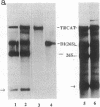
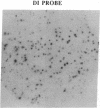
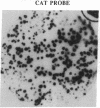
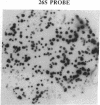
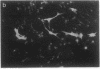
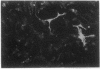
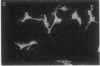
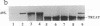Abstract
Sindbis virus, the type member of the alpha-viruses, is an enveloped virus containing a nonsegmented positive-strand RNA genome. We show that the nonstructural and the structural genes can function to produce infectious virus particles when they are expressed on two different RNA segments. The nonstructural genes are translated from an RNA in which the structural genes have been replaced by the chloramphenicol acetyltransferase gene [Xiong, C., Levis, R., Shen, P., Schlesinger, S., Rice, C. M. & Huang, H. V. (1989) Science 243, 1188-1191]. The structural genes are encoded in a defective-interfering RNA but are translated from a subgenomic RNA. Both segments contain the cis-acting sequences required for replication and packaging and are copackaged. This type of genome provides a model for an ancestral intermediate between alphaviruses and the multipartite positive-strand RNA viruses of plants. These different viruses show sequence similarities in their replicative proteins and are thought to have evolved from a common ancestor.
Full text
PDF

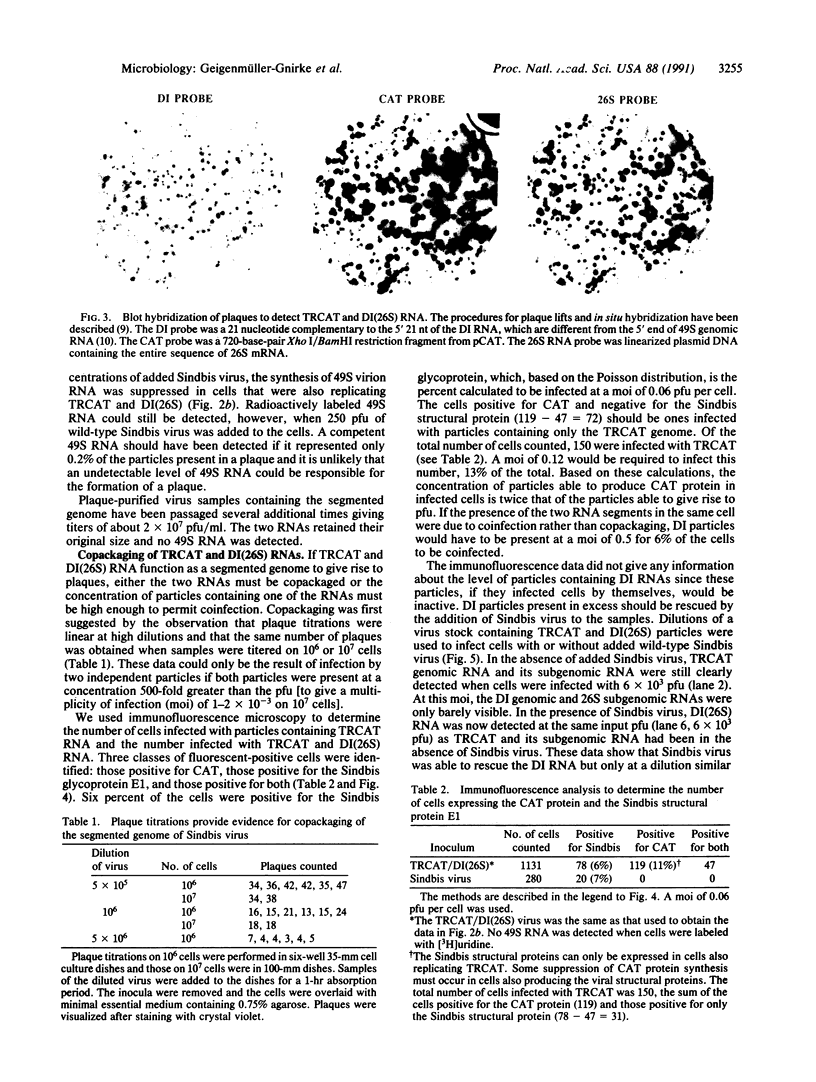
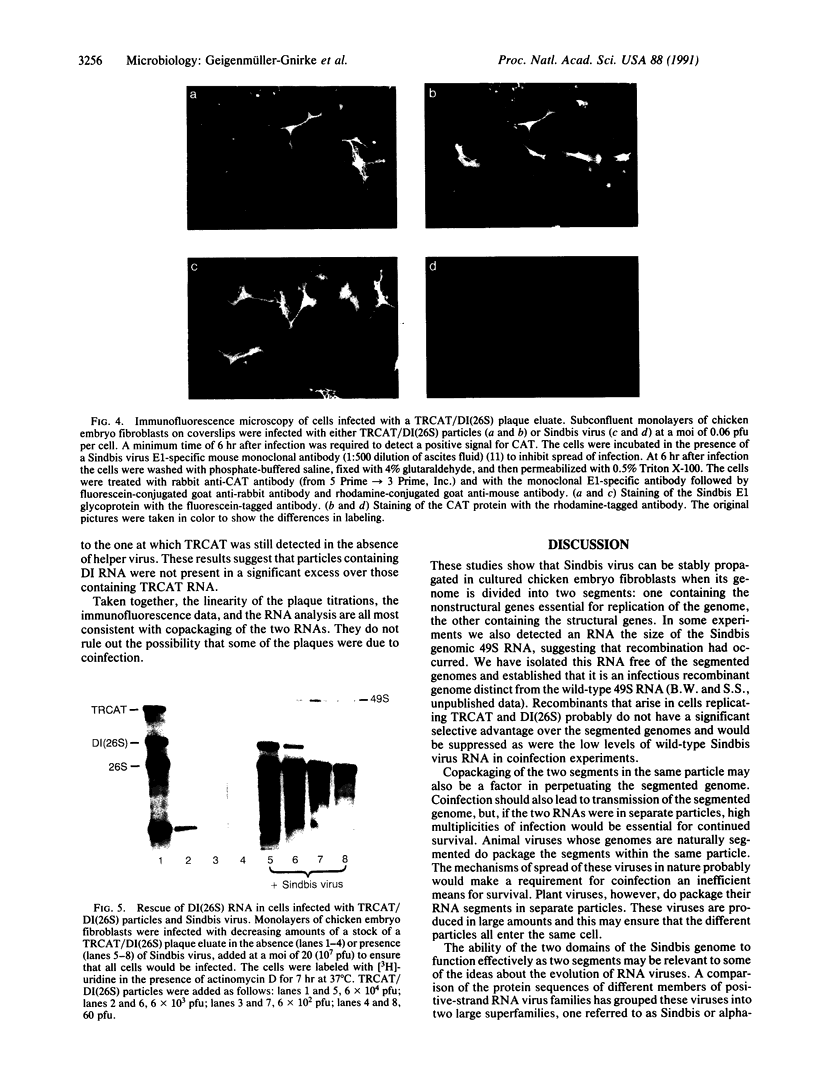

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., Strauss E. G., Rice C. M., Strauss J. H., Haseloff J., Zimmern D. Sindbis virus proteins nsP1 and nsP2 contain homology to nonstructural proteins from several RNA plant viruses. J Virol. 1985 Feb;53(2):536–542. doi: 10.1128/jvi.53.2.536-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner P. L., Gadek T. R., Holm M., Roman R., Chan H. W., Wenz M., Northrop J. P., Ringold G. M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbach R. Genome similarities between plant and animal RNA viruses. Microbiol Sci. 1987 Jul;4(7):197–202. [PubMed] [Google Scholar]
- Haseloff J., Goelet P., Zimmern D., Ahlquist P., Dasgupta R., Kaesberg P. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4358–4362. doi: 10.1073/pnas.81.14.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Huang H., Schlesinger S. Engineered defective interfering RNAs of Sindbis virus express bacterial chloramphenicol acetyltransferase in avian cells. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4811–4815. doi: 10.1073/pnas.84.14.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Schlesinger S., Huang H. V. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J Virol. 1990 Apr;64(4):1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Weiss B. G., Tsiang M., Huang H., Schlesinger S. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell. 1986 Jan 17;44(1):137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- Li G. P., Rice C. M. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J Virol. 1989 Mar;63(3):1326–1337. doi: 10.1128/jvi.63.3.1326-1337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe S. S., Schlesinger S. RNAs from two independently isolated defective interfering particles of Sindbis virus contain a cellular tRNA sequence at their 5' ends. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3279–3283. doi: 10.1073/pnas.80.11.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaljohn A. L., Kokubun K. M., Cole G. A. Protective monoclonal antibodies define maturational and pH-dependent antigenic changes in Sindbis virus E1 glycoprotein. Virology. 1983 Oct 15;130(1):144–154. doi: 10.1016/0042-6822(83)90124-1. [DOI] [PubMed] [Google Scholar]
- Strauss J. H., Strauss E. G. Evolution of RNA viruses. Annu Rev Microbiol. 1988;42:657–683. doi: 10.1146/annurev.mi.42.100188.003301. [DOI] [PubMed] [Google Scholar]
- Weiss B., Nitschko H., Ghattas I., Wright R., Schlesinger S. Evidence for specificity in the encapsidation of Sindbis virus RNAs. J Virol. 1989 Dec;63(12):5310–5318. doi: 10.1128/jvi.63.12.5310-5318.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C., Levis R., Shen P., Schlesinger S., Rice C. M., Huang H. V. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science. 1989 Mar 3;243(4895):1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]