Supplemental Digital Content is available in the text.
Keywords: alcohol, aldehyde dehydrogenase 2, cumulative risk, prediction model, upper aerodigestive tract cancer
Abstract
Alcohol consumption and the aldehyde dehydrogenase 2 (ALDH2) polymorphism are associated with the risk of upper aerodigestive tract cancer, and a significant gene–environment interaction between the two has been confirmed in a Japanese population. To aid the development of a personalized prevention strategy, we developed a risk-prediction model and estimated absolute risks stratified by a combination of the ALDH2 genotype and alcohol consumption. We carried out two age-matched and sex-matched case–control studies: one (630 cases and 1260 controls) for model derivation and the second (654 cases and 654 controls) for external validation. On the basis of data from the derivation study, a prediction model was developed by fitting a conditional logistic regression model using the following predictors: age, sex, smoking, drinking, and the ALDH2 genotype. The risk model, including a combination of the ALDH2 genotype and alcohol consumption, provided high discriminatory accuracy and good calibration in both the derivation and the validation studies: C statistics were 0.82 (95% confidence interval 0.80–0.84) and 0.83 (95% confidence interval 0.81–0.85), respectively, and the calibration plots of both studies remained close to the ideal calibration line. Cumulative risks were obtained by combining odds ratios estimated from the risk model with the age-specific incidence rate and population size. For heavy drinkers with a heterozygous genotype, the cumulative risk at age 80 was above 20%. In contrast, risk in the other groups was less than 5%. In conclusion, modification of alcohol consumption according to the ALDH2 genotype will have a major impact on upper aerodigestive tract cancer prevention. These findings represent a simple and practical model for personalized cancer prevention.
Introduction
In 2012, more than one million cases of upper aerodigestive tract cancer (UATC), comprising cancers of the oral cavity, pharynx, larynx, and esophagus, were newly diagnosed worldwide, and ∼10% of all cancer deaths were attributed to UATC (Ferlay et al., 2013). In Japan, UATC was the seventh most common cancer, with 45 439 new cases in 2011 (Matsuda et al., 2013). Although the efficacy of medical and surgical treatment of cancer has improved markedly, the 5-year relative survival rate for UATC remains unchanged since the 1990s in the Japanese population; the rate for oral cavity and pharyngeal cancer is about 53% and that for esophageal cancer is about 32% (Matsuda et al., 2011). This background suggests that treatment alone is unlikely to solve the problem of UATC and that efforts should also be directed toward the establishment of personalized prevention strategies with implementation at the population level.
The impact of alcohol drinking on the risk of UATC has been established (Cogliano et al., 2011). In Japan, alcohol consumption in the adult population has not changed much in the past two decades. The average yearly consumption between 2008 and 2010 in Japan was 7.2 l per capita, which was slightly higher than the average of the world (World Health Organization, 2014). Evidence suggests that a plausible candidate for the carcinogenic effect of ethanol is not ethanol itself, but acetaldehyde (Boffetta and Hashibe, 2006), the primary metabolite of ethanol, which is further metabolized mainly by aldehyde dehydrogenase 2 (ALDH2). In East Asian populations, the ALDH2 gene shows a polymorphism (rs671, Glu504Lys) that modulates individual differences in acetaldehyde-oxidizing capacity (Yoshida et al., 1984; Bosron and Li, 1986; Li et al., 2006). As the ALDH2 Lys allele encodes a catalytically inactive subunit, individuals with the Lys allele show a marked increase in blood acetaldehyde after alcohol ingestion (Mizoi et al., 1994), and as a result carry a high risk of UATC (Yokoyama et al., 1998; Boccia et al., 2009). Moreover, the Lys allele has been confirmed to increase susceptibility to UATC among drinkers, particularly heavy drinkers. We previously showed for the first time a strong gene–environment interaction between the ALDH2 genotype and alcohol drinking on the risk of esophageal cancer (Matsuo et al., 2001), and subsequent studies, including our own, showed the same phenomenon in UATC (Yokoyama et al., 2006; Asakage et al., 2007; Hiraki et al., 2007).
Early identification of populations at high risk of UATC is important for UATC prevention and will facilitate intensive targeted prevention in individuals at high risk. Although a few prediction models for esophageal cancer have been developed for clinical settings (Yokoyama et al., 2008; Collins and Altman, 2013; Thrift et al., 2013), no prediction model for practical prevention settings has been developed as yet. Our first aim was to develop a risk-prediction model using established risk factors, which we hoped would be useful as a personalized prevention strategy. For this, we carried out two age-matched and sex-matched case–control studies: the first for model derivation and the second for external validation. As predictors, the model included alcohol drinking, the ALDH2 genotype, and cigarette smoking – already established as a preventable exposure associated with UATC (Cogliano et al., 2011) – as these enable reliable stratification by simple lifestyle questions and genotyping.
Our second aim was to estimate absolute risks stratified by level of alcohol consumption in consideration of the ALDH2 genotype using the estimates from the risk model. This would enable us to present more easily graspable information that may be effective in motivating individuals to reduce their alcohol intake. By evaluating a combination of the ALDH2 genotype and alcohol drinking, we would be able to encourage individuals to modify their drinking behavior, specifically on the basis of their genotype.
Materials and methods
Study population
In the derivation case–control study, the case participants were 630 patients with no previous history of cancer who were histologically diagnosed with UATC (365 with head and neck cancer and 265 with esophageal cancer) between January 2001 and December 2005 at Aichi Cancer Center Hospital in Nagoya, Japan. Participants in the derivation study were recruited within the framework of the Hospital-based Epidemiologic Research Program at Aichi Cancer Center (HERPACC)-2. Details of the program are provided elsewhere (Hamajima et al., 2001) and brief descriptions of the program are provided in the Appendix. UATC was defined according to the following codes of the International Classification of Diseases for Oncology, 3rd ed. (ICD-O-3): oral cavity and oropharynx (C00.3–C00.9, C01.9, C02.0–C02.4, C03, C04, C05.0–C05.2, C06, C09, C10), hypopharynx (C12, C13), oral cavity–oropharynx–hypopharynx not otherwise specified (C02.8, C02.9, C05.8, C05.9, C14), larynx (C32), and esophagus (C15). Malignant neoplasms of the salivary glands (C07, C08), nasopharynx (C11), nasal (C30), and paranasal sinuses (C31) were excluded as they have quite distinct etiologies. The controls were 1260 first-visit outpatients during the same period who had no cancer and no history of neoplasia. Noncancer status was confirmed by medical examinations, including radiographic examinations. Those who were suspected of having UATC were first examined by physical or endoscopic inspection and subsequently radiographically, if indicated. Controls were selected randomly and frequency-matched by age (±4 years) and sex (male, female) at a case–control ratio of 1 : 2.
Participants for the validation case–control study were recruited from HERPACC-3; this was carried out between November 2005 and March 2013 under an enrollment framework equivalent to that of HERPACC-2. A total of 654 UATC cases (309 with head and neck cancer, 328 with esophageal cancer, and 17 with cancer of both sites) and 654 individually age-matched (±2 years) and sex-matched (male, female) noncancer controls were recruited. Inclusion criteria for controls in the validation study were similar to those in the derivation study.
All participants in both studies provided written informed consent, completed a self-administered questionnaire, and provided blood. The present studies were approved by the Institutional Ethical Committee of Aichi Cancer Center.
Genotyping procedure
DNA was extracted from the buffy coat fraction using a DNA Blood mini kit (Qiagen, Tokyo, Japan). Genotyping for rs671 (ALDH2 Glu504Lys) was based on TaqMan Assays (Applied Biosystems, Foster City, California, USA).
Evaluation of environmental factors
Information on cumulative smoking and alcohol consumption was collected by a self-administered questionnaire. Responses were checked by trained interviewers. Cumulative smoking was evaluated as pack-years, calculated by multiplying the number of packs consumed per day by the number of years of smoking, and then classified into three categories: never (pack-years=0), light-moderate (0<pack-years<20), and heavy (20≤pack-years). Alcohol consumption was classified into four categories: never, moderate, high-moderate, and heavy. Those who seldom or never drank were defined as never drinkers. Moderate drinking was defined as consumption on 4 days or fewer per week; high-moderate drinking as consumption on 5 days or more per week of less than 46 g ethanol on each occasion; and heavy drinking as consumption on 5 days or more per week of more than 46 g ethanol on each occasion.
Statistical analysis
All analyses were carried out using STATA, version 13 (Stata Corporation, College Station, Texas, USA). We considered two-sided P values of less than 0.05 as statistically significant. Discrepancies between expected and observed genotype and allele frequencies in the controls were assessed in accordance with the Hardy–Weinberg equilibrium using the χ2-test.
Model construction
On the basis of data from HERPACC-2 (derivation study), we developed three risk-prediction models, a genetic, environmental, and inclusive model, by fitting conditional logistic regression models. In addition to age and sex, each model included the following factors: the genetic model included the ALDH2 genotype; the environmental model included cumulative smoking and alcohol consumption; and the inclusive model included cumulative smoking and a combination of the ALDH2 genotype and alcohol consumption. The categories of cumulative smoking (never, light-moderate, and heavy), alcohol consumption (never, moderate, high-moderate, and heavy), and the ALDH2 genotype (Glu/Glu, Glu/Lys, and Lys/Lys) were introduced as dummy variables. The combination of the ALDH2 genotype and alcohol consumption was assessed by adding interaction terms to the risk models. Missing data were coded using dummy variables.
Assessment of the performance of prediction models
The performance of the prediction models was assessed in a derivation study (as ‘internal validation’) and in a validation study (as ‘external validation’) using standard methods to measure discriminative ability and calibration. A model’s discrimination indicates how accurately it can distinguish between individuals with and without the outcome. Calibration reflects the precision of how close the predicted probabilities are to the observed probabilities.
Discriminative ability was assessed by the value of the area under the receiver operating characteristic (ROC) curve (AUC), which is also known as the concordance (c) statistic. In the ROC, the y-axis shows sensitivity and the x-axis shows the false-positive rate. The AUC values were compared using the method of DeLong et al. (1988).
Model calibration was assessed by the Hosmer–Lemeshow goodness-of-fit statistic and calibration plots. Participants were grouped by decile of predicted probability. The Hosmer–Lemeshow statistic was computed from a χ2-test comparing the observed frequencies with the predicted frequencies in the10 groups; a nonsignificant value indicates good calibration, whereas a significant P-value indicates disagreement between the predicted and the observed outcomes. In a calibration plot, the mean predicted probability was plotted against the mean observed probability for each decile. Ideally, the predicted probability equals the observed probability; thus, perfect predictions should be on the 45° line (Steyerberg, 2009). In addition, we estimated the slope of the calibration plots. With perfect calibration, a calibration slope equals 1. A slope below 1.0 reflects overfitting of a model, which indicates the need to shrink the regression coefficients (Steyerberg, 2009).
Cumulative risk estimation by risk strata
The two case–control studies were combined to stratify patients into different risk groups by a combination of the ALDH2 genotype and alcohol drinking. We constructed a conditional logistic regression model on the basis of the combined data of the two case–control studies in exactly the same way as the inclusive model development because calculation on a larger sample size improves coefficient precision. The odds ratio (OR) and 95% confidence interval (CI) were calculated for each risk group compared with never drinkers with the Glu/Glu genotype.
Accordingly, we estimated the cumulative risks using a method already adopted in several studies (Peto et al., 2000; Brennan et al., 2006; Bosetti et al., 2008) (see Appendix). The ORs were combined with the prevalence of each subgroup in the control and the age-specific population size of 2007, the middle year of the study period. Combining these with the age-specific incidence rate of UATC of 2007 produced the age-specific absolute rates in the different subgroups. We then calculated cumulative rates (C) for the different subgroups by adding age-specific absolute rates and finally estimated cumulative risk by age 80 years using the standard formula: 100×[1−exp (–5×C/105)]. Cumulative risk can be interpreted as the probability that an individual will develop UATC before the age of 80 years in the absence of competing causes of death. The age-specific incidence rate of UATC and population size in 2007 were published by the National Cancer Center, Japan (Matsuda et al., 2013).
Results
A total of 3198 participants were included in the analysis: 1890 (630 cases and 1260 controls) in the derivation study and 1308 (654 cases and 654 controls) in the validation study. Table 1 shows the distribution of cases and controls by background characteristics. Distribution of smoking status, alcohol consumption, and the ALDH2 genotype differed markedly between cases and controls in both studies. Genotype frequencies among controls did not deviate from the values predicted from the Hardy–Weinberg equilibrium. The participant characteristics by cancer site are presented in Supplementary Table 1 (Supplemental digital content 1, http://links.lww.com/EJCP/A46).
Table 1.
Participant characteristics
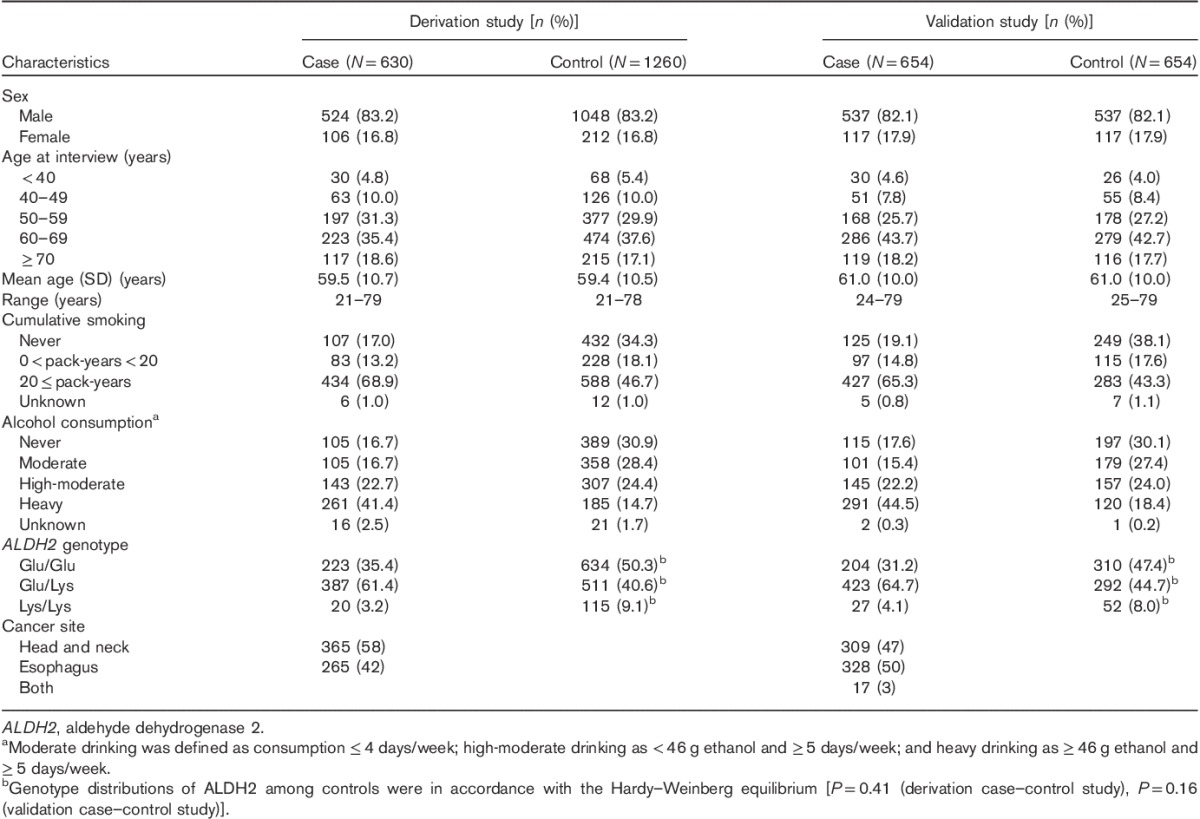
Table 2 shows the discriminative abilities of the three risk models in the derivation study and validation study. Fig. 1 shows ROC curves in the three risk models. The ROC curves by cancer site are shown in Supplementary Fig. 1 (Supplemental digital content 2, http://links.lww.com/EJCP/A47). As shown in Table 2 and Fig. 1, the discriminatory abilities of the three risk models in the validation study were similar to those in the derivation study. In both the derivation and the validation studies, the inclusive model provided excellent discrimination of UATC and esophageal cancer and acceptable discrimination in head and neck cancer (Hosmer and Lemeshow, 2000), with AUC values around 0.8 in UATC, 0.7 in head and neck cancer and 0.9 in esophageal cancer. In addition, AUC values of the environmental and inclusive models in the two studies were significantly different.
Table 2.
C statistic (95% confidence interval) of each risk model in derivation and validation studies
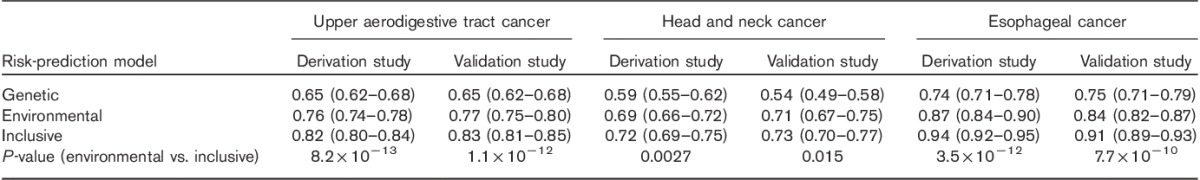
Fig. 1.
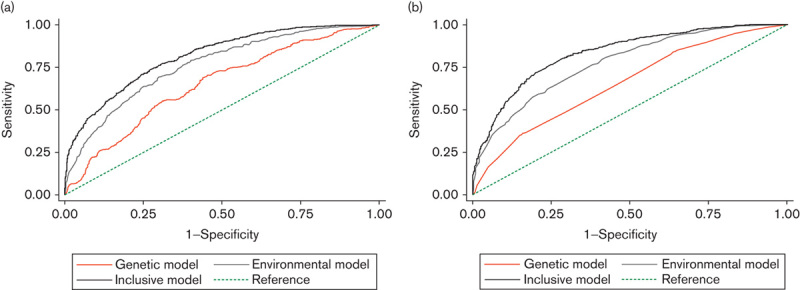
Receiver operating characteristic curves in the three risk models for upper aerodigestive tract cancer in the derivation study (a) and the validation study (b). The straight dashed line with an area under the curve of 50% is the reference. The upper black curved line represents the inclusive risk model, the gray line represents the environmental model, and the orange line represents the genetic model.
As shown in Supplementary Table 2 (Supplemental digital content 3, http://links.lww.com/EJCP/A48) no Hosmer–Lemeshow tests of the inclusive model were statistically significant, except two for data sets of UATC and esophageal cancer in the validation study (P=0.005 and P<0.001, respectively). The calibration plots of the inclusive model remained close to the ideal calibration line throughout the risk spectrum in all data sets of both studies (Fig. 2 and Supplementary Fig. 2, Supplemental digital content 4, http://links.lww.com/EJCP/A49), and all of their calibration slopes were close to 1.0 (Supplementary Table 2, Supplemental digital content 3, http://links.lww.com/EJCP/A48).
Fig. 2.
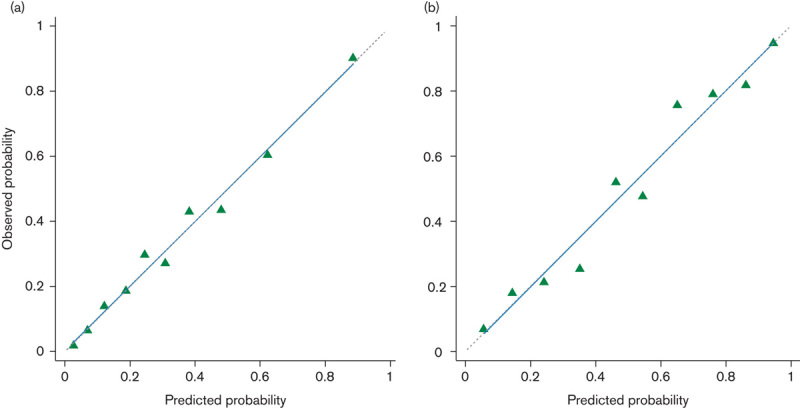
Calibration plots for the inclusive model for upper aerodigestive tract cancer. Predicted and observed probabilities within deciles of predicted probability in the derivation study (a) and the validation study (b). The straight dashed line (45° line) represents the ideal calibration line and the solid blue line shows the linear calibration line for the risk model.
The ORs for each study are presented in Table 3. In both studies, the highest OR was observed in heavy drinkers with the Glu/Lys genotype and P values for interaction were significant. Table 4 shows the OR and cumulative risk by age 80 for a combination of the ALDH2 genotype and alcohol consumption in combined data sets of derivation and validation studies. The cumulative risk for heavy drinkers with the Glu/Lys genotype by age 80 was very high: risks for UATC, head and neck cancer, and esophageal cancer were 20.2, 6.1, and 15.9%, respectively, versus respective values for the other subgroups of <4.6, <2.0, and <4.1%, respectively. Figure 3 shows the marked increase in the cumulative risk for heavy drinkers with the Glu/Lys genotype in comparison with the other subgroups, particularly after the age of around 50 (the graph of cumulative risk by cancer site; Supplementary Fig. 3, Supplemental digital content 5, http://links.lww.com/EJCP/A50).
Table 3.
Odds ratios for a combination of the ALDH2 genotype and alcohol consumption
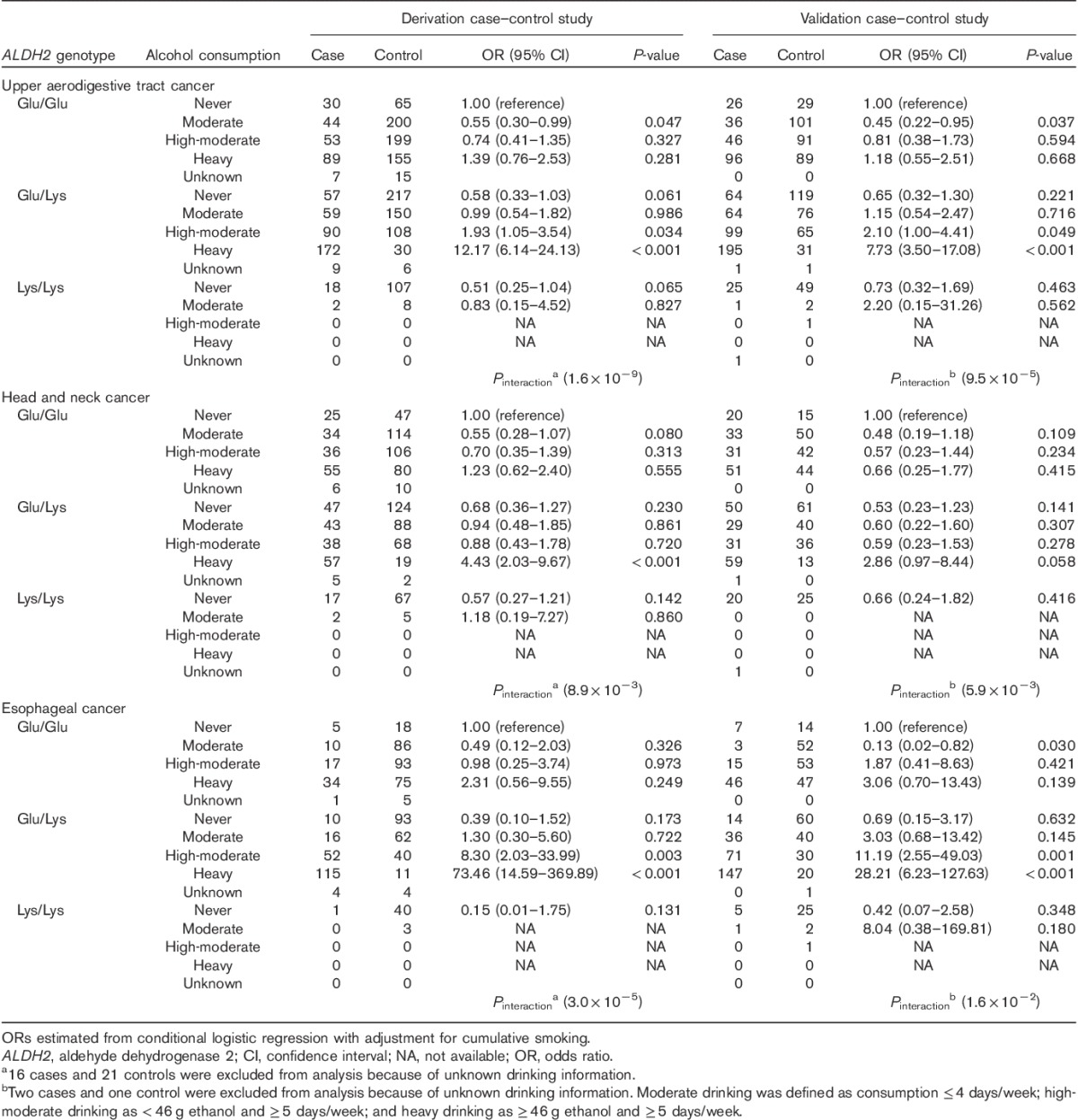
Table 4.
Odds ratios and cumulative risks by the age of 80 years for a combination of ALDH2 genotype and alcohol consumption by combined data sets of derivation and validation studies
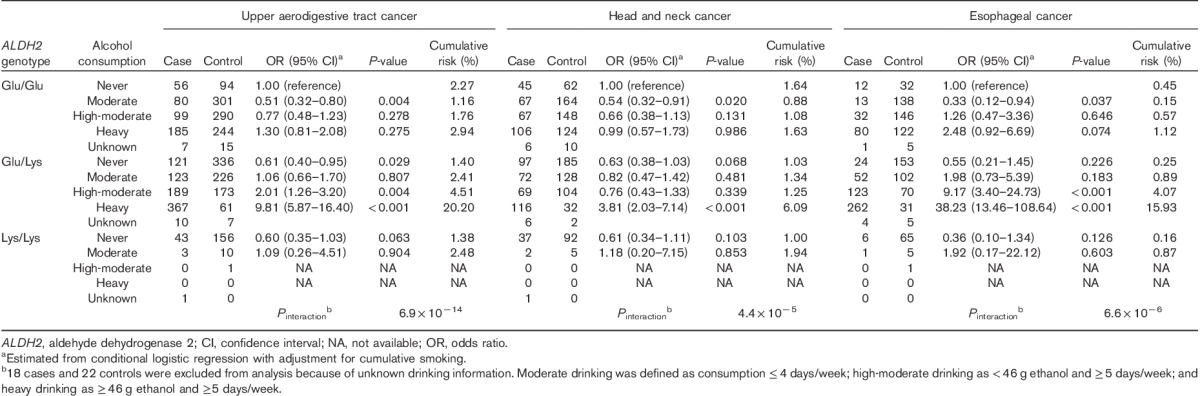
Fig. 3.
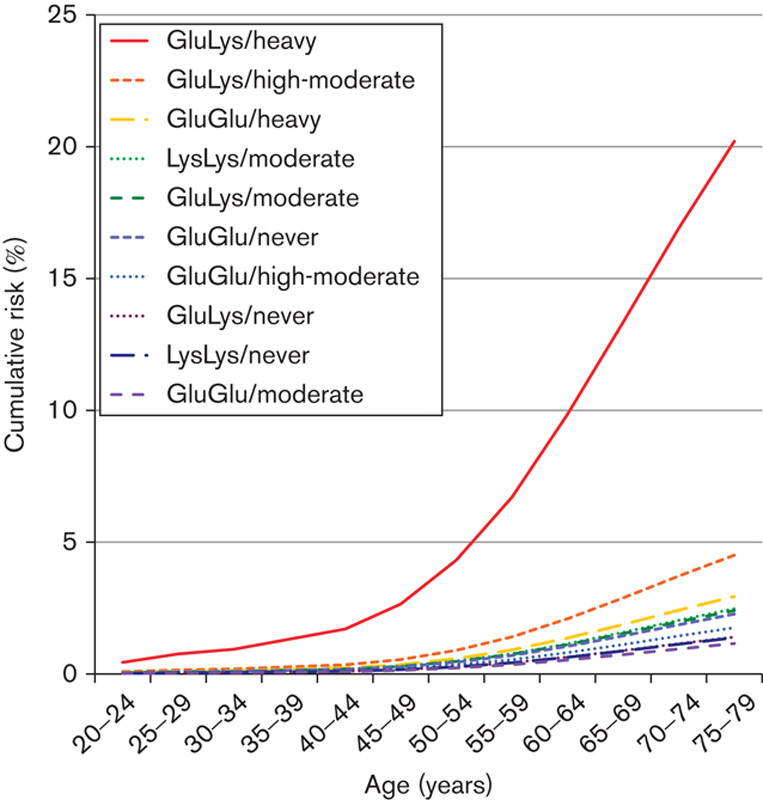
Cumulative risk (%) for upper aerodigestive tract cancer for each risk group (ALDH2 genotype/alcohol consumption) at various ages up to the age of 80 years, estimated using the age-specific incidence rate and population size in 2007. ALDH2, aldehyde dehydrogenase 2.
Discussion
In the derivation case–control study (HERPACC-2), we developed genetic, environmental, and inclusive risk-prediction models using the established risk predictors of age, sex, smoking, alcohol consumption, and the ALDH2 genotype. Compared with the other models, the inclusive model, with a combination of the ALDH2 genotype and alcohol consumption, showed excellent discriminatory ability and good calibration. This was confirmed by external validation in the other participant data set (HERPACC-3). Further, we estimated cumulative risk by means of ORs calculated on the basis of the combined data of the two studies. In the analysis, we found that heavy drinkers with the ALDH2 Glu/Lys genotype had a very high cumulative risk by the age of 80 years at 20.2% for UATC, 6.1% for head and neck cancer, and 15.9% for esophageal cancer. These results indicate that the inclusive model can stratify individuals into different risk categories accurately. We speculate that the presentation of cumulative risk by these risk categories might be highly persuasive in inducing a reduction in alcohol intake.
Animal studies suggest that circulating ethanol-derived acetaldehyde causes esophageal DNA damage and that the extent of damage is influenced by ALDH2 gene impairment (Yukawa et al., 2014). Mizoi et al. (1994) reported that individuals with the Lys/Lys genotype showed markedly higher acetaldehyde levels after ethanol intake than those with the Glu/Lys genotype, who in turn showed about six times the level of those with the Glu/Glu genotype. Interestingly, however, individuals with the Lys/Lys genotype had a relatively low risk of UATC because the Lys/Lys genotype is strongly associated with nondrinking (Matsuo et al., 2006). Any consideration of the carcinogenic impact of the Lys allele on UATC prevention should take alcohol consumption into account.
We assessed the effectiveness of the inclusive model with respect to the interaction between ALDH2 genotype and alcohol consumption using the AUC value, the Hosmer–Lemeshow test, and calibration plots and slope. The significant result of the Hosmer–Lemeshow test in the UATC and esophageal cancer data sets in the validation study was considered to be because of specific disadvantages of the Hosmer–Lemeshow test. As often noted (Steyerberg, 2009; Abbasi et al., 2012; Allison, 2013), the Hosmer–Lemeshow test is strongly influenced by sample size and number of groups and should therefore be interpreted with caution. For example, when we randomly sampled 50% of participants in the UATC data set of the validation study and ran the same test, the P-value was not significant. Judging from its excellent discriminatory abilities and good calibration on the basis of graphical inspection, we conclude that the inclusive model performs well in identifying individuals at very high risk of future UATC.
Some differences were observed between cancers of the head and neck and cancers of the esophagus. Consistent with previous studies (Oze et al., 2010; Anantharaman et al., 2011), alcohol-associated risk was not as strong for head and neck cancer as it was for esophageal cancer. In fact, multicenter case–control studies in Western populations showed that alcohol drinking in the absence of smoking conferred a relatively small (compared with smoking alone) or no apparent risk of head and neck cancer (Hashibe et al., 2009; Anantharaman et al., 2011). In contrast to studies in Western countries, where almost all populations had the Glu/Glu genotype, studies in Asian populations can easily evaluate the combined effect of the functional ALDH2 genotype and alcohol drinking and investigate details of the alcohol-associated risk of UATC in consideration of genetic factors. Even though the combined effect of the ALDH2 genotype and alcohol drinking is relatively low in head and neck cancer (Tables 2–4, Supplementary Figs 1–3, Supplemental digital content 2, http://links.lww.com/EJCP/A47, Supplemental digital content 4, http://links.lww.com/EJCP/A49, Supplemental digital content 5, http://links.lww.com/EJCP/A50), this study showed that the risk was nevertheless highest for heavy drinkers with the Glu/Lys genotype. In addition, among moderate drinkers with the Glu/Glu genotype, ORs of less than one were observed in both head and neck and esophageal cancers. This inverse association should be interpreted carefully and clarified in a larger study.
To our knowledge, the cumulative risk of UATC by subgroup of alcohol consumption or ALDH2 genotype has not been investigated. Given previous risk communication findings that absolute risk formats promoted better patient understanding of probabilistic information than relative risk formats (Zipkin et al., 2014), presentation of cumulative risk in place of relative risk information for each risk group is more suitable in prevention settings and will motivate individuals to reduce their alcohol intake. In addition, our evaluation of cumulative risk highlighted the very high risk for heavy drinkers with the Glu/Lys genotype. Public health efforts should target such heavy drinkers with the Glu/Lys genotype, with prevention efforts aimed at reducing their alcohol exposure. Further, frequent screening of these individuals will considerably enable the early diagnosis of UATC.
Our study has several methodological strengths and limitations, which are described in the Appendix.
Conclusion
Our study showed that a risk model developed using established predictors, including a combination of the ALDH2 genotype and alcohol drinking, had high discriminatory accuracy and good calibration. This model might be a promising way of stratifying individuals into different risk groups. On the basis of their surprisingly high cumulative risk, heavy drinkers with the ALDH2 Glu/Lys genotype should be targeted for prevention efforts aimed at reducing alcohol consumption.
Supplementary Material
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (www.eurjcancerprev.com).
Acknowledgements
The authors thank Dr Carlo La Vecchia for his constructive comments on this manuscript.
This study was supported by a National Cancer Center Research and Development Fund (25-A-14 and 27-A-XX), a grant-in-aid for Scientific Research (grant number: 26253041) from the Ministry of Education, Culture, Sports, Science and Technology, JST (from Dr Ito), and a grant-in-aid for the Third-Term Comprehensive Ten-Year Strategy for Cancer Control from the Ministry of Health, Labor and Welfare of Japan.
Conflicts of interest
There are no conflicts of interest.
Appendix
Brief description of the Hospital-based Epidemiologic Research Program at the Aichi Cancer Center
Participants in the derivation study were recruited within the framework of the Hospital-based Epidemiologic Research Program at Aichi Cancer Center (HERPACC)-2. Details of the program are provided elsewhere (Hamajima et al., 2001). Briefly, 23 408 first-visit outpatients between January 2001 and November 2005 were asked to provide blood, in addition to information on lifestyle factors. Among the participants, 22 727 (97.1%) completed the questionnaire satisfactorily and were enrolled in HERPACC. Each patient was asked about his or her lifestyle when healthy and before his or her current symptoms developed. We previously showed that the general lifestyle of cancer-free outpatients was in accord with that of a general population selected randomly from the electoral roll of Nagoya city, confirming the feasibility of their inclusion as controls in epidemiological studies (Inoue et al., 1997).
Estimation of cumulative risk
Cumulative risk was calculated using methods similar to those described elsewhere (Peto et al., 2000; Brennan et al., 2006; Bosetti et al., 2008). The following steps summarize the main measures used to obtain the cumulative risk.
ri is relative risk for the ith subgroup of the ALDH2 genotype stratified by alcohol consumption (step 1),
where the subgroups are 1=Glu/Glu and never drinker; 2=Glu/Glu and moderate drinker; 3=Glu/Glu and high-moderate drinker; 4=Glu/Glu and heavy drinker; 5=Glu/Lys and never drinker; 6=Glu/Lys and moderate drinker; 7=Glu/Lys and high-moderate drinker; 8=Glu/Lys and heavy drinker; 9=Lys/Lys and never drinker; 10=Lys/Lys and moderate drinker; 11=Lys/Lys and high-moderate drinker; and 12=Lys/Lys and heavy drinker.
pi is the risk group prevalence of controls in the ith subgroup.
pj is the proportion of population size in the jth age group.
where age categories are as follows: 20–24, 25–29, 30–34, 35–39, 40–44, 45–49, 50–54, 55–59, 60–64, 65–69, 70–74, and 75–79 years.
hj is the age-specific incidence rate (step 4).
aij is the absolute rate in the (i,j)th cell, and aij=(fj×ri) (step 6).
Ci is the cumulative rate, and Ci=ΣjRjaij (step 7).
where Rj is the width of the jth age category in years.
Briefly, to estimate the cumulative risk, we first need to calculate the relative risks in the subgroups of a combination of the ALDH2 genotype and alcohol consumption (step 1). In our study, relative risks of upper aerodigestive tract cancer were estimated by means of the odds ratios using conditional logistic regression, with the Glu/Glu genotype and never drinkers forming the reference subgroup.
The next step was to calculate the proportion of controls in each subgroup and for each age group (step 2) by multiplying the risk group prevalence of controls and the age-specific population size of 2007 under the assumption that drinking distribution stratified by ALDH2 genotype of the population was represented by that observed among study controls.
The third step was to estimate common factors combining the relative risk (step 1) for the different subgroups with the age-specific prevalence of the subgroups among study controls (step 2), thus obtaining the quantities denoted (step 3).
By combining the age-specific cancer incidence rates (step 4) with the common factors (step 3), we obtained the proportions given (step 5). Multiplying these proportions by the relative risks for the different subgroups produced the age-specific absolute risks in the different subgroups (step 6).
Next, we calculated the cumulative rates (step 7) for the different subgroups by adding age-specific absolute rates, and then finally estimated the cumulative risks by the age of 80 years using the standard formula (step 8). Cumulative risk may be interpreted as the probability that an individual will develop upper aerodigestive tract cancer before the age of 80 years in the absence of competing causes of death.
Strength and limitations of the present study
Our study has several methodological strengths. First, it was carried out within the framework of the HERPACC study, which has enrolled a very large number of patients with 95% response rates to the completion of questionnaires. Second, potential confounding by age and sex was considered by matching. Third, given that our allele frequencies were comparable with those reported previously in public databases, such as HapMap JPT (http://www.ncbi.nlm.nih.gov/snp), bias in the distribution of the selected polymorphism was likely negligible. Fourth, our study was consistent with a previous study carried out in Japanese men by Yokoyama et al. (2008). They used a similar approach to develop a risk model for esophageal cancer at screening in a clinical setting, on the basis of alcohol drinking, ALDH2 genotype, smoking, and intake of vegetables and fruit. The AUC value of their model was 0.86, whereas the AUC value of our model on the basis of age, sex, alcohol drinking, smoking, and ALDH2 genotype was 0.9.
Several potential limitations also warrant mention. First, values collected with a self-administered questionnaire and considered potential confounding factors might have been inaccurate. Second, the cumulative risks do not reflect age-specific alcohol consumption. As the present study was an age-matched case–control study, age-specific alcohol consumption in the controls could not be taken into account. Third, the sample size in the present study was modest, particularly when stratified by risk. This explains why we did not increase the number of risk groups by adding cumulative smoking categories. More personalized evaluation that includes other predictors would require a larger sample size. Fourth, external validation was performed using a data set collected within the same framework as the derivation study. External validation by other investigators or multi-site testing would provide more convincing validity of the model.
References
- Abbasi A, Peelen LM, Corpeleijn E, van der Schouw YT, Stolk RP, Spijkerman AM, et al. (2012). Prediction models for risk of developing type 2 diabetes: systematic literature search and independent external validation study. BMJ 345:e5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison P. (2013). Why I don’t trust the Hosmer–Lemeshow test for logistic regression. Statistical horizons. Available at: http://www.statisticalhorizons.com/hosmer-lemeshow. [Accessed 15 October 2015].
- Anantharaman D, Marron M, Lagiou P, Samoli E, Ahrens W, Pohlabeln H, et al. (2011). Population attributable risk of tobacco and alcohol for upper aerodigestive tract cancer. Oral Oncol 47:725–731. [DOI] [PubMed] [Google Scholar]
- Asakage T, Yokoyama A, Haneda T, Yamazaki M, Muto M, Yokoyama T, et al. (2007). Genetic polymorphisms of alcohol and aldehyde dehydrogenases, and drinking, smoking and diet in Japanese men with oral and pharyngeal squamous cell carcinoma. Carcinogenesis 28:865–874. [DOI] [PubMed] [Google Scholar]
- Boccia S, Hashibe M, Gallì P, De Feo E, Asakage T, Hashimoto T, et al. (2009). Aldehyde dehydrogenase 2 and head and neck cancer: a meta-analysis implementing a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev 18:248–254. [DOI] [PubMed] [Google Scholar]
- Boffetta P, Hashibe M. (2006). Alcohol and cancer. Lancet Oncol 7:149–156. [DOI] [PubMed] [Google Scholar]
- Bosetti C, Gallus S, Peto R, Negri E, Talamini R, Tavani A, et al. (2008). Tobacco smoking, smoking cessation, and cumulative risk of upper aerodigestive tract cancers. Am J Epidemiol 167:468–473. [DOI] [PubMed] [Google Scholar]
- Bosron WF, Li TK. (1986). Genetic polymorphism of human liver alcohol and aldehyde dehydrogenases, and their relationship to alcohol metabolism and alcoholism. Hepatology 6:502–510. [DOI] [PubMed] [Google Scholar]
- Brennan P, Crispo A, Zaridze D, Szeszenia-Dabrowska N, Rudnai P, Lissowska J, et al. (2006). High cumulative risk of lung cancer death among smokers and nonsmokers in Central and Eastern Europe. Am J Epidemiol 164:1233–1241. [DOI] [PubMed] [Google Scholar]
- Cogliano VJ, Baan R, Straif K, Grosse Y, Lauby-Secretan B, El Ghissassi F, et al. (2011). Preventable exposures associated with human cancers. J Natl Cancer Inst 103:1827–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins GS, Altman DG. (2013). Identifying patients with undetected gastro-oesophageal cancer in primary care: external validation of QCancer® (Gastro-Oesophageal). Eur J Cancer 49:1040–1048. [DOI] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL. (1988). Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845. [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. (2013). GLOBOCAN 2012 v1.0, cancer incidence and mortality worldwide: IARC CancerBase No. 11 International Agency for Research on Cancer, Lyon, France. Available at: http://globocan.iarc.fr/Default.aspx. [Accessed 15 October 2015].
- Hamajima N, Matsuo K, Saito T, Hirose K, Inoue M, Takezaki T, et al. (2001). Gene-environment interactions and polymorphism studies of cancer risk in the Hospital-based Epidemiologic Research Program at Aichi Cancer Center II (HERPACC-II). Asian Pac J Cancer Prev 2:99–107. [PubMed] [Google Scholar]
- Hashibe M, Brennan P, Chuang SC, Boccia S, Castellsague X, Chen C, et al. (2009). Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev 18:541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraki A, Matsuo K, Wakai K, Suzuki T, Hasegawa Y, Tajima K. (2007). Gene-gene and gene-environment interactions between alcohol drinking habit and polymorphisms in alcohol-metabolizing enzyme genes and the risk of head and neck cancer in Japan. Cancer Sci 98:1087–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmer DW, Lemeshow S. (2000). Appliedlogistic regression. New York: John Wiley & Sons, in; 156–164. [Google Scholar]
- Inoue M, Tajima K, Hirose K, Hamajima N, Takezaki T, Kuroishi T, Tominaga S. (1997). Epidemiological features of first-visit outpatients in Japan: comparison with general population and variation by sex, age, and season. J Clin Epidemiol 50:69–77. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang D, Jin W, Shao C, Yan P, Xu C, et al. (2006). Mitochondrial aldehyde dehydrogenase-2 (ALDH2) Glu504Lys polymorphism contributes to the variation in efficacy of sublingual nitroglycerin. J Clin Invest 116:506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Ajiki W, Marugame T, Ioka A, Tsukuma H, Sobue T, Research Group of Population-Based Cancer Registries of Japan (2011). Population-based survival of cancer patients diagnosed between 1993 and 1999 in Japan: a chronological and international comparative study. Jpn J Clin Oncol 41:40–51. [DOI] [PubMed] [Google Scholar]
- Matsuda A, Matsuda T, Shibata A, Katanoda K, Sobue T, Nishimoto H, Japan Cancer Surveillance Research Group (2013). Cancer incidence and incidence rates in Japan in 2007: a study of 21 population-based cancer registries for the Monitoring of Cancer Incidence in Japan (MCIJ) project. Jpn J Clin Oncol 43:328–336. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Hamajima N, Shinoda M, Hatooka S, Inoue M, Takezaki T, Tajima K. (2001). Gene-environment interaction between an aldehyde dehydrogenase-2 (ALDH2) polymorphism and alcohol consumption for the risk of esophageal cancer. Carcinogenesis 22:913–916. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Wakai K, Hirose K, Ito H, Saito T, Tajima K. (2006). Alcohol dehydrogenase 2 His47Arg polymorphism influences drinking habit independently of aldehyde dehydrogenase 2 Glu487Lys polymorphism: analysis of 2,299 Japanese subjects. Cancer Epidemiol Biomarkers Prev 15:1009–1013. [DOI] [PubMed] [Google Scholar]
- Mizoi Y, Yamamoto K, Ueno Y, Fukunaga T, Harada S. (1994). Involvement of genetic polymorphism of alcohol and aldehyde dehydrogenases in individual variation of alcohol metabolism. Alcohol Alcohol 29:707–710. [PubMed] [Google Scholar]
- Oze I, Matsuo K, Hosono S, Ito H, Kawase T, Watanabe M, et al. (2010). Comparison between self-reported facial flushing after alcohol consumption and ALDH2 Glu504Lys polymorphism for risk of upper aerodigestive tract cancer in a Japanese population. Cancer Sci 101:1875–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto R, Darby S, Deo H, Silcocks P, Whitley E, Doll R. (2000). Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case–control studies. BMJ 321:323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyerberg EW. (2009). Clinical prediction models. New York: Springer; 270–279. [Google Scholar]
- Thrift AP, Kendall BJ, Pandeya N, Whiteman DC. (2013). A model to determine absolute risk for esophageal adenocarcinoma. Clin Gastroenterol Hepatol 11:138–144.e2. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2014). Global status report on alcohol and health 2014. Geneva: WHO.
- Yokoyama A, Muramatsu T, Ohmori T, Yokoyama T, Okuyama K, Takahashi H, et al. (1998). Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis 19:1383–1387. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Kato H, Yokoyama T, Igaki H, Tsujinaka T, Muto M, et al. (2006). Esophageal squamous cell carcinoma and aldehyde dehydrogenase-2 genotypes in Japanese females. Alcohol Clin Exp Res 30:491–500. [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Yokoyama A, Kumagai Y, Omori T, Kato H, Igaki H, et al. (2008). Health risk appraisal models for mass screening of esophageal cancer in Japanese men. Cancer Epidemiol Biomarkers Prev 17:2846–2854. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Huang IY, Ikawa M. (1984). Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci USA 81:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa Y, Ohashi S, Amanuma Y, Nakai Y, Tsurumaki M, Kikuchi O, et al. (2014). Impairment of aldehyde dehydrogenase 2 increases accumulation of acetaldehyde-derived DNA damage in the esophagus after ethanol ingestion. Am J Cancer Res 4:279–284. [PMC free article] [PubMed] [Google Scholar]
- Zipkin DA, Umscheid CA, Keating NL, Allen E, Aung K, Beyth R, et al. (2014). Evidence-based risk communication: a systematic review. Ann Intern Med 161:270–280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (www.eurjcancerprev.com).


