydrogenase (LDH) and lack of nuclear matrix protein 84 (NMP84). C: nuclear protein yield, as demonstrated by NMP84.
Fig. 5.
A: nuclear PGC-1α. B: cytosolic PGC-1α. All samples for a given subject were run on the same gel. All samples were processed in parallel. *P < 0.05 from preexercise.
DISCUSSION
The main findings of this study were that PGC-1α mRNA expression, as well as expression of several other mitochondrial biogenesis-related genes, is blunted following exercise in a hot environment, coinciding with decreased binding of CREB, MEF2, and FoxO1 to the PGC-1α promoter region. However, translocation of PGC-1α protein from the cytosol into the nucleus following exercise was unaffected by the temperature intervention. These data demonstrate somewhat of a paradox, as the presence of PGC-1α in the nucleus should allow it to exert its cotranscriptional activity. Yet, several genes coactivated by PGC-1α, including ERRα, MEF2, and NRF-1, were downregulated following exercise in the heat. Overall, these data indicate that acute exercise in the heat has a deleterious effect on transcription factor binding at the PGC-1α promoter, which could potentially inhibit mitochondrial biogenesis.
PGC-1α mRNA expression following acute exercise has previously been shown to be intensity dependent (7, 28). Based on these findings alone, it may be expected that PGC-1α mRNA would increase to a greater degree following exercise in the heat, as at a given workload relative exercise intensity (V̇o2) is higher in the heat. Indeed, there was a higher relative exercise intensity at the clamped workload in the heat in the present study, as demonstrated by higher V̇o2, HR, and reported RPE. However, the differences in relative exercise intensity in the present study were relatively small and certainly did not augment PGC-1α expression in the heat. Rather, these data support earlier findings of blunted PGC-1α mRNA expression following exercise in the heat (41).
The potential mechanism through which PGC-1α mRNA expression is blunted following exercise in the heat was previously unknown. To explore potential avenues through which this may occur, chromatin immunoprecipitation was carried out to assess binding of the transcription factors CREB, ATF2, MEF2, and FoxO1 to the promoter region of the PGC-1α gene. Binding of CREB, MEF2, and FoxO1 was all reduced following exercise in the heat. CREB binding was lower both immediately post- and 3 h postexercise in the hot trial than the control. CREB has been shown to be activated in response to cold temperatures through the β3-adrenergic receptor (5, 34); however, it is currently unknown whether this pathway would be further downregulated in response to hot temperatures. If so, it may explain the lower amount of CREB binding observed in the hot trial. MEF2 and FoxO1 binding was increased postexercise in the control trial and not in the hot trial. However, neither MEF2 nor FoxO1 binding was different between the two trials 3 h postexercise. Heat stress has been shown to upregulate Akt activity in rats (51) and swine skeletal muscle satellite cells (11). Akt-mediated phosphorylation excludes FoxO1 from the nucleus, therefore inhibiting its DNA binding (42). This mechanism of FoxO1 nuclear export could explain decreased FoxO1 binding postexercise in the hot trial. By 3 h postexercise, FoxO1 binding was not different between the trials, as participants had been recovering at room temperature. This finding may indicate an altered time course of normally observed PGC-1α expression following exercise in the heat. It is possible that rather than a blunting of mitochondrial biogenesis-related gene expression, there is a delay in their onset, and further time-course investigation is warranted. Additionally, it is unknown whether the decreased transcription factor binding observed in the present study is a general cellular phenomenon whereby all transcription factor-DNA interatction is reduced or specific to these transcription factors.
In response to heat stress, a wide variety of chaperone proteins, including several heat-shock proteins, are upregulated as a part of the well-documented cellular heat shock response (12, 31). These proteins act to ensure the correct folding of proteins and stabilize protein structure (12, 31). The heat shock response is coordinated by heat-shock factor 1 (HSF1), a transcription factor that induces the expression of many heat-shock genes (2). It has recently been shown that PGC-1α acts as a direct transcriptional repressor of HSF1 in C2C12 myotubes and mice skeletal muscle (29). It is possible that the cell prioritizes the heat shock response immediately following an exercise bout in a hot environment in an attempt to return to thermal homeostasis before the induction of mitochondrial biogenesis via this PGC-1α-related mechanism. This mechanism would work to prevent the misfolding of mitochondrial proteins and prevent the formation of dysfunctional mitochondria. Thus the mechanism described here may be protective and ensure that dysfunctional mitochondria are not assembled under conditions of heat stress. It is uncertain if the inhibition of markers of mitochondrial biogenesis described here persist after repeated exposure to heat stress, when thermotolerance is enhanced due to greater expression of heat shock proteins (20).
At rest, the majority of PGC-1α protein resided in the cytosol, and following exercise, there was an increase in nuclear PGC-1α, indicating a translocation of PGC-1α into the nucleus. While there was not a corresponding decrease in cytosolic PGC-1α, this could be explained by the fact that with approximately four times as much PGC-1α in the cytosol, a comparatively small decrease in concentration is less likely to be detected than the ∼20% increase in the nuclear fraction. The observed subcellular location of PGC-1α at rest and following exercise is in agreement with previous work (14, 25). While the present study displays a smaller increase in nuclear PGC-1α postexercise than previously reported [∼20% in the present study vs. ∼32% reported by Gurd et al. (14) and ∼54% reported by Little et al. (25)], this may be due to slight differences in the exercise protocol or training status of the participants.
While the translocation of PGC-1α to the nucleus was not impaired by exercise in the heat, the transcription of several of its downstream targets, including ERRα, MEF2, and NRF-1, were downregulated following exercise in the heat. Additionally, while GABPA expression was not downregulated, it was blunted following exercise in the heat compared with control conditions. PGC-1α coactivates ERRα and GABPA, which each contain binding sites in their own promoter regions (21, 30), which results in an autoregulatory loop. Additionally, while MEF2 modulates transcription of PGC-1α, it also increases transcription of MEF2, creating an additional autoregulatory loop (23). With PGC-1α present in the nucleus, it would be expected that transcription of each of these targets would increase, which was observed in the control condition. However, following exercise in the heat, this relationship was disrupted. One potential mechanism, although not assessed in this study, though which this paradox may be explained is through the deacytelase SIRT1. SIRT1 deacytelates PGC-1α, activating it (36). SIRT1 activity is increased with exercise and linked to increased mitochondrial biogenesis (13, 14). Recently, SIRT1 has been shown to decline after heat shock in HCT116 cells (50). While it is unknown how SIRT1 activity responds to heat stress in a human model, the potential for a decrease in SIRT1 activity following exercise in the heat would explain the blunted transcription of transcription factors downstream of PGC-1α, even when PGC-1α is present in the nucleus.
It is uncertain from the current data whether the blunting of mitochondrial biogenesis-related gene expression following exercise in the heat is truly a deleterious effect, or if a time-delayed, but intact, response of these genes is observed over a recovery period of greater than 3 h. It remains possible that through either the heat shock response or a separate unknown mechanism, the time-course of mitochondrial biogenesis-related gene expression is impacted by exercise in the heat, rather than blunted. One potential mechanism for the observed blunting of mitochondrial biogenesis-related genes following exercise in the heat is a decrease in SIRT1 activity, although the response of SIRT1 activity to exercise in the heat has not been categorized in a human model. Additionally, the chronic impact of exercise in the heat on mitochondrial biogenesis should be explored. The data collection period in this study took place during the fall and winter months, when participants were unlikely to be acclimated to exercising in the heat. It is unknown whether the acute response observed in the present study will lead to reduced mitochondrial biogenesis over a period of exercise training in the heat or whether there will be some type of adaptation, or “muscular heat acclimation” that allows for the acute response of mitochondrial biogenesis-related gene expression to return to a more normal profile. This possibility is especially intriguing given the finding that translocation PGC-1α protein into the nucleus, where it can exert its coactivating activity, was unaffected by exercise in the heat compared with room temperature, despite the observed blunting of mitochondrial biogenesis-related gene expression. While athletic performance has long been understood to benefit from heat acclimation during training, these findings may further underscore the importance of acclimation to the heat to attain beneficial muscular adaptations in athletic and clinical populations alike.
In conclusion, through decreased binding of CREB, MEF2, and FoxO1 to the promoter region of the PGC-1α, gene expression of PGC-1α, as well as expression of several other mitochondrial biogenesis-related genes, is blunted following exercise in the heat compared with a room temperature control. This blunting effect occurred despite no differences in the translocation of PGC-1α into the nucleus following exercise between the hot and control conditions, thus indicating that a mechanism independent of PGC-1α nuclear translocation is involved in the observed blunting of mitochondrial biogenesis-related gene expression. Together, these data suggest that acute exercise in the heat either blunts or delays the process of mitochondrial biogenesis, particularly in non-heat-acclimated individuals.
GRANTS
This publication was made possible by National Institute for General Medical Science (NIGMS) Grant 5P20GM103427, a component of the National Institutes of Health (NIH) and its contents are the sole responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. Additional funding was provided by a Graduate Research and Creative Activity Grant from the University of Nebraska at Omaha Office for Research and Creative Activity.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
M.W.H., R.J.S., and D.R.S. conception and design of research; M.W.H., R.J.S., J.L.K., and D.R.S. performed experiments; M.W.H., R.J.S., J.L.K., and D.R.S. analyzed data; M.W.H., R.J.S., and D.R.S. interpreted results of experiments; M.W.H. prepared figures; M.W.H. drafted manuscript; M.W.H., R.J.S., J.L.K., and D.R.S. edited and revised manuscript; M.W.H., R.J.S., J.L.K., and D.R.S. approved final version of manuscript.
ACKNOWLEDGEMENTS
We thank Terrence Laursen, Matthew Bubak, Nicholas Dinan, and David Taylor La Salle for assistance with data collection. Additionally, we thank Dr. Kris Berg, Dr. Jeffery French, and Dr. Nicholas Stergiou for reviewing and providing feedback on the manuscript.
REFERENCES
- 1.Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem 280: 19587–19593, 2005. [DOI] [PubMed] [Google Scholar]
- 2.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem 80: 1089–1115, 2011. [DOI] [PubMed] [Google Scholar]
- 3.Azad MA, Kikusato M, Maekawa T, Shirakawa H, Toyomizu M. Metabolic characteristics and oxidative damage to skeletal muscle in broiler chickens exposed to chronic heat stress. Comp Biochem Physiol A Mol Integr Physiol 155: 401–406, 2010. [DOI] [PubMed] [Google Scholar]
- 4.Borg GA. Perceived exertion: a note on “history” and methods. Med Sci Sports 5: 90–93, 1973. [PubMed] [Google Scholar]
- 5.Boss O, Bachman E, Vidal-Puig A, Zhang CY, Peroni O, Lowell BB. Role of the beta(3)-adrenergic receptor and/or a putative beta(4)-adrenergic receptor on the expression of uncoupling proteins and peroxisome proliferator-activated receptor-gamma coactivator-1. Biochem Biophys Res Commun 261: 870–876, 1999. [DOI] [PubMed] [Google Scholar]
- 6.Cho Y, Hazen BC, Russell AP, Kralli A. Peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. J Biol Chem 288: 25207–25218, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egan B, Carson BP, Garcia-Roves PM, Chibalin AV, Sarsfield FM, Barron N, McCaffrey N, Moyna NM, Zierath JR, O'Gorman DJ. Exercise intensity-dependent regulation of peroxisome proliferator-activated receptor coactivator-1 mRNA abundance is associated with differential activation of upstream signalling kinases in human skeletal muscle. J Physiol 588: 1779–1790, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr 93: 884S-90, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest 116: 615–622, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fosslien E. Mitochondrial medicine–molecular pathology of defective oxidative phosphorylation. Ann Clin Lab Sci 31: 25–67, 2001. [PubMed] [Google Scholar]
- 11.Gao CQ, Zhao YL, Li HC, Sui WG, Yan HC, Wang XQ. Heat stress inhibits proliferation, promotes growth, and induces apoptosis in cultured Lantang swine skeletal muscle satellite cells. J Zhejiang Univ Sci B 16: 549–559, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gidalevitz T, Prahlad V, Morimoto RI. The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harb Perspect Biol 3: pii: a009704, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurd BJ. Deacetylation of PGC-1alpha by SIRT1: importance for skeletal muscle function and exercise-induced mitochondrial biogenesis. Appl Physiol Nutr Metab 36: 589–597, 2011. [DOI] [PubMed] [Google Scholar]
- 14.Gurd BJ, Yoshida Y, McFarlan JT, Holloway GP, Moyes CD, Heigenhauser GJ, Spriet L, Bonen A. Nuclear SIRT1 activity, but not protein content, regulates mitochondrial biogenesis in rat and human skeletal muscle. Am J Physiol Regul Integr Comp Physiol 301: R67–R75, 2011. [DOI] [PubMed] [Google Scholar]
- 15.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci USA 100: 7111–7116, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413: 179–183, 2001. [DOI] [PubMed] [Google Scholar]
- 18.Holloszy JO, Booth FW. Biochemical adaptations to endurance exercise in muscle. Annu Rev Physiol 38: 273–291, 1976. [DOI] [PubMed] [Google Scholar]
- 19.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51: 2944–2950, 2002. [DOI] [PubMed] [Google Scholar]
- 20.Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol (1985) 92: 2177–2186, 2002. [DOI] [PubMed] [Google Scholar]
- 21.Laganiere J, Tremblay GB, Dufour CR, Giroux S, Rousseau F, Giguere V. A polymorphic autoregulatory hormone response element in the human estrogen-related receptor alpha (ERRalpha) promoter dictates peroxisome proliferator-activated receptor gamma coactivator-1alpha control of ERRalpha expression. J Biol Chem 279: 18504–18510, 2004. [DOI] [PubMed] [Google Scholar]
- 22.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1: 361–370, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418: 797–801, 2002. [DOI] [PubMed] [Google Scholar]
- 24.Ling C, Poulsen P, Carlsson E, Ridderstrale M, Almgren P, Wojtaszewski J, Beck-Nielsen H, Groop L, Vaag A. Multiple environmental and genetic factors influence skeletal muscle PGC-1alpha and PGC-1beta gene expression in twins. J Clin Invest 114: 1518–1526, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Little JP, Safdar A, Cermak N, Tarnopolsky MA, Gibala MJ. Acute endurance exercise increases the nuclear abundance of PGC-1α in trained human skeletal muscle. Am J Physiol Regul Integr Comp Physiol 298: R912–R917, 2010. [DOI] [PubMed] [Google Scholar]
- 26.Liu CT, Brooks GA. Mild heat stress induces mitochondrial biogenesis in C2C12 myotubes. J Appl Physiol (1985) 112: 354–361, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-delta delta C(T)] method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 28.Mathai AS, Bonen A, Benton CR, Robinson DL, Graham TE. Rapid exercise-induced changes in PGC-1alpha mRNA and protein in human skeletal muscle. J Appl Physiol (1985) 105: 1098–1105, 2008. [DOI] [PubMed] [Google Scholar]
- 29.Minsky N, Roeder RG. Direct link between metabolic regulation and the heat-shock response through the transcriptional regulator PGC-1alpha. Proc Natl Acad Sci USA 112: E5669-5678, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, Willy PJ, Schulman IG, Heyman RA, Lander ES, Spiegelman BM. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA 101: 6570–6575, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morimoto RI. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp Quant Biol 76: 91–99, 2011. [DOI] [PubMed] [Google Scholar]
- 32.Pedersen BL, Baekgaard N, Quistorff B. Muscle mitochondrial function in patients with type 2 diabetes mellitus and peripheral arterial disease: implications in vascular surgery. Eur J Vasc Endovasc Surg 38: 356–364, 2009. [DOI] [PubMed] [Google Scholar]
- 33.Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. J Physiol 546: 851–858, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839, 1998. [DOI] [PubMed] [Google Scholar]
- 35.Quanjer PH, Tammeling GJ, Cotes JE, Pedersen OF, Peslin R, Yernault JC. Lung volumes and forced ventilatory flows. Report Working Party Standardization of Lung Function Tests, European Community for Steel and Coal Official Statement of the European Respiratory Society. Eur Respir J Suppl 16: 5–40, 1993. [PubMed] [Google Scholar]
- 36.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett 582: 46–53, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakaki K, Tashiro K, Kuhara S, Mihara K. Response of genes associated with mitochondrial function to mild heat stress in yeast Saccharomyces cerevisiae. J Biochem 134: 373–384, 2003. [DOI] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108, 2008. [DOI] [PubMed] [Google Scholar]
- 39.Siri WE. Body composition from fluid spaces and density: analysis of methods. 1961. Nutrition 9: 480-91; discussion 480, 492, 1993. [PubMed] [Google Scholar]
- 40.Skulachev VP, Longo VD. Aging as a mitochondria-mediated atavistic program: can aging be switched off? Ann NY Acad Sci 1057: 145–164, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Slivka DR, Dumke CL, Tucker TJ, Cuddy JS, Ruby B. Human mRNA response to exercise and temperature. Int J Sports Med 33: 94–100, 2012. [DOI] [PubMed] [Google Scholar]
- 42.Southgate RJ, Bruce CR, Carey AL, Steinberg GR, Walder K, Monks R, Watt MJ, Hawley JA, Birnbaum MJ, Febbraio MA. PGC-1alpha gene expression is down-regulated by Akt- mediated phosphorylation and nuclear exclusion of FoxO1 in insulin-stimulated skeletal muscle. FASEB J 19: 2072–2074, 2005. [DOI] [PubMed] [Google Scholar]
- 43.Sun Y, Cheng J, Mitchelson KR. PCR DNA-array profiling of DNA-binding transcription factor activities in adult mouse tissues. Methods Mol Biol 687: 319–331, 2011. [DOI] [PubMed] [Google Scholar]
- 44.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene 354: 169–180, 2005. [DOI] [PubMed] [Google Scholar]
- 46.Wallace DC, Shoffner JM, Trounce I, Brown MD, Ballinger SW, Corral-Debrinski M, Horton T, Jun AS, Lott MT. Mitochondrial DNA mutations in human degenerative diseases and aging. Biochim Biophys Acta 1271: 141–151, 1995. [DOI] [PubMed] [Google Scholar]
- 47.Wright DC, Han DH, Garcia-Roves PM, Geiger PC, Jones TE, Holloszy JO. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1alpha expression. J Biol Chem 282: 194–199, 2007. [DOI] [PubMed] [Google Scholar]
- 48.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98: 115–124, 1999. [DOI] [PubMed] [Google Scholar]
- 49.Xiong S, Salazar G, Patrushev N, Alexander RW. FoxO1 mediates an autofeedback loop regulating SIRT1 expression. J Biol Chem 286: 5289–5299, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang H, Yan B, Liao D, Huang S, Qiu Y. Acetylation of HDAC1 and degradation of SIRT1 form a positive feedback loop to regulate p53 acetylation during heat-shock stress. Cell Death Dis 6: e1747, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshihara T, Naito H, Kakigi R, Ichinoseki-Sekine N, Ogura Y, Sugiura T, Katamoto S. Heat stress activates the Akt/mTOR signalling pathway in rat skeletal muscle. Acta Physiol (Oxf) 207: 416–426, 2013. [DOI] [PubMed] [Google Scholar]