Abstract
Candidemia is the fourth most common cause of bloodstream infection, with a high mortality rate of up to 40%. Identification of host genetic factors that confer susceptibility to candidemia may aid in designing adjunctive immunotherapeutic strategies. We hypothesized that variation in immune genes may predispose to candidemia. We analyzed 118,989 SNPs across 186 loci known to be associated with immune-mediated diseases in the largest candidemia cohort to date of 217 patients of European ancestry and a group of 11,920 controls. The significant associations were validated by comparison with a disease-matched control group. We observed significant association between candidemia and SNPs in the CD58 (P = 1.97×10−11; OR = 4.68), LCE4A-C1orf68 (P = 1.98×10−10; OR = 4.25) and TAGAP (P = 1.84×10−8; OR = 2.96) loci. Individuals carrying two or more risk alleles had an increased risk for candidemia of 19.4-fold compared to individuals carrying no risk allele. While latent cornified envelope (LCE) genes contribute to mucosal integrity, the role of CD58 and TAGAP in host defense is unknown. Studies using transcriptomics, pathway analysis, and immunological validation showed that CD58 plays a role in the recognition and phagocytosis of Candida by macrophages, while TAGAP was involved in Candida-induced cytokine production. TAGAP-deficient mice were more susceptible to systemic Candida infection. We identified three novel genetic risk factors for candidemia, which we subsequently validated for their role in antifungal host defense.
Introduction
Candidemia is the fourth most common bloodstream infection1, with known risk factors such as neutropenia, mucosal barrier injury, transplantation, immunosuppressive drugs, intravascular catheters, and extended intensive care unit (ICU) stay2, 3, 4. Despite the availability of potent antifungal drugs, the mortality of patients with candidemia remains high (up to 37–44%)5, 6. It has therefore been proposed that only intensified patient care, using risk assessment and adjuvant immunotherapy, may improve the outcome7.
The host immune status is crucial for the outcome of Candida infections, and identifying genetic variation in immune genes that confer susceptibility to Candida infection may aid in designing effective preventive strategies. Several small-scale candidate gene association studies suggest a role in candidemia risk for single nucleotide polymorphisms (SNPs) in Toll-like receptors (TLR-1, TLR-2, TLR-3, and TLR-4), interleukins (IL-12B and IL-10), and lymphoid protein tyrosine phosphatase PTPN223. Interestingly, approximately ten monogenetic disorders have been reported to be associated with chronic mucocutaneous candidiasis and almost all are caused by defects in genes of the immune system3. Intriguingly, common SNPs in eight out of these ten monogenetic disorder genes (AIRE, CARD9, STAT1, STAT3, TYK2, CD25, IL17RA, IL17F) are also associated with susceptibility to different immune-mediated diseases (NHGRI GWAS catalog8), implying that genes necessary for immune regulation are strong candidates in determining susceptibility to fungal infections.
Here we report the first genome-wide screen of around 200,000 SNPs in 186 loci in the largest candidemia cohort to date. By using the Immunochip SNP array9, we identified three novel genetic risk factors for candidemia that were validated using transcriptomics, pathway analysis, and immunological studies.
Results
Immunochip-wide association analysis identified three candidemia susceptibility loci
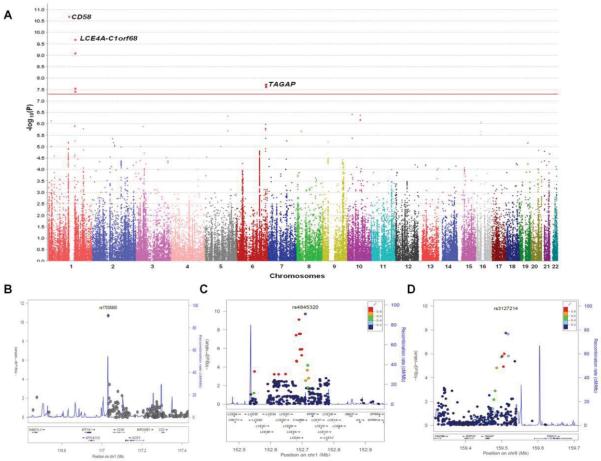
After filtering the immunochip data using standard quality parameters, we obtained 118,989 SNPs from the 217 candidemia cases and 11,920 healthy controls, which we analyzed using logistic regression analysis. The results of this analysis revealed significant association (P < 5 × 10−8) to three independent loci with candidemia (Fig. 1A and Fig. S9). The top SNPs from these three loci were rare in healthy controls, with risk allele frequencies below 2%, whereas the risk allele frequencies were above 5% in candidemia cases (Table 1). The top-associated SNP, rs17035850 (P=1.97×10−11; odds ratio = 4.68) is located in a block of linkage disequilibrium (LD) of around 30 kb at 1p13.1 which contains the CD58 gene (Fig. 1B and Fig. S10), two long non-coding RNAs (lncRNA), RP5-1086K13.1 and RP4-655J12.4, and a pseudogene NAP1L4P1 (Fig. S7). The second hit was with rs4845320 (P=1.98×10−10; odds ratio = 4.25), which lies in an LD block of 150 kb at 1q21.3 (Fig. 1C and Fig. S11) that contains a cluster of genes encoding late cornified envelope (LCE) proteins10. The third significantly associated SNP rs3127214 (P=1.84×10−8; odds ratio = 2.96) is located at the 5 prime end of TAGAP (Fig. 1D and Fig. S12), encoding T-cell activation RhoGTPase activating protein11, in an LD block of 120 kb at 6q25.3.
Fig. 1.
(A) Manhattan plot showing the genome-wide p values of association with candidemia. The y axis represents the −log10 p values of 118,989 SNPs and their chromosomal positions are shown on the x axis. The horizontal red line shows the genome-wide significant threshold of p < 5×10−8. Regional association plots at (B) 1p13.1, (C) 1q21.3, and (D) 6q25.3. The p values of genotyped SNPs are plotted as −log10 values against their physical chromosomal positions (hg19). Estimated recombination rates from the 1000Genomes European population show the local LD structure. Inset, the colors of the SNPs indicate LD with the top-associated SNP according to a scale from r2 = 0 to r2 = 1 based on pairwise r2 values from the 1000Genomes European population. Lower panel, gene annotations from the University of California Santa Cruz genome browser (hg19).
Table 1.
Significantly associated loci with candidemia that were validated using disease-matched controls
| Case | Controls | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs ID | Chr | Cohorts | AA | AB | BB | MAF | AA | AB | BB | MAF | P* | OR (95% CI) | Gene |
| rs17035850 | 1 | Cases Vs population-based controls | 3 | 16 | 198 | 0.051 | 0 | 277 | 11642 | 0.012 | 1.97 × 10−11 | 4.68 (2.98–7.35) | CD58 |
| Cases Vs disease-matched controls | 3 | 16 | 198 | 0.051 | 0 | 8 | 138 | 0.027 | 0.15 | 1.58 (0.66–3.78) | |||
| rs12025416 | 1 | Cases Vs population-based controls | 11 | 57 | 147 | 0.184 | 179 | 2643 | 9097 | 0.126 | 0.00035 | 1.57 (1.22–2.01) | CD58 |
| Cases Vs disease-matched controls | 11 | 57 | 147 | 0.184 | 1 | 32 | 111 | 0.118 | 0.022 | 1.69 (1.08–2.56) | |||
| rs4845320 | 1 | Cases Vs population-based controls | 3 | 16 | 198 | 0.051 | 2 | 290 | 11627 | 0.012 | 1.98 × 10−10 | 4.25 (2.72–6.65) | LCE4A-C1orf68 |
| Cases Vs disease-matched controls | 3 | 16 | 198 | 0.051 | 0 | 3 | 143 | 0.010 | 0.008 | 4.09 (1.21–13.85) | |||
| rs3127214 | 6 | Cases Vs population-based controls | 3 | 23 | 191 | 0.067 | 11 | 522 | 11386 | 0.023 | 1.84 × 10−8 | 2.96 (2.03–4.33) | TAGAP |
| Cases Vs disease-matched controls | 3 | 23 | 191 | 0.067 | 0 | 8 | 137 | 0.027 | 0.026 | 2.51 (1.13–5.55) | |||
P values are derived from logistic regression analysis by including MDS components as covariates. Chr, chromosome; OR, odds ratio; CI, confidence interval.
Validation and replication of three associated SNPs
Since we used a large population-based control cohort for discovering candidemia susceptibility loci, we tested whether these associations could be confirmed using 146 disease-matched controls (Table 2). We observed a significant difference between cases and controls at two loci (LCE4A-C1orf68 and TAGAP), whereas at the CD58 locus we observed a trend of association (Table 1). This latter effect could be explained by the small number of controls and/or low frequency of the risk allele at rs17035850 (Table 1). The top CD58 SNP (rs17035850) is a rare variant with minor allele frequency (MAF) of 0.012 and the second CD58 SNP (rs12025416) is a frequent one with MAF of 0.13. We assessed the pair-wise LD between these two CD58 SNPs in our population-based controls. We observed low correlation (R2 = 0.09) whereas a high D prime (D'> 0.96) between these two SNPs indicating the existence of rare risk haplotype carrying the risk alleles of these two SNPs. Therefore, we also tested for association at the second CD58 top SNP, rs12025416, which is more frequent, and we observed a significant association with susceptibility to candidemia (P=0.022). Therefore, these results suggest that the observed associations are true genetic associations.
Table 2.
Clinical characteristics of the candidemia cohort.
| Variable | Controls | Patients | p-value |
|---|---|---|---|
| Mean age [SD] (years) | 60.2 [17.5] | 54.7 [20.2] | 0.01 |
|
| |||
| Male Gender (%) | 49.3 | 64.0 | 0.008 |
|
| |||
| Immunocompromised State (%) | 36.3 | 61.7 | <0.0001 |
|
| |||
| Hematopoietic stem cell transplantation (%) | 0 | 2.8 | 0.06 |
|
| |||
| Solid organ transplant (%) | 0.7 | 15.4 | <0.0001 |
|
| |||
| Active Malignancy* (%) | 21.9 | 35.0 | 0.008 |
| Solid Tumor | 12.3 | 26.0 | |
| Leukemia | 6.2 | 5.7 | |
| Lymphoma | 4.1 | 4.0 | |
|
| |||
| Chemotherapy within past 3 months (%) | 12.3 | 18.9 | 0.11 |
|
| |||
| Neutropenia (ANC <500 cells/mm3) (%) | 2.7 | 10.3 | 0.008 |
|
| |||
| HIV-infected (%) | 0 | 0 | - |
|
| |||
| Surgery within past 30 days (%) | 56.2 | 49.1 | 0.21 |
|
| |||
| Receipt of Total Parenteral Nutrition (%) | 3.4 | 21.5 | <0.0001 |
|
| |||
| Dialysis dependent (%) | 4.1 | 11.6 | 0.02 |
|
| |||
| Acute Renal Failure (%) | 15.8 | 31.6 | 0.001 |
|
| |||
| Liver Disease (%) | 2.8 | 19.4 | <0.0001 |
|
| |||
| Intensive Care Unit Admission within past 14 days | 34.2 | 54.4 | 0.0003 |
|
| |||
| Candida spp. ** (%) | 43 | ||
| albicans | 27 | ||
| glabrata | 16 | ||
| parapsilosis | 13 | ||
| tropicalis | 3 | ||
| krusei | 4 | ||
| other Candida spp. | |||
|
| |||
| Baseline Serum Creatinine, Mean [SD] (mg/dL) | 1.3 [1.0] | 1.6 [1.4] | 0.008 |
|
| |||
| Baseline WBC count, Mean [SD] (cells/mm3) | 10.6 [7.8] | 13.5 [13.7] | 0.02 |
subjects could have more than one
18 subjects had >1 species isolated.
A few patients had positive cultures with more than one Candida species, explaining the slightly higher sum of percentages than 100%.
To further evaluate these associations genetically, we carried out replication analysis by genotyping the four SNPs in two independent candidemia cohorts (one of African-American 75 patients and one of 27 European patients from Switzerland). Because of the small size of these additional cohorts, we found no successful replication in two independent cohorts, although we observed only a trend of association for rs4845320 SNP in the TAGAP locus in the African-American cohort (Table S1). Since the Swiss candidemia replication cohort included matched-controls, we performed a joint analysis of the discovery cohort with the matched-controls and the Swiss candidemia replication cohort. This analysis revealed an improved association for two loci: for rs12025416 in the CD58 locus (P=0.015), and rs4845320 in the LCE4A-C1orf68 locus (P=0.0036) with the same allelic direction and without any evidence for heterogeneity between the two cohorts (Table S2).
Functional annotation of Candidemia associated SNPs
In order to understand how the candidemia associated SNPs from these three loci, we intersected the three top SNPs as well as their proxies (R2 ≥ 0.8 and D prime = 1) with functional information from ENCODE using HaploReg tool12. We found seven, nineteen and five SNPs to be in high LD with rs17035850, rs4845320 and rs3127214 SNPs, respectively (Table S3). All SNPs that were in LD with the three candidemia top SNPs were located in the non-coding region and not within exons of protein coding genes. Some of these SNPs were overlapping with ENCODE characterized regulatory regions such as enhancers and/or DNAse hypersensitive sites suggesting that these SNPs may regulate gene expression. Next, we tested whether these SNPs affect expression of nearby genes using publicly available blood eQTL data from 5000 samples13. However, we found no significant eQTLs for these SNPs.
Interestingly, the LD block around the top-associated SNP, rs17035850 contains not only CD58 protein-coding gene, but also non-coding genes, namely RP5-1086K13.1, RP4-655J12.4, and a pseudogene NAP1L4P1 (Fig. S7). Since many lncRNAs are co-regulated with their protein-coding gene in cis, there is a possibility that RP5-1086K13.1 lncRNA is co-regulating CD58. To test this possibility, we extracted all co-regulated genes with RP5-1086K13.1 using GeneNetwork database. We found that RP5-1086K13.1 is significantly co-regulated with CD58 compared to any other genes in the human genome (Table S4). This observation suggest the possibility that even if the SNP affect the expression of RP5-1086K13.1 that may in turn affect the CD58 expression levels.
Association of CD58 and TAGAP SNPs with severity of disease
In addition to increasing susceptibility to candidemia, these polymorphisms identified above may also influence the severity of the disease. Indeed, assessment of the effect of these SNPs revealed that CD58 SNP rs17035850 associated with persistent fungemia defined as positive blood cultures for more than 5 days despite adequate therapy (P=0.005), while TAGAP SNP rs3127214 associated with disseminated disease in the organs (P=0.017) (see also Table S5).
Transcriptome and pathway analyses in CD58- and TAGAP-deficient macrophages
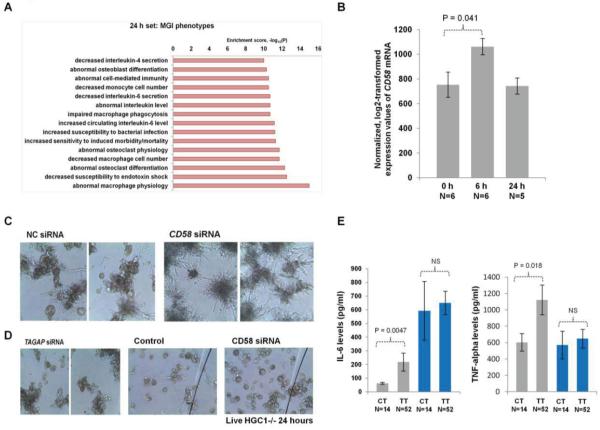
Practically nothing is known regarding the roles of CD58 and TAGAP in the antifungal host defense. In order to identify the antifungal host defense mechanisms influenced by these two genes, we assessed genome-wide transcriptional changes in wild-type, and macrophages in which the expression of CD58 and TAGAP was strongly reduced by siRNA transfection at 6 and 24 h upon Candida infection. Efficient down-regulation of CD58 mRNA was obtained by siRNA transfection, whereas only a mild effect was seen on TAGAP mRNA levels (Fig. S3). Genome-wide transcriptional changes in wild-type and CD58-deficient macrophages showed a total of 169 (at 6h) and 93 (at 24h) transcripts with at least a 1.25-fold differential expression (Fig.S4). Enrichment analysis showed that the differentially expressed genes are enriched for innate immune function, regulation of cytokine production, and general cellular responses to bacterial infections (Fig. S5). Furthermore, we investigated whether mutations in these differentially expressed genes show any common phenotypes in the mouse using the Mouse Genome Informatics phenotype data integrated in the GeneNetwork database. We indeed observed a significant enrichment for genes, where mutations in these genes may cause altered levels of interleukin-6 (IL-6) and TNF-alpha secretion, and impaired macrophage phagocytosis (Fig. 2A). These data suggest that altered expression levels of CD58 may regulate Candida phagocytosis on the one hand, and indirectly the IL-6 and TNF-alpha secretion on the other hand.
Fig. 2.
(A) Common phenotypes enrichment in the mouse using the Mouse Genome Informatics (MGI) phenotype data of the 93 genes differentially expressed at 24 hours between control and CD58 siRNA cells. (B) CD58 gene expression levels in wild-type macrophages at 0 hour, 6 hour and 24 hour upon Candida infection were extracted from the microarray experiment. (C) Human monocyte-derived macrophages were transfected with control and CD58 siRNA, respectively, for 48 hours. Macrophages were co-incubated with live C. albicans for an additional 24 hours. Representative microscopic photos of the Candida outgrowth through the macrophage are shown. (D) First panel: Macrophages were transfected with control and TAGAP siRNA, respectively, for 48 hours. Macrophages were co-incubated with live C. albicans for an additional 24 hours. Second panel: Control and CD58 siRNA transected macrophages were co-incubated with live C. albicans Δhgc1strain for an additional 24 hours. Representative microscopic photos of the Candida outgrowth through the macrophage are shown. (E) Human monocyte-derived macrophages from 66 healthy volunteers with different CD58 SNP rs12025416 genotypes were stimulated with Candida for 24 hours (grey bars) or with LPS for 24 hours (blue bars) and the supernatant was collected for IL-6 and TNF-α measurements. Average values of Candida induced IL-6 levels among CT individuals is 62.68 and among TT is 218.19; whereas for LPS it is 592.71 in CT group and 650.26 in TT group. Average values of Candida induced TNF-alpha levels among CT individuals is 603.57 and among TT is 1122.88; whereas for LPS it is 570.02 in CT group and 648.36 in TT group. The error bars indicate standard error of the mean (SEM). The correlation between cytokine production and genotypes was tested by the Wilcoxon rank sum test. NS; not significant.
Functional validation of CD58 for anti-Candida host defense
We next tested the CD58 mRNA levels in macrophages at 6 and 24 h upon Candida infection using microarray data. We observed a significant up-regulation of CD58 in response to Candida infection at 6 h (Fig. 2B; P=0.04), whereas no difference was seen at 24 h suggesting CD58 as a early-response gene in host defense against Candida. To validate the functional role of CD58 in anti-Candida response further, we investigated the phenotypes of the macrophage with silenced CD58. It is demonstrated that yeast-to-hyphae transition is one of the virulence factor for Candida to escape macrophage phagocytosis14. As expected, live Candida through germ-tube formation could escape macrophage phagocytosis in the control siRNA transfected group as well as in TAGAP siRNA group. Strikingly, a massive fungal outgrowth with extensive fungal hyphae formation was observed in the CD58 siRNA transfected group (Fig. 2C), showing that CD58 is important for Candida phagocytosis and inhibition of germination. To validate this possibility further, the colocalization of CD58 and Candida in the phagosome was examined by the fluorescence microscopy. Upon Candida phagocytosis, a clear recruitment of CD58 (green) around the calcofluor white-labeled Candida (blue) was observed, indicating that the physical colocalization of CD58 and Candida during the phagocytosis (Fig. S8). When a yeast-locked Δhgc1 C. albicans strain was used, there was no defect in the control of fungal growth in cells transfected with CD58 siRNA (Fig. 2D).
Cytokine-genotype correlation validate the CD58-mediated IL-6 and TNF-α production
As the transcriptome and pathway analyses in CD58-deficient macrophages implicated altered levels of IL-6 and TNF-α secretion, we tested the role of CD58 SNPs with these cytokine levels. The cytokines IL-6 and TNF-α were quantified from macrophages stimulated with either LPS or Candida. The top CD58 SNP, rs17035850, is a rare SNP with very low risk allele frequency. Therefore, we tested the second CD58 top SNP, rs12025416 that is more frequent, for association with cytokine levels. Functional genetic validation showed that CD58 SNP rs12025416 genotypes modulated cytokine production, where the risk allele C was associated with lower levels of Candida-stimulated IL-6 and TNF-α (Fig. 2E, P = 0.0047 and P = 0.018, respectively). In contrast, we found no association with LPS-stimulated IL-6 and TNF-α levels (Fig. 2E), confirming the specific role of CD58 polymorphisms in response to Candida infection.
Functional validation of TAGAP for anti-Candida host defense
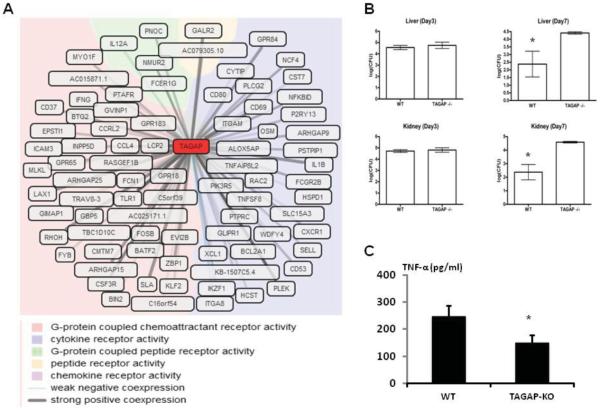
Because TAGAP siRNA inhibition in primary macrophages was not efficient, we interrogated the GeneNetwork database to predict TAGAP function based on co-expression data extracted from approximately 80,000 Affymetrix microarray experiments (see www.genenetwork.nl/genenetwork). Several GO molecular functions such as chemoattractant receptor activity and cytokine production were strongly influenced by TAGAP (Fig. 3A). To validate these pathways, the role of TAGAP was tested in an in vivo disseminated candidiasis model in Tagap-deficient mice. While the fungal loads did not differ between control and knock-out mice early during infection, the Tagap−/− mice were incapable of clearing the fungus from the organs at the later stages of infection (Fig. 3B). In addition, splenocytes isolated from Tagap−/− mice produced significantly less TNF-α compared with control animals (Fig. 3C).
Fig. 3.
(A) The co-expression analysis (GeneNetwork) to predict the potential role of TAGAP. Coexpressed genes are colored based on their GO molecular function. (B) Wild-type and TAGAP −/− mice were infected i.v. with C. albicans blastoconidia (5×105 CFU/mouse). Subgroups of animals were killed on days 3 and 7 of infection, and fungal outgrowth was assessed in both liver (upper panel) and kidneys (lower panel). The results are pooled data from at least 5 mice per group (mean±SEM). (C) Splenocytes isolated from wild-type and TAGAP −/− mice were stimulated with C. albicans for 24 hours, after which TNF-α was measured by ELISA in the supernatant (mean±SEM).
Discussion
This first genome-wide association study assessing genetic susceptibility to a fungal infection identified three novel risk factors for susceptibility to candidemia, namely CD58, LCE4A-C1orf68 locus and TAGAP. Carrying at least 2 or more risk alleles from these loci increases the risk for candidemia by more than 19-fold (Fig. S6). Using transcriptomic analysis and immunological validation, we identified unknown roles of CD58 and TAGAP in the host defense against Candida spp.
Genetic association studies are extremely challenging in systemic fungal infections due to the inherent difficulty of the relatively low number of patients available. The difficulty of enrolling a large number of candidemia patients is exemplified by the fact that earlier cohorts had no more than 60 patients15, 16. In order to surpass these difficulties, we took several steps. Firstly, we increased the number of patients by combining patients of European descent from Duke University and Radboud University Nijmegen Medical Center. Secondly, for the discovery analysis we used a large cohort of 11,920 population-based controls. Analyzing large number of population-based controls against a small number of cases could potentially yield spurious associations due to population substructure. Therefore, we strictly relied on confirming the significant findings by comparing the patients with the underlying disease-matched control cohort. Additional validation studies were performed in independent cohorts. Thirdly, we focused on genes and gene-regions known to be involved in immune-mediated diseases by using the Immunochip array. Finally, we confirmed the biological significance of our findings by immunological and functional genomics experiments.
Our findings have important implications. On the one hand, the 19-fold increased risk of developing candidemia in individuals carrying at least 2 or more risk alleles opens the possibility to use these SNPs to classify patients at-risk, and identify individuals who could benefit from prophylactic antifungal treatment. On the other hand, we have identified novel pathways of host defense mechanisms against fungi that contribute both to a better understanding of the host defense, and which may also be used for designing future immunotherapeutic strategies. In this respect, the unexpected identification of CD58 as an important factor mediating Candida phagocytosis and the inhibition of germination on the one hand, and modulation of Candida-specific cytokine production on the other hand, is an important step towards understanding the pathogenesis of the infection. CD58 is known as a member of the immunoglobulin superfamily17 and mediates adhesion and activation of T lymphocytes18. It has been involved in the host defense against viral infections such as hepatitis B19. The role identified here in inhibiting fungal germination at the level of the phagosome is unexpected and sheds light on a novel function of this molecule.
The role of LCE4A-C1orf68 locus for anti-Candida host defense could have been anticipated from its involvement in the barrier function of the epithelium, as Candida colonization of the mucosae is one of the main risk factors for candidemia in at-risk patients20. In contrast, nothing was known regarding a potential role for TAGAP in antifungal host defense. Using a GeneNetwork microarray database and co-expression analysis, and immunological validation in mice with a genetic defect in Tagap, we demonstrate its role in Candida-induced inflammation and antifungal host defense.
Interestingly, the genes we have identified as being involved in the susceptibility to candidemia are also involved in the genetic susceptibility to immune-mediated diseases. Polymorphisms in CD58 have been reported to increase susceptibility to multiple sclerosis (MS)21 and rheumatoid arthritis (RA)22, LCE4A-C1orf68 locus variants are associated with RA23 and psoriasis24, and TAGAP polymorphisms influence several autoinflammatory diseases22, 25, 26, 27, 28. These data point to strong similarities between the immune-mediated mechanisms involved in the host defense against fungal pathogens and those for immune-mediated pathology. This hypothesis is strengthened by the associations described between anti-Candida specific antibodies and Crohn's disease29. Similar shared relationships have been proposed for other pathologies such as leprosy and Crohn's disease27, and it has even been hypothesized that the genetic susceptibility to autoimmune diseases in modern human populations was shaped by evolutionary pressure exerted by infections30, 31.
In conclusion, this first, unbiased, genetic association study in a fungal infection demonstrates the potential of functional genomics approaches to identify novel risk factors in infections even in clinical conditions in which a limited number of patients are available. Our study highlights genetic variants in three novel antifungal host defense mechanisms that increase susceptibility to candidemia.
Materials and Methods
Study populations
Discovery cohort
Adult Candidemia patients were enrolled after informed consent at the Duke University Hospital (DUMC, Durham, North Carolina, USA) and Radboud University Nijmegen Medical Centre (RUNMC, Nijmegen, the Netherlands). Disease matched controls were enrolled after informed consent at the Duke University Hospital (DUMC, Durham, North Carolina, USA). The study was approved by the Institutional Review Boards at each study centre, and enrolment occurred between January 2003 and January 200932, 33. Patients must have had at least one positive blood culture for a Candida species. A total of 217 patients of European ancestry (36 of the Netherlands origin and 181 of the USA origin) were included in the study (for clinical characteristics, see Table 2).
Two different control groups were employed. One, consisting of 11,920 population-based individuals of European ancestry25, 34, was used for the discovery phase of the study. The second control group consisted of 146 candidiasis-free but otherwise matched patients. Non-infected patients were recruited from the same hospital wards as infected patients so that comorbidities and clinical risk factors for infection were similar. This second control group was used to confirm the candidate SNPs identified in the discovery phase. The infected subjects at DUMC were followed prospectively for up to 12 weeks following diagnosis of candidemia to determine their clinical outcome. Disseminated infection was defined as the presence of Candida spp. at normally sterile body sites other than the bloodstream or urine. Persistent fungemia was defined as ≥ 5 days of persistently positive blood cultures.
Validation cohorts
Two independent cohorts of patients with candidemia were used for validation studies. Firstly, a cohort of 75 patients of African-American descent with candidemia was recruited at DUMC, and 54 patients of African-American descent without candidemia from the same wards served as a disease-matched control group. Secondly, a cohort of 69 European surgical ICU patients, including 29 cases of invasive candidiasis (without candidemia) and 40 non-infected controls were recruited for the Funginos study, as described elsewhere35.
Genotyping and quality control
The case and the control samples of the discovery cohort were genotyped using the Immunochip according to Illumina's protocols25. We applied SNP quality control filters to exclude SNPs with (a) a low call rate (< 99%), (b) a Hardy-Weinberg equilibrium p < 0.01 in control samples only, and (c) a minor allele frequency of < 0.01. In the end, 118,989 SNPs were used for case-control analysis. We also excluded 54 samples with a low genotyping rate (< 98%) and 40 ethnic outlier samples based on multidimensional scaling (MDS) analysis (Supplemental Fig. 1)34. We included 217 candidemia cases and 11,920 controls of Caucasian descent in the discovery phase of the case-control association analysis. The replication cohorts were genotyped at four SNPs using Competitive Allele Specific PCR (KASP™) system according to manufacturer's protocol (LGC Genomics; http://www.lgcgenomics.com, formerly KBioscience). The KASP allele-specific forward primers and common reverse primer were designed by Kraken™ assay design and workflow management software (LGC Genomics, formerly KBioscience). Results were analyzed on KlusterKaller software (LGC Genomics, formerly KBioscience) according to standard protocols and quality controls.
Statistical analysis
In the discovery phase, the associations between Immunochip SNPs and candidemia susceptibility was tested by logistic-regression after adjusting for the first four components from the MDS analysis using PLINKv1.0736. A strong inflation in immunochip studies has been observed as the selection of SNPs is biased towards only loci associated with immune-mediated traits37. Therefore, we considered SNPs that map to non-immune regions but are present on immunochip to calculate the inflation factor. Comparison of the genetic inflation factor of all SNPs (λ = 1.22) to the genetic inflation factor of non-immune SNPs (λ = 1.102) indicated that there was little population stratification effect (Supplemental Fig 2). We considered p < 5×10−8 as the threshold for significant association. The association to the top-associated SNPs within the validation and replication cohorts was tested using the Cochran-Armitage trend test and meta-analysis was conducted using the Mantel-Haenszel method. Heterogeneity across the two cohorts was examined using the Breslow-Day test.
We tested the cumulative effects of three risk SNPs on candidemia risk among individuals carrying either 1 or 2 and more risk alleles. The odds ratios were calculated relative to the individuals with no risk alleles for the three SNPs.
CD58/TAGAP knockdown and phagocytosis experiments
Human monocyte-derived macrophages were derived as described previously38. Macrophages from five different volunteers were transfected with CD58 siRNA (L-004538-00-0005), TAGAP siRNA (L-008711-01-0005) or control siRNA (D-001810-10-20) by Dharmafect 4 for 2 days (Thermo Scientific). Total RNA was isolated at 6 hours and 24 hours, and global gene expression was profiled using Illumina Human HT-12 Expression BeadChip39. Differentially expressed genes by at least 1.25-fold between control and CD58 siRNA cells were subjected to pathway enrichment analysis using GeneNetwork analysis40. After siRNA transfection, macrophages were exposed to live C. albicans at multiplicity of infection (MOI)= 1 for 24 hours, after which the phagocytosis and fungal outgrowth was determined by microscopy. The role of fungal germination was assessed by using the yeast-locked Hgc1-deficient C. albicans strain (provided by Dr. Bernhard Hube, Jena University). Cytokine concentrations were determined by ELISA.
In vitro macrophage stimulation assays
The effect of SNPs in CD58 on cytokine production was studied in monocyte-derived macrophages isolated from a cohort of 66 healthy Europeans. Macrophages were incubated at 37°C for 24 hours with RPMI culture medium, lipopolysaccharide (LPS, 10 ng/ml, Sigma-Aldrich, MO, USA), or heat-killed C. albicans yeasts or hyphae (1×106 microorganisms/ml). Cytokines were measured by ELISA (R&D Systems, MN, USA), and the correlation between cytokine production and genotypes was tested by the Wilcoxon rank sum test.
Systemic C. albicans infection in TAGAP-deficient mice
C57BL/6J and Tagap loss-of-function female mice41 (8–12 weeks) were used for assessing their susceptibility to C. albicans. The experiments were approved by the Ethics Committee on Animal Experiments of the University of Athens. Mice were injected with live C. albicans blastoconidia 5×107 cfu/mouse. The fungal loads in the liver and kidneys were assessed by microbiological dilution plating on days 3 and 7 after infection. Cytokine production capacity was assessed after stimulation of splenocytes (1×105/well) with C. albicans (1×106 microorganisms/well).
Supplementary Material
Acknowledgements
This work was supported by the Netherlands Organization for Scientific Research (NWO-VENI grant 916.10.135 to LF), the European Research Council (Consolidator Grant, ERC-310372 to MN, and Advanced Grant, ERC-671274 to CW), and the Dutch Digestive Diseases Foundation (MLDS WO11-30 to CW). Research reported in this publication was also supported by the National Institutes of Health under award number K23AI51537 (to MDJ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Mathieu Platteel, Astrid Maatman, and Gosia Trynka for assisting in the RNA and DNA analysis, and array experiments (Immunochip and gene expression). We also thank Thierry Calandra, Jacques Bille, Frederic Tissot, Frederic Lamoth, Christina Orasch, Philippe Eggimann (Lausanne), Chloë Kaech, Martin Siegemund, Ursula Flückiger (Basel), Stefan Zimmerli (Bern) and all other members of the Funginos group involved in the validation study. We thank Jackie Senior for editing the final version of the manuscript.
Footnotes
Contributions M.G.N, C.W. and V.K. conceived the study. V.K. analysed the genetic data and microarray data. S.C., T.S.P., S.P.S., F.L.V., J.W.M. and L.A.B.J. summarised all immunological assays. M.G.N., C.W., J.R.P. and M.D.J. provided the samples for discovery phase. A.W., E.G.B., P.Y.B., and O.M. provided samples for replication phase. J.K., S.W. and L.F. performed the pathway enrichment analyses. R.X., B.J.K., and O.M. contributed in interpreting the results. M.G.N. and C.W. directed the study. All authors co-wrote the paper.
Competing financial interests The authors declare no competing financial interests.
References
- 1.Rangel-Frausto MS. The epidemiology of bacterial sepsis. Infectious disease clinics of North America. 1999;13:299–312. vii. doi: 10.1016/s0891-5520(05)70076-3. [DOI] [PubMed] [Google Scholar]
- 2.Bassetti M, Righi E, Tumbarello M, Di Biagio A, Rosso R, Viscoli C. Candida infections in the intensive care unit: epidemiology, risk factors and therapeutic strategies. Expert review of anti-infective therapy. 2006;4:875–885. doi: 10.1586/14787210.4.5.875. [DOI] [PubMed] [Google Scholar]
- 3.Smeekens SP, van de Veerdonk FL, Kullberg BJ, Netea MG. Genetic susceptibility to Candida infections. EMBO molecular medicine. 2013;5:805–813. doi: 10.1002/emmm.201201678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tortorano AM, et al. Epidemiology of candidaemia in Europe: results of 28-month European Confederation of Medical Mycology (ECMM) hospital-based surveillance study. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2004;23:317–322. doi: 10.1007/s10096-004-1103-y. [DOI] [PubMed] [Google Scholar]
- 5.Leroy O, et al. Epidemiology, management, and risk factors for death of invasive Candida infections in critical care: a multicenter, prospective, observational study in France (2005–2006) Critical care medicine. 2009;37:1612–1618. doi: 10.1097/CCM.0b013e31819efac0. [DOI] [PubMed] [Google Scholar]
- 6.Moran C, Grussemeyer CA, Spalding JR, Benjamin DK, Jr., Reed SD. Comparison of costs, length of stay, and mortality associated with Candida glabrata and Candida albicans bloodstream infections. American journal of infection control. 2010;38:78–80. doi: 10.1016/j.ajic.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Veerdonk FL, Kullberg BJ, Netea MG. Adjunctive immunotherapy with recombinant cytokines for the treatment of disseminated candidiasis. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2012;18:112–119. doi: 10.1111/j.1469-0691.2011.03676.x. [DOI] [PubMed] [Google Scholar]
- 8.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson B, et al. Late cornified envelope family in differentiating epithelia--response to calcium and ultraviolet irradiation. The Journal of investigative dermatology. 2005;124:1062–1070. doi: 10.1111/j.0022-202X.2005.23699.x. [DOI] [PubMed] [Google Scholar]
- 11.Mao M, et al. T lymphocyte activation gene identification by coregulated expression on DNA microarrays. Genomics. 2004;83:989–999. doi: 10.1016/j.ygeno.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids research. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nature genetics. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcil A, Harcus D, Thomas DY, Whiteway M. Candida albicans killing by RAW 264.7 mouse macrophage cells: effects of Candida genotype, infection ratios, and gamma interferon treatment. Infection and immunity. 2002;70:6319–6329. doi: 10.1128/IAI.70.11.6319-6329.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van der Graaf CA, et al. Toll-like receptor 4 Asp299Gly/Thr399Ile polymorphisms are a risk factor for Candida bloodstream infection. European cytokine network. 2006;17:29–34. [PubMed] [Google Scholar]
- 16.Woehrle T, et al. Pathogen specific cytokine release reveals an effect of TLR2 Arg753Gln during Candida sepsis in humans. Cytokine. 2008;41:322–329. doi: 10.1016/j.cyto.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Selvaraj P, Plunkett ML, Dustin M, Sanders ME, Shaw S, Springer TA. The T lymphocyte glycoprotein CD2 binds the cell surface ligand LFA-3. Nature. 1987;326:400–403. doi: 10.1038/326400a0. [DOI] [PubMed] [Google Scholar]
- 18.Dustin ML, Springer TA. T-cell receptor cross-linking transiently stimulates adhesiveness through LFA-1. Nature. 1989;341:619–624. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- 19.Ryckman KK, et al. Host genetic factors and vaccine-induced immunity to HBV infection: haplotype analysis. PloS one. 2010;5:e12273. doi: 10.1371/journal.pone.0012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hostetter MK. Adhesins and ligands involved in the interaction of Candida spp. with epithelial and endothelial surfaces. Clinical microbiology reviews. 1994;7:29–42. doi: 10.1128/cmr.7.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nature genetics. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 22.Raychaudhuri S, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nature genetics. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uddin M, Sturge M, Rahman P, Woods MO. Autosome-wide copy number variation association analysis for rheumatoid arthritis using the WTCCC high-density SNP genotype data. The Journal of rheumatology. 2011;38:797–801. doi: 10.3899/jrheum.100758. [DOI] [PubMed] [Google Scholar]
- 24.Zhang XJ, et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nature genetics. 2009;41:205–210. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- 25.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nature genetics. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyre S, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nature genetics. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar V, Wijmenga C, Withoff S. From genome-wide association studies to disease mechanisms: celiac disease as a model for autoimmune diseases. Seminars in immunopathology. 2012;34:567–580. doi: 10.1007/s00281-012-0312-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Standaert-Vitse A, et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn's disease. Gastroenterology. 2006;130:1764–1775. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Zhernakova A, et al. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am J Hum Genet. 2010;86:970–977. doi: 10.1016/j.ajhg.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Netea MG, Wijmenga C, O'Neill LA. Genetic variation in Toll-like receptors and disease susceptibility. Nature immunology. 2012;13:535–542. doi: 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
- 32.Rosentul DC, et al. Genetic variation in the dectin-1/CARD9 recognition pathway and susceptibility to candidemia. The Journal of infectious diseases. 2011;204:1138–1145. doi: 10.1093/infdis/jir458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson MD, et al. Cytokine gene polymorphisms and the outcome of invasive candidiasis: a prospective cohort study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;54:502–510. doi: 10.1093/cid/cir827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smeekens SP, et al. Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans. Nature communications. 2013;4:1342. doi: 10.1038/ncomms2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tissot F, et al. Beta-Glucan Antigenemia Anticipates Diagnosis of Blood Culture-Negative Intra-Abdominal Candidiasis. American journal of respiratory and critical care medicine. 2013 doi: 10.1164/rccm.201211-2069OC. [DOI] [PubMed] [Google Scholar]
- 36.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nature genetics. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng SC, et al. The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. Journal of leukocyte biology. 2011;90:357–366. doi: 10.1189/jlb.1210702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu J, et al. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS genetics. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar V, et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS genetics. 2013;9:e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauer H, Willert J, Koschorz B, Herrmann BG. The t complex-encoded GTPase-activating protein Tagap1 acts as a transmission ratio distorter in mice. Nature genetics. 2005;37:969–973. doi: 10.1038/ng1617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.