Abstract
Throughout the animal kingdom, p53 genes function to restrain mobile elements and recent observations indicate that transposons become derepressed in human cancers. Together, these emerging lines of evidence suggest that cancers driven by p53 mutations could represent ‘transpospoathies’, i.e. disease states linked to eruptions of mobile elements. The transposopathy hypothesis predicts that p53 acts through conserved mechanisms to contain transposon movement and, in this way, prevents tumor formation. How transposon eruptions provoke neoplasias is not well understood but, from a broader perspective, this hypothesis also provides an attractive framework to explore unrestrained mobile elements as inciters of late-onset idiopathic disease.
Keywords: Drosophila, human cancers, mouse cancer models, p53, piRNAs, retrotransposons, mobile elements, zebrafish
“The past is already written. The ink is dry.”
-Game of Thrones
Introduction
The p53 gene family occupies central positions in stress response networks throughout the animal kingdom. In humans, p53 is altered in most cancers and implicated in age-related diseases. As transcription factors, products encoded by this gene family mediate selective activation and repression of targets to specify adaptive responses. However, despite extensive characterization, precisely how they act to suppress tumors and mitigate age-related disease remains poorly understood. Since p53 genes are broadly conserved, ancestral properties of these genes offer promising routes towards understanding functions of p53 that become deranged in human diseases. Leveraging genetic models to interrogate p53 function in vivo, we discovered that p53 normally acts to contain retrotransposons [1], which are mobile elements broadly implicated in sporadic and heritable human disease. Furthermore, in complementation studies, normal human p53 genes could restrain transposons but mutant alleles from cancer patients were disabled for this activity, suggesting that p53 mitigates disease, in part, by suppressing the movement of transposons. Consistent with this, unrestrained retrotransposons were detected in p53-driven mouse and human cancers. We consider these new findings within the context of common misconceptions and frame the implications within a novel hypothesis that links defective p53 to eruptions of mobile elements and potential ‘transposopathies’.
Three common myths about p53
As one of the most highly studied and cited genes, p53 has attracted compelling discoveries and numerous controversies. But, along with this celebrity status, p53 research is also branded with three fables that have likely precluded a thorough exploitation of p53 biology in the clinic.
Myth 1: We know how p53 acts to suppress tumors
Mutations in p53 define perhaps the most common class of genetic culprits seen in neoplastic disease, occurring in over half of human cancers. Hence, it is commonly postulated that p53 needs to be disabled in order for cancers to form and, for this reason, it is intensively studied and commonly featured in the cancer research community. However, despite impressive advances, unexplained mysteries have confounded efforts to articulate a definitive account for how this single gene specifies tumor suppression. Underscoring this point, no single effector or combination of effectors has replicated p53− cancer phenotypes when genetically tested and, in fact, mice that are triply mutated for p21, noxa and puma remained surprisingly tumor free despite failures in apoptosis and proliferation checkpoints [2]. Likewise, mice harboring an acetylation-defective engineered p53 allele also remained tumor free despite the combined loss of p53-mediated cell-cycle arrest, apoptosis and senescence [3]. Hence, p53 is clearly able to suppress tumor formation without the need for these canonical effectors and their associated responses. So, given this inconvenient truth, how does p53 actually prevent cancers?
Myth 2: p53 is a conventional tumor suppressor
Though commonly cited as a text book example for tumor suppressor genes, p53 does not truly qualify as a poster child for this class of genes. Unlike conventional tumor suppressors, which show biallelic inactivation, the mutation spectrum seen in patient tumors is heavily skewed toward missense alterations of the full length protein [4]. These mutants were conventionally thought to act as dominant negatives (by poisoning tetrameric complexes) but, in most cancers, these variants exist in trans to a complete deletion at the alternate locus [5, 6], revealing patterns entirely at odds with dominant negative models [7–9]. Consistent with this, knock-in mice ‘humanized’ with p53 cancer alleles produced more severe and more diverse cancer types compared to nulls [10–12], reflecting gain-of-function oncogenic properties that are clearly distinct from dominant negative activity [13]. Furthermore, p53 mutant proteins are frequently stabilized in cancers and these two properties seem to be fundamentally linked, perhaps through disruption of a negative feedback loop involving MDM2, an E3 ligase responsible for p53 turnover [7, 9]. However, accumulation of p53 per se appears to be a manifestation of the transformed state rather than a primary cause of it, since p53 stabilization does not occur when normal tissues are sampled from Li Fraumeni patients (germ line p53 mutants) or from humanized ‘knock-in’ mutant mice [14]. Taken together, genetic lessons from humans and mice establish that, strictly speaking, p53 is not a conventional tumor suppressor gene. Instead, it seems that cellular transformation involves loss of growth suppression, encoded by wild type p53, together with poorly understood gain-of-function activity conferred by p53 point mutants [14].
Myth 3: p53 evolved to prevent tumors
The evolutionary appearance of p53 genes was previously thought to coincide with the emergence of multicellular organisms [15]. However, more recent phyletic evidence shows that p53 was present in unicellular protists as well as vertebrates and invertebrates [16] suggesting that p53 genes have evolutionary roots predating tumor formation by hundreds of millions of years [15, 17–19]. Hence, p53 was initially molded by selective pressures unrelated to cancer and only later became co-opted for tumor suppression in long-lived animals. If true, this inference suggests that attempts to identify ancient p53 effectors could open promising routes towards understanding adaptive functions of p53 that, when deranged, cause human diseases [15, 19–21].
Common ancestry in the p53 network
Before the emergence of adaptations for tumor prevention, what were the ancient functions of p53? Are these fundamentally relevant for tumor suppression by p53 today? And, why do missense variants dominate the allelic spectrum seen in patient tumors? Since p53 genes are broadly conserved, evolutionary principles suggest that ancient foundations supporting this tumor suppression network could advance answers to these questions. Like mammalian counterparts, p53 genes in flies and worms act to specify adaptive responses to damage and promote genome stability (reviewed in [15, 21, 22]). As transcription factors, fly p53 and human p53 bind to identical sequence elements [23–25] but perhaps the most definitive evidence for functional conservation comes from studies on ‘humanized p53 fly strains”, where human p53 genes were used to rescue defective phenotypes caused by mutations in the fly counterpart [26]. Upstream regulators in these networks are also conserved. For example, in both flies and mammals, the Chk2 kinase activates p53 [27]. Likewise, during meiotic recombination [28] the action of Spo11 exposed an intrinsic physiological role for p53 in meiosis in both flies and mice [28] raising the possibility that tumor-suppressive functions were co-opted from primordial activities coupled to DNA breaks during genetic recombination [15, 28]. Abnormal growth often provokes p53 activity in mammals [29, 30] and, likewise, unscheduled growth (e.g. RasV12) or failed differentiation similarly provokes p53 activity in flies [31], indicating that ancestral pathways are involved. Moreover, these unappreciated mechanisms are very likely conserved but they do not involve ARF or MDM2, since both are absent in flies. Similarly, important parallels occur at the level of downstream target genes. For example, prominent p53 target effectors shared in common between flies and mammals include IAP antagonists, death receptors and ribonucleotide reductase [15, 32–35].
Mobile elements provoke p53 action
In the lab, genotoxic drugs and ionizing radiation are routinely used to trigger p53 responses but none of these represent ‘natural’ agents that we would expect ancestral organisms to normally encounter. Reasoning that mobile elements might qualify as physiologically relevant genome destabilizers, we tested whether transposon activity might provoke p53 function. To do this, we tested p53 biosensors in mutants defective for the piRNA network, an ancient system that curbs retrotransposons in the animal germline. When effectors in this pathway are mutated, generalized derepression of these mobile elements is observed [36, 37] and, in association with this, we observed persistent p53 activity [31]. Notably, this constitutive p53 activity occurred without challenge by exogenous stressors and, consistent with this, robust genetic interactions between p53 and components of the piRNA pathway were also detected [1]. Together, these observations established that retrotransposons can provoke functional p53 activity, raising the possibility that these mobile elements could have shaped primordial p53 networks.
p53 contains mobile elements in model systems and in cancers
Active mobile elements consistently triggered p53 [31], raising the possibility that p53 might do more than just sense mobile elements and could perhaps act to contain them. To test this idea, we profiled multiple classes of retrotransposons in p53− flies and, strikingly, all were dysregulated, with some exhibiting quite profound derepression when p53 was absent. Likewise, similarly striking patterns occurred in p53 mutant zebrafish challenged with a synthetic transposon enabling us to directly score de novo integration events [1]. Consistent with this, p53 has also been shown to negatively regulate transcription of repetitive elements and endogenous retroviruses in cultured cell lines [38–40]. Together, these findings establish that containing mobile elements is a broadly conserved function of the p53 genes in short-lived model systems and cultured somatic cell lines [41].
Does human p53 encode this same genoprotective activity and, if so, could this also help explain how the human gene acts to suppress tumor formation? To address this question, we engineered strains that replace the fly p53 gene with human alleles, producing a collection of flies that are, in effect, ‘humanized’ for p53 variants most commonly seen in cancer patients. In this complementation platform, human p53 restrained mobile elements and corrected dysregulated transposon phenotypes seen in p53− animals but, remarkably, all five cancer-associated alleles were disabled for this activity despite expressing comparable levels of protein. Hence, suppressing transposons is a general property of p53 genes that extends well beyond short-lived animals and includes humans. Furthermore, since cancer-associated alleles were commonly defective for this function, these observations raise the hypothesis that p53 tonically restrains transposons through conserved mechanisms that, if defective, could drive tumors. If true, associations between p53 mutations and elevated retroelements should be exposed in cancers. Consistent with this prediction, robust derepression of L1 and IAP retrotransposons was specifically linked to p53− driven cancers in mice [1]. Likewise, in human RNA seq data sets, elevated L1 retrotroelement RNAs were strongly associated with p53− status in colon cancers [1] and, importantly, these links were specific to L1 elements, since no associations emerged between p53− status and either low complexity repeats or simple repeats in these same data sets [1]. Similarly, in Wilms tumors specimens, L1 retroelement eruptions clearly stratified with p53 mutations [1].
p53-driven cancers as transposopathies
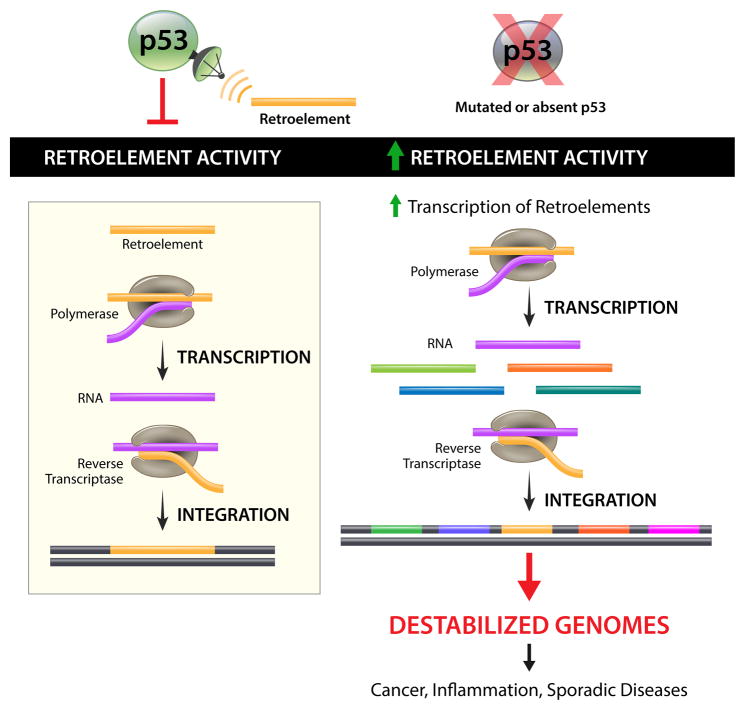
The observations in Wylie et al. [1] raise an attractive model, which views p53 as a restraint against mobile elements. In this scenario p53 is a custodian of mobile elements, guarding against “transposopathies” by surveilling transposons and containing their activity (see Figure 1). Accordingly, failures in p53 function foster conditions permissive for transposon eruptions that either promote tumor formation or predispose tissues toward neoplastic growth. This “transposopathy hypothesis” is attractive for numerous reasons. First, it offers a new and testable mechanism for p53-mediated tumor suppression which, in the real world, may collaborate with well known modalities that involve checkpoints and apoptosis. Second, it helps explain the inherent instability of cancer genomes, since hyperactive transposons encourage destabilizing events, including chromosomal rearrangements. Third, defective containment of mobile elements is consistent with mounting evidence for transposon movement in cancer genomes [42, 43]. Fourth, given that retroelements incite inflammatory responses [44–50] it opens plausible explanations for intimate links seen between cancers and inflammation. And finally, the transposopathy hypothesis offers a compelling candidate for the mysterious ancestral function encoded by p53 genes that may have been coopted for tumor suppression.
Figure 1.
p53 senses and represses retrotransposons. Retroelements move through an RNA intermediate, are reverse transcribed, and integrate into the genome and can cause mutations and genomic instability. These mobile elements are incited to move by exogenous stressors and DNA breaks associated with meiotic recombination [63]. The p53 tumor suppressor senses retrotransposon movement and acts to restrain these mobile elements. When p53 is mutated, retroelement activity is dramatically increased [1], which could potentially drive tumorigenesis and other transposopathies.
How does p53 act to contain transposons?
Throughout the animal kingdom p53 can trigger stress-dependent apoptotic responses [15] so, perhaps p53 acts by purging cells that have experienced transposons eruptions. Though attractive, this is not a satisfactory explanation since p53 restrains retrolelements in ways that are clearly uncoupled from apoptotic responses. This inference is supported by multiple observations. First, virtually no programmed cell death occurs in the germ line cells of the Drosophila ovary, where we know p53 acts [51]. Second, the rare apoptotic events that are occasionally seen in this organ are unaffected by p53 status [31]. Third, p53-dependent effects in zebrafish were seen prior the onset of programmed cell death [1]. Fourth, other mutants defective for stress-induced death (e.g. chk2−) do not exhibit transposon derepression [1]. These observations point to primary mechanisms that are unrelated to cell death which, if overwhelmed, could possibly engage apoptosis as a secondary, fail-safe mechanism.
So, if purging cells that have experienced retroelement eruptions is not the primary explanation, how does p53 actually restrain mobile elements? Some of the most compelling clues in this regard come from studies where synthetic retroelements were injected into zebrafish embryos and then profiled for epigenetic histone modifications. In wild type fish, robust H3K9me3 marks were deposited at 5′ regulatory sequences but these same repressive marks were starkly absent when examined in p53− mutants. Hence, it appears that p53 normally instructs epigenetic features that control retrotransposon activity [1]. Furthermore, this could plausibly involve direct mechanisms, since p53 can associate with at least one histone methyltransferase [52] and p53 binding sites have been reported in human LINE 1 retroelements [53].
p53 might also collaborate with the piRNA system to contain the action of retrotransposons and several lines of evidence from the Drosophila model support this possibility. For example, p53 genetically interacts with a pivotal catalytic component of the piRNA network, referred to as Aubergine [36, 37, 54, 55]. Moreover, similar to other mutants of the piRNA biogenesis pathway [56], loss of p53 caused abnormal accumulation of piRNA precursor RNAs, suggesting that p53 acts at steps impacting the biogenesis of piRNAs.
Future discoveries will elaborate precisely how transposon eruptions contribute to neoplasias. Do new integrations impact pivotal genes? Do they incite cancers or exacerbate them? Or perhaps transposon RNAs and DNAs are themselves are pathogenic? Recent work has highlighted the role of accumulated cytosolic LINE derived DNAs in the inflammatory pathology driving cardiomyopathy in a mouse model [57–61]. Likewise, p53 can promote inflammatory responses that suppress viral replication [62]. Future studies could also resolve how p53 detects active mobile elements and illuminate p53-dependent effectors that restrain transposons. As part of the ancestral landscape in this network, these findings too are likely to generalize and could themselves present compelling opportunities for demystifying the p53 pathway. For example, because a deranged protein can impact networks in ways that are distinct from simply removing them, they may help explain why missense p53 mutations are predominant in cancers. Finally, it will also be important to determine whether other p53 subfamilies (e.g. p63 and p73) similarly restrain mobile elements.
The transposopathy hypothesis as a framework for sporadic disease
Unlike most genetic material, transposons can mobilize to new genomic locations. In the healthy state, they are effectively contained through mechanisms that are only partially understood. The advent of new deep sequencing technologies is enabling a new appreciation of the scale at which these elements can impact somatic genomes and the extent to which de novo integrations are tolerated. Conceivably, if unrestrained, the cumulative effects of dysregulated mobile elements could erode genome function, prompting distinct histopathologies in different tissue types. Here we have proposed that in stem cells (or other cells with proliferative potential) the p53− state is permissive for transposon eruptions that predispose toward neoplastic growth. So what might happen if mobile elements become unrestrained in cells that lack proliferative potential, such as muscles or neurons? In this scenario, as the function of genomes in terminally differentiated cells erodes, it seems plausible that sporadic, age-related syndromes could occur. Given this, we believe the transposopathy hypothesis also offers an attractive framework to explore dysregulated mobile elements as inciting events that could provoke late-onset idiopathic disease in otherwise healthy individuals.
Conclusions
Restraining mobile elements is a broadly conserved function of p53 that likely constitutes a key mechanism of cancer prevention by this tumor suppressor. We propose that p53-driven cancers, and perhaps other diseases, are incited by transposon eruptions manifesting as transposopathies when the genomes of diseased cells are examined. If verified, the transposopathy hypothesis offers exciting implications for developing new diagnostic tools and and new therapeutic agents for disease treatment.
Acknowledgments
This work was supported by grants to AW (NRSA 1F31CA189691-01), AEJ (ACS 128847-PF-15-160-01-DDC), and JMA (NIH R01GM072124, R01GM115682 and the Welch Foundation I-1865).
References
- 1.Wylie A, Jones AE, D’Brot A, Lu WJ, et al. p53 genes function to restrain mobile elements. Genes Dev. 2016;30:64–77. doi: 10.1101/gad.266098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valente LJ, Gray DH, Michalak EM, Pinon-Hofbauer J, et al. p53 Efficiently Suppresses Tumor Development in the Complete Absence of Its Cell-Cycle Inhibitory and Proapoptotic Effectors p21, Puma, and Noxa. Cell reports. 2013 doi: 10.1016/j.celrep.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Li T, Kon N, Jiang L, Tan M, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–83. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michalovitz D, Halevy O, Oren M. p53 mutations: gains or losses? J Cell Biochem. 1991;45:22–9. doi: 10.1002/jcb.240450108. [DOI] [PubMed] [Google Scholar]
- 5.Soussi T, Lozano G. p53 mutation heterogeneity in cancer. Biochem Biophys Res Commun. 2005;331:834–42. doi: 10.1016/j.bbrc.2005.03.190. [DOI] [PubMed] [Google Scholar]
- 6.Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1:233–40. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 7.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes & development. 2012;26:1268–86. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–58. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vousden KH, Prives C. P53 and prognosis: new insights and further complexity. Cell. 2005;120:7–10. doi: 10.1016/j.cell.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 10.Olive KP, Tuveson DA, Ruhe ZC, Yin B, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–60. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Lang GA, Iwakuma T, Suh YA, Liu G, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–72. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Hanel W, Marchenko N, Xu S, Yu SX, et al. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20:898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 14.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–13. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 15.Lu WJ, Amatruda JF, Abrams JM. p53 ancestry: gazing through an evolutionary lens. Nat Rev Cancer. 2009;9:758–62. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- 16.King N, Westbrook MJ, Young SL, Kuo A, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–8. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu WJ, Abrams JM. Lessons from p53 in non-mammalian models. Cell Death Differ. 2006 doi: 10.1038/sj.cdd.4401922. [DOI] [PubMed] [Google Scholar]
- 18.Belyi VA, Ak P, Markert E, Wang H, et al. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol. 2010;2:a001198. doi: 10.1101/cshperspect.a001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutcliffe JE, Brehm A. Of flies and men; p53, a tumour suppressor. FEBS Lett. 2004;567:86–91. doi: 10.1016/j.febslet.2004.03.122. [DOI] [PubMed] [Google Scholar]
- 20.Herzog G, Joerger AC, Shmueli MD, Fersht AR, et al. Evaluating Drosophila p53 as a model system for studying cancer mutations. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.417980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu WJ, Abrams JM. Lessons from p53 in non-mammalian models. Cell Death Differ. 2006;13:909–12. doi: 10.1038/sj.cdd.4401922. [DOI] [PubMed] [Google Scholar]
- 22.Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–5. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- 23.Brodsky MH, Nordstrom W, Tsang G, Kwan E, et al. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–13. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 24.Jin S, Martinek S, Joo WS, Wortman JR, et al. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:7301–6. doi: 10.1073/pnas.97.13.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ollmann M, Young LM, Di Como CJ, Karim F, et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- 26.D’Brot A, Kurtz P, Regan E, Jakubowski B, et al. A platform for interrogating cancer-associated p53 alleles. Oncogene. 2016 doi: 10.1038/onc.2016.190. [DOI] [PubMed] [Google Scholar]
- 27.Peters M, DeLuca C, Hirao A, Stambolic V, et al. Chk2 regulates irradiation-induced, p53-mediated apoptosis in Drosophila. Proc Natl Acad Sci U S A. 2002;99:11305–10. doi: 10.1073/pnas.172382899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu WJ, Chapo J, Roig I, Abrams JM. Meiotic recombination provokes functional activation of the p53 regulatory network. Science. 2010;328:1278–81. doi: 10.1126/science.1185640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, et al. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–7. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 30.Meek DW. Mechanisms of switching on p53: a role for covalent modification? Oncogene. 1999;18:7666–75. doi: 10.1038/sj.onc.1202951. [DOI] [PubMed] [Google Scholar]
- 31.Wylie A, Lu WJ, D’Brot A, Buszczak M, et al. p53 activity is selectively licensed in the Drosophila stem cell compartment. Elife. 2014;3:e01530. doi: 10.7554/eLife.01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akdemir F, Christich A, Sogame N, Chapo J, et al. p53 directs focused genomic responses in Drosophila. Oncogene. 2007;26:5184–93. doi: 10.1038/sj.onc.1210328. [DOI] [PubMed] [Google Scholar]
- 33.Brodsky MH, Weinert BT, Tsang G, Rong YS, et al. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol. 2004;24:1219–31. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brodsky MH, Sekelsky JJ, Tsang G, Hawley RS, et al. mus304 encodes a novel DMA damage checkpoint protein required during Drosophila development. Genes & Development. 2000;14:666–78. [PMC free article] [PubMed] [Google Scholar]
- 35.Jin S, Kalkum M, Overholtzer M, Stoffel A, et al. CIAP1 and the serine protease HTRA2 are involved in a novel p53-dependent apoptosis pathway in mammals. Genes Dev. 2003;17:359–67. doi: 10.1101/gad.1047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 37.Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191:905–13. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leonova KI, Brodsky L, Lipchick B, Pal M, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E89–98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang NT, Yang WK, Huang HC, Yeh KW, et al. The transcriptional activity of HERV-I LTR is negatively regulated by its cis-elements and wild type p53 tumor suppressor protein. J Biomed Sci. 2007;14:211–22. doi: 10.1007/s11373-006-9126-2. [DOI] [PubMed] [Google Scholar]
- 40.Levine AJ, Ting DT, Greenbaum BD. P53 and the defenses against genome instability caused by transposons and repetitive elements. Bioessays. 2016;38:508–13. doi: 10.1002/bies.201600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leonova KI, Brodsky L, Lipchick B, Pal M, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A. 2013;110:E89–98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riordan JD, Dupuy AJ. Domesticated transposable element gene products in human cancer. Mob Genet Elements. 2013;3:e26693. doi: 10.4161/mge.26693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tubio JM, Li Y, Ju YS, Martincorena I, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345:1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balada E, Ordi-Ros J, Vilardell-Tarres M. Molecular mechanisms mediated by human endogenous retroviruses (HERVs) in autoimmunity. Rev Med Virol. 2009;19:273–86. doi: 10.1002/rmv.622. [DOI] [PubMed] [Google Scholar]
- 45.Baudino L, Yoshinobu K, Morito N, Santiago-Raber ML, et al. Role of endogenous retroviruses in murine SLE. Autoimmun Rev. 2010;10:27–34. doi: 10.1016/j.autrev.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kassiotis G. Endogenous retroviruses and the development of cancer. J Immunol. 2014;192:1343–9. doi: 10.4049/jimmunol.1302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perl A, Fernandez D, Telarico T, Phillips PE. Endogenous retroviral pathogenesis in lupus. Curr Opin Rheumatol. 2010;22:483–92. doi: 10.1097/BOR.0b013e32833c6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruprecht K, Mayer J, Sauter M, Roemer K, et al. Endogenous retroviruses and cancer. Cell Mol Life Sci. 2008;65:3366–82. doi: 10.1007/s00018-008-8496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol. 2014;15:415–22. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peterson JS, Bass BP, Jue D, Rodriguez A, et al. Noncanonical cell death pathways act during Drosophila oogenesis. Genesis. 2007;45:396–404. doi: 10.1002/dvg.20306. [DOI] [PubMed] [Google Scholar]
- 52.Shi X, Kachirskaia I, Yamaguchi H, West LE, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–46. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris CR, Dewan A, Zupnick A, Normart R, et al. p53 responsive elements in human retrotransposons. Oncogene. 2009;28:3857–65. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353–9. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 56.Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, et al. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010;24:2499–504. doi: 10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones RB, Garrison KE, Wong JC, Duan EH, et al. Nucleoside analogue reverse transcriptase inhibitors differentially inhibit human LINE-1 retrotransposition. PLoS One. 2008;3:e1547. doi: 10.1371/journal.pone.0001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morita M, Stamp G, Robins P, Dulic A, et al. Gene-targeted mice lacking the Trex1 (DNase III) 3′-->5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–27. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–98. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 61.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–86. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 62.Menendez D, Shatz M, Resnick MA. Interactions between the tumor suppressor p53 and immune responses. Curr Opin Oncol. 2013;25:85–92. doi: 10.1097/CCO.0b013e32835b6386. [DOI] [PubMed] [Google Scholar]
- 63.Brouha B, Meischl C, Ostertag E, de Boer M, et al. Evidence consistent with human L1 retrotransposition in maternal meiosis I. American journal of human genetics. 2002;71:327–36. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]