Abstract
Use of viral vectors to deliver therapeutic genes to the central nervous system holds promise for the treatment of neurodegenerative diseases and neurotrauma. Adeno-associated viral (AAV) vectors encoding brain-derived neurotrophic factor (BDNF) or ciliary derived neurotrophic factor (CNTF) promote the viability and regeneration of injured adult rat retinal ganglion cells. However, these growth-inducing transgenes are driven by a constitutively active promoter, thus we examined whether long-term AAV-mediated secretion of BDNF or CNTF affected endogenous retinal gene expression. One year after the intravitreal injection of AAV-green fluorescent protein (GFP), bi-cistronic AAV-BDNF-GFP or AAV-CNTF-GFP, mRNA was extracted and analyzed using custom 96 well polymerase chain reaction arrays. Of 93 test genes, 56% showed significantly altered expression in AAV-BDNF-GFP and/or AAV-CNTF-GFP retinas compared with AAV-GFP controls. Of these genes, 73% showed differential expression in AAV-BDNF versus AAV-CNTF injected eyes. To focus on retinal ganglion cell changes, quantitative polymerase chain reaction was undertaken on mRNA (16 genes) obtained from fixed retinal sections in which the ganglion cell layer was enriched. The sign and extent of fold changes in ganglion cell layer gene expression differed markedly from whole retinal samples. Sustained and global alteration in endogenous mRNA expression after gene therapy should be factored into any interpretation of experimental/clinical outcomes, particularly when introducing factors into the central nervous system that require secretion to evoke functionality.
Introduction
Replication-deficient adeno-associated virus (AAV) or lentivirus vectors are powerful tools for delivering therapeutic genes into nerve cells, providing targeted supply of neuroprotective and/or neurodegenerative molecules to the dysfunctional central or peripheral nervous systems. Viral vectors can be used to introduce genes that encode proteins utilized by the transduced population itself, for example, delivery of genes encoding neurotransmitter-producing enzymes or lysosomal enzymes. Alternatively, vectors can be used to deliver secretable proteins such as neurotrophic factors, which can have an effect not only on transduced cells but also on neighboring nontransduced cell populations.1,2
Some clinical trials have shown efficacy,3–7 showing that the gene therapy approach holds promise for the treatment of neurodegenerative diseases of the brain and spinal cord,8–10 retina,11 glioblastomas,12 and has the potential to enhance repair after neurotrauma to central13,14 and peripheral15,16 nervous systems. AAV has been used to deliver nerve growth factor (NGF) in patients with Alzheimer’s disease, neurturin or glial cell-derived neurotrophic factor in Parkinson’s disease, and insulin-like growth factor 1 in amyotrophic lateral sclerosis.7 However, some of the therapeutic benefits obtained to date using these secreted proteins have been less than optimal, perhaps due to problems with vector delivery/dose and insufficient tissue coverage, immunological reaction, or lack of specificity of the vector/growth factor for appropriate cell types.5,7,17–19
There may be additional factors that influence functional outcomes following vector-mediated growth factor delivery. Using the retina and retinal ganglion cells (RGCs) as a model for central nervous system (CNS) trauma, we previously reported that intraocular injection of AAV (serotype 2) encoding brain-derived neurotrophic factor (BDNF) increased RGC viability after optic nerve injury, while AAV encoding a secretable form of the cytokine ciliary neurotrophic factor (CNTF) enhanced both cell survival and long-distance regeneration of RGC axons.2,20–22 Typical of most AAV studies, we used a strong constitutive promoter to drive transgene expression in the retina, effects that were sustained for over 1 year.23 Others have also provided evidence, in a variety of CNS neurons, for ongoing expression of neurotrophic factors for many months after transduction.1,17,24 Clinical trials in ophthalmology and neurology are ongoing, yet comparatively less is known about the impact of prolonged overexpression of growth factors on the phenotype of neurons and other cells in CNS tissues.
To address this important issue, we first quantified changes to the dendritic fields of regenerating RGCs 5–8 months after intraocular injection of AAV-BDNF or AAV-CNTF.25 There were measurable changes in dendritic architecture in transduced as well as nontransduced RGC populations, changes that could potentially affect connectivity and function. In this study, we set out to determine whether long-term constitutive expression of BDNF or CNTF also alters endogenous gene expression in retinal tissue. Normal adult rats received a single, unilateral intravitreal injection of AAV-green fluorescent protein (GFP) (control vector), or bi-cistronic AAV-BDNF-GFP or AAV-CNTF-GFP. One year later, mRNA was extracted from unfixed retinal tissue and analyzed using custom 96 well polymerase chain reaction (PCR) arrays containing selected genes likely to be influenced by BDNF and/or CNTF. To characterize changes in RGCs more selectively, the subpopulation of retinal neurons most affected by invitreal AAV2 injection,26,27 quantitative PCR (qPCR) on 16 genes was performed using mRNA obtained from fixed retinal sections in which almost all tissue except the ganglion cell layer (GCL) and nerve fiber layer (NFL) had been removed. In summary, widespread, often large, transgene-specific changes in endogenous gene expression were found in retinal tissue, indicating a sustained and complex response to the secreted proteins for at least 1 year after vector administration. The implications of these data for vector design and translational therapies to treat CNS disorders are discussed.
Results
Profiling of gene expression by PCR array for unfixed whole retinal tissue
Retinal mRNA expression in uninjected right eye versus AAV-GFP injected eye.
The right eye is not strictly a control because of the sympathetic ophthalmia phenomenon,28 nonetheless, it was of interest to compare expression in retinal tissue from AAV-GFP eyes with tissue from noninjected eyes 1 year after transduction. For this comparison, results were analyzed using the Qiagen RT2 Profiler PCR Array Data Analysis Rotor-Gene Q Template v1.0 Microsoft excel spreadsheet (see “Methods”). Expression in three noninjected right eyes was compared with five left eye AAV-GFP samples. Seven genes (Creb1, Crebp, Dag1, Itgb1, Nos1, Raf1, and Tfgb1) showed small but significant (P < 0.05, t-test) differences. It is not clear if these changes relate to the injection procedure, the presence of the vector itself or the expression of GFP, but it reinforces the importance of using bi-cistronic BDNF or CNTF vectors that also express GFP in experimental groups when AAV-GFP is used as a control group.
Comparison of gene expression in nasal versus temporal retina—AAV injected eyes.
Numerous earlier studies from our lab have shown that, after intravitreal AAV injection into the lower temporal region of the eye, there are quantifiable differences in the level of transduction in temporal compared with nasal retina.2,23,29,30 In most cases, and based on post-IRES GFP expression amplified using GFP immunohistochemistry, a single injection of 4 µl of a bi-cistronic AAV2 vector results in unequivocal transduction of RGCs in 35–45% of the retina. To determine if this translates into differential changes in endogenous gene expression we compared mRNA levels in nasal and temporal samples for each AAV group. Comparison of the AAV-GFP nasal (n = 5) and AAV-GFP temporal (n = 5) samples revealed significant (P < 0.05, t-test) nasal-temporal differences in 5 of the 93 genes: Arg1, Gria1, Nos1, Nrp1, and Rhoa. In retinas from eyes injected with AAV-CNTF-GFP (nasal n = 5, temporal n = 5) only Tgfb1 differed significantly between nasal and temporal samples. Similarly in AAV-BDNF-GFP retinas (nasal n = 5, temporal n = 5) only Sort1 showed a significant difference in expression. These small differences in gene expression between nasal and temporal retina suggest a strong and widespread paracrine effect from the transduced cells, which in most instances affected gene expression in the nontransduced cell population on the opposite side of the eye as much as the transduced cell population in temporal retina.
Comparison between retinas treated with AAV-GFP, AAV-CNTF-GFP or AAV BDNF-GFP.
Of the 279 comparisons of nasal versus temporal transcript expression, relatively small fold differences were seen in only seven genes, and no gene was consistently altered across the three AAV groups. Therefore, we averaged the two (replicate) expression values for each retina and used these values for further analysis and comparison between each of the treatment groups AAV-GFP (n = 5), AAV-BDNF-GFP (n = 5) and AAV-CNTF-GFP (n = 5). Note here that the nonparametric Mann–Whitney (M-W) comparisons are those used to illustrate results and specifically to compare gene expression between BDNF and CNTF treatment groups (Figures 1 2 3 4 and Figure 6).
Figure 1.
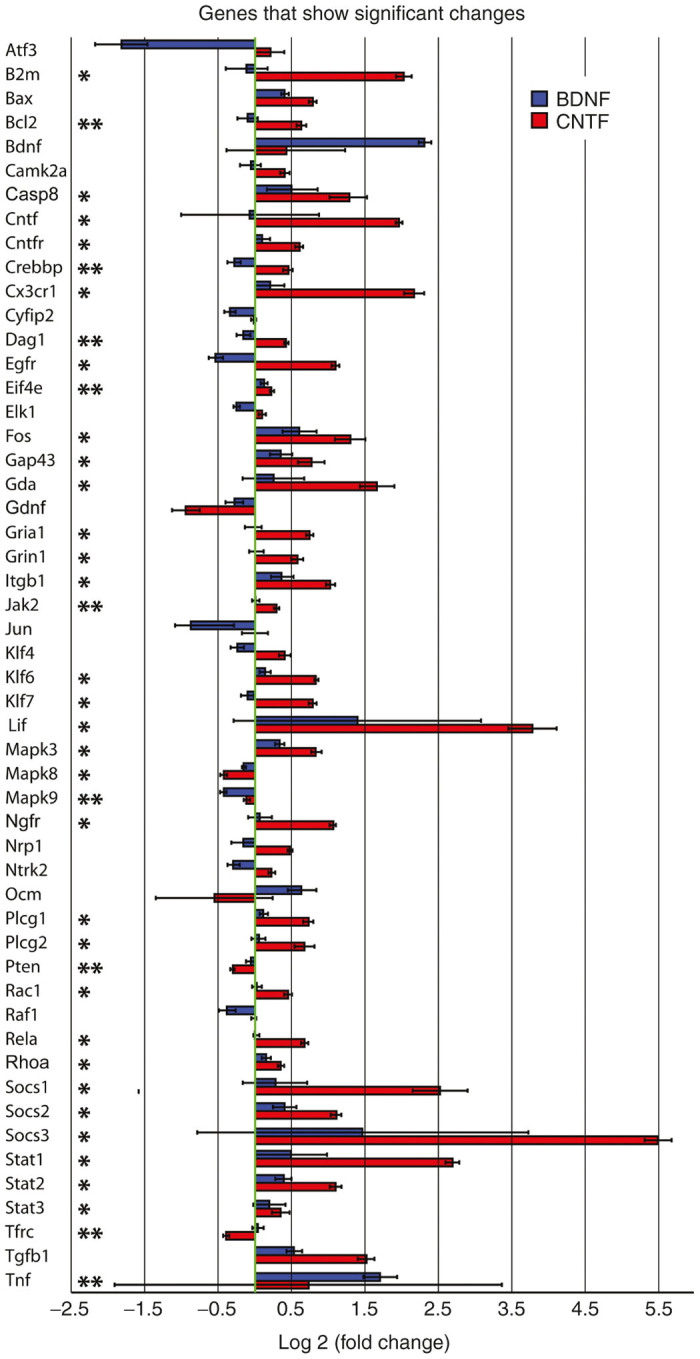
All genes that showed significant expression differences (P < 0.05) between retinal tissue from AAV-GFP injected eyes and tissue from either one or both of the two AAV experimental treatment groups. The bars indicate relative up or down log2 fold changes in expression in AAV-BDNF (blue) and AAV-CNTF (red) injected eyes when compared with AAV-GFP (green) injected eyes. AAV-GFP expression was normalized to zero (green line). The asterisks indicate Mann–Whitney U-test significant differences between AAV-CNTF-GFP and AAV-BDNF-GFP injected eyes (**P < 0.05 and *P < 0.01). AAV, adeno-associated viral; BDNF, brain-derived neurotrophic factor; CNTF, ciliary derived neurotrophic factor; GFP, green fluorescent protein.
Figure 2.
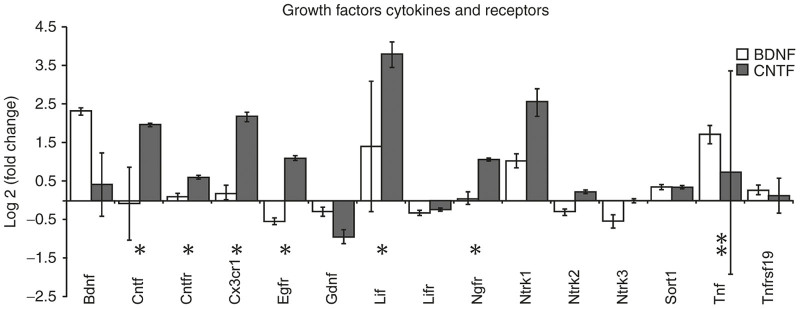
Genes within the Qiagen functional gene grouping of “growth factors, cytokines, and receptors”. The bars indicate relative up or down log2 fold changes in expression in AAV-BDNF (white) and AAV-CNTF (dark) injected eyes when compared with AAV-GFP injected eyes. AAV-GFP expression was normalized to zero. The asterisks indicate Mann–Whitney U-test significant differences between AAV-CNTF-GFP and AAV-BDNF-GFP injected eyes (**P < 0.05 and *P < 0.01). AAV, adeno-associated viral; BDNF, brain-derived neurotrophic factor; CNTF, ciliary derived neurotrophic factor; GFP, green fluorescent protein.
Figure 3.
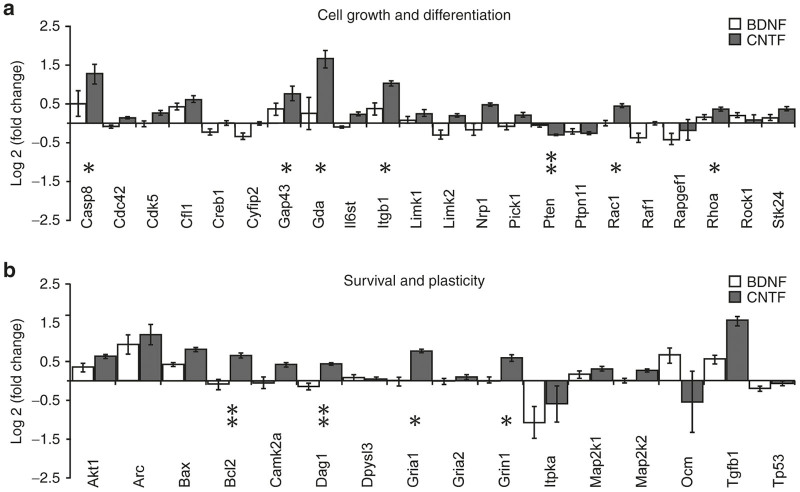
(a) Genes within the Qiagen functional gene grouping of “cell growth and differentiation”. (b) Genes within the Qiagen functional gene grouping of “survival and plasticity”. For both “a” and “b”, the bars indicate relative up or down log2 fold changes in expression in AAV-BDNF (white) and AAV-CNTF (dark) injected eyes when compared with AAV-GFP injected eyes. AAV-GFP expression was normalized to zero. The asterisks indicate Mann–Whitney U-test significant differences between AAV-CNTF-GFP and AAV-BDNF-GFP injected eyes (**P < 0.05 and *P < 0.01). AAV, adeno-associated viral; BDNF, brain-derived neurotrophic factor; CNTF, ciliary derived neurotrophic factor; GFP, green fluorescent protein.
Figure 4.
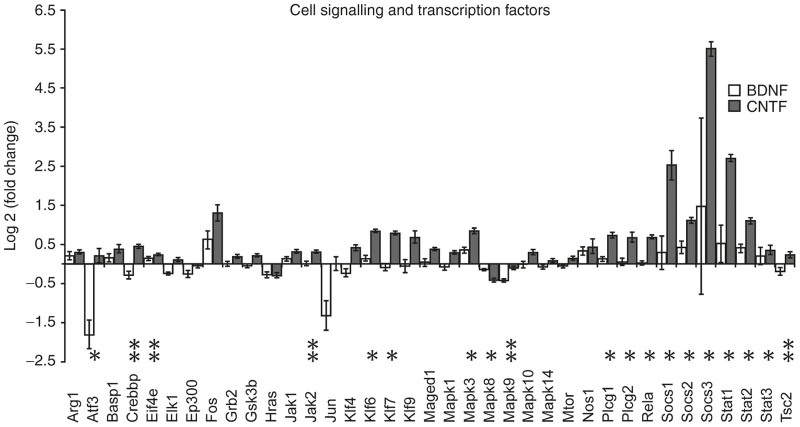
Genes within the Qiagen functional gene grouping of “cell signaling and transcription factors”. The bars indicate relative up or down log2 fold changes in expression in AAV-BDNF (white) and AAV-CNTF (dark) injected eyes when compared with AAV-GFP injected eyes. AAV-GFP expression was normalized to zero. The asterisks indicate Mann–Whitney U-test significant differences between AAV-CNTF-GFP and AAV-BDNF-GFP injected eyes (**P < 0.05 and *P < 0.01). AAV, adeno-associated viral; BDNF, brain-derived neurotrophic factor; CNTF, ciliary derived neurotrophic factor; GFP, green fluorescent protein.
Figure 6.
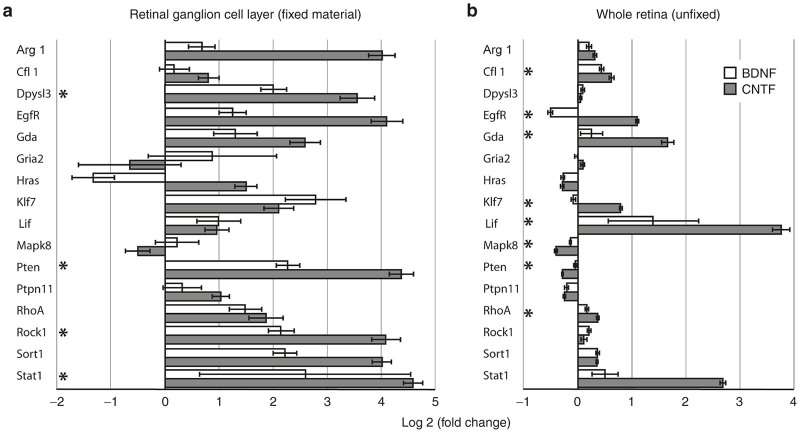
qPCR analysis of gene expression restricted to the enriched retinal ganglion cell layer (a, fixed material) compared with expression in unfixed whole retina (b). The bars indicate relative up or down log2 fold changes in expression in AAV-BDNF (white) and AAV-CNTF (dark) injected eyes when compared with AAV-GFP injected eyes. AAV-GFP expression was normalized to zero. Note significant differences in gene expression between the two samples. AAV, adeno-associated viral; BDNF, brain-derived neurotrophic factor; CNTF, ciliary derived neurotrophic factor; GFP, green fluorescent protein; qPCR, quantitative polymerase chain reaction.
All genes that showed significant expression differences (Kruskal-Wallis - K-W, P < 0.05) between retinal tissue from AAV-GFP injected eyes and tissue from either one or both of the two AAV experimental treatment groups are displayed in Figure 1. The bars indicate relative up or down log2 fold changes in expression in AAV-BDNF-GFP (blue) and AAV-CNTF-GFP (red) injected eyes when compared with AAV-GFP (green) injected eyes. Of the 93 genes tested, expression of 52 genes was significantly altered relative to AAV-GFP, after 1 year of a single intraocular application of the AAV encoding either BDNF or CNTF. Importantly, we observed high expression of the Cntf gene in the AAV-CNTF-GFP samples and high expression of Bdnf gene in the AAV-BDNF-GFP samples, serving as a positive control for the array analyses.
Three times as many genes were altered after CNTF compared with BDNF treatment. Most of these altered genes exhibited an increase in expression, the changes usually greater in retinas exposed to AAV-CNTF-GFP. Down-regulation of genes was occasionally evident, usually in tissue from AAV-BDNF-GFP injected eyes (Figure 1). In most cases, the changes in mRNA expression in retinal tissue 1 year after exposure to AAV-BDNF-GFP or AAV-CNTF-GFP were in the same direction but of different magnitude (e.g., Cntfr, Cx3cr1, Gap43, Gdnf, Klf6, Mapk9, Pten, Rhoa, Socs1, and Stat1). More rarely the changes were in different directions (e.g., Bcl2, Egfr, and Klf7). Overall, significantly different effects of CNTF and BDNF on endogenous retinal gene expression were seen in almost three quarters of the 52 genes with altered expression (M-W, P < 0.05, asterisks, Figure 1).
Ninety of the tested genes were categorized into four groups based on Qiagen functional gene groupings: (i) growth factors, cytokines, and receptors; (ii) cell growth and differentiation; (iii) survival and plasticity and (iv) cell signaling and transcription factors. Relative expression of genes compared with normalized levels in AAV-GFP injected eyes is shown in Figures 2 3 4 5. Of the 15 genes in the growth factor, cytokine and receptor cluster (Figure 2), 10 showed expressions significantly different from normalized AAV-GFP values (Figure 1), and the majority of these were differentially regulated in retinas exposed to BDNF versus CNTF. With the exception of Tnf, which showed substantial variance, increased expression was greatest in AAV-CNTF-GFP injected eyes. Relative to AAV-GFP controls, long-term exposure to BDNF did not significantly alter the expression of Cntf, Cntfr, Cx3cr1 or Ngfr genes, but Ntrk2 and Ntrk3 were down-regulated.
Figure 5.
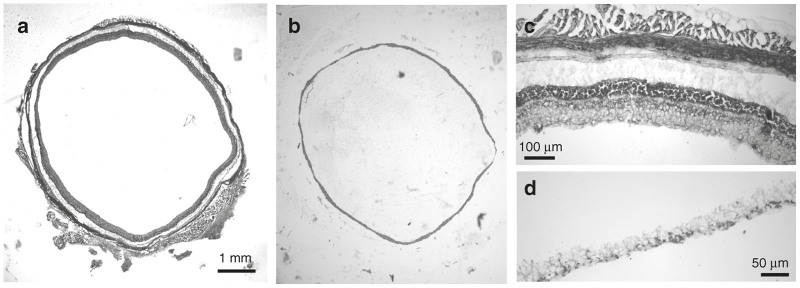
(a) Section of retina on a slide, prior to removal of all but the retinal ganglion cell layer (b). (c) high power view of cross-section through retina. (d) retinal ganglion cell layer and nerve fiber layer after removal of other retinal layers. Scale bars as shown.
Twenty-two genes were allocated to the cell growth and differentiation group (Figure 3a), but in this case expression of only 10 differed from GFP controls, and seven of these displayed differential expression in retinal tissue from eyes injected with AAV-BDNF-GFP versus AAV-CNTF-GFP. These genes were Casp8, Gap43, Gda, Itgb1, Pten, Rac1, and Rhoa. Again, increased expression was most commonly seen after CNTF treatment. There was significant down-regulation of Cyfip2, Limk2, and Raf1 in AAV-BDNF-GFP injected eyes, and down-regulation of Pten after chronic CNTF but not BDNF exposure. One year after AAV exposure, expression of 7 out of 15 “survival and plasticity” genes remained altered in retinal tissue (Figure 3b), but in only three genes (Bcl2, Gria2, and Grin1) did expression significantly differ between BDNF and CNTF eyes.
The largest Qiagen functional cluster comprised genes associated with cell signaling and transcription factors (38 genes, Figure 4). Twenty-two of these genes showed significantly different expression from AAV-GFP controls (Figure 1), and in 18 cases, the expression levels differed in retinal tissue from AAV-BDNF-GFP versus AAV-CNTF-GFP injected eyes. Increased levels of mRNA were most common, once again, usually in retinas from AAV-CNTF-GFP injected eyes. Genes sensitive to CNTF but not BDNF included Dag1, Eif4e, Klf4, Klf6, Klf7, Klf9, Plcg1, Plcg2, and Rela. Genes significantly down-regulated by BDNF relative to control AAV-GFP were Atf3, Crebpp, Jun, and Mapk9. Overall, these data indicate ongoing and substantial changes in the regulation of cellular function 1 year after initial exposure to AAV mediated release of neurotrophic factors.
Data were also examined using Gene Ontology (GO) software (http://go.princeton.edu).31 We used GO to identify gene products defined in terms of their associated biological processes. A biological process is the series of events accomplished by one or more ordered assemblies of molecular functions. The 12 differentially expressed genes from AAV-BDNF-GFP eyes and 45 differentially expressed genes from AAV-CNTF-GFP injected eyes were used for analysis. The genes that were differentially regulated compared with AAV-GFP controls were found to affect a wide-range of processes and functions. After AAV-BDNF-GFP or AAV-CNTF-GFP exposure, 11 and 12 biological processes respectively were linked to changes in at least 50% of the differentially expressed retinal genes (Table 1). Processes are ranked from top to bottom reflecting the total number of differentially regulated genes captured by each process. The nature of these biological processes was remarkably similar between groups, and included likely effects on signal transduction, cell differentiation, response to stress, biosynthesis and metabolism, and cell death.
Table 1. Data were examined using Gene Ontology (GO) analysis software to identify gene products defined in terms of their associated biological processes. Only biological processes linked to changes in at least 50% of the differentially expressed retinal genes are shown. Figures in brackets indicate number of altered genes in each category.
| AAV-BDNF-GFP (12 genes) | AAV-CNTF-GFP (45 genes) |
|---|---|
| Signal transduction (12) | Anatomical structure development (41) |
| Cell differentiation (11) | Signal transduction (38) |
| Anatomical structure development (11) | Cell differentiation (36) |
| Response to stress (10) | Response to stress (34) |
| Cellular protein modification process (10) | Transport (27) |
| Biosynthetic process (9) | Cell death (27) |
| Cellular nitrogen compound metabolic process (9) | Biosynthetic process (26) |
| Cell death (9) | Cellular nitrogen compound metabolic process (25) |
| Cell proliferation (8) | Cell proliferation (24) |
| Transport (7) | Cellular protein modification process (24) |
| Cellular component assembly (6) | Locomotion (24) |
| Immune system process (23) |
AAV, adeno-associated viral; BDNF, brain-derived neurotrophic factor; CNTF, ciliary derived neurotrophic factor; GFP, green fluorescent protein.
Further qPCR analysis of gene expression in the enriched retinal GCL
Finally, we wished to assess the long-term effect of vector mediated expression of CNTF or BDNF on altered gene expression that was more localized to the retinal GCL, where most of the AAV transduction occurred.26,27 To do this we isolated the GCL and NFL from fixed frozen retinal sections (Figure 5) and then quantitatively analyzed the expression of 16 genes (see “Methods” for details). These genes were selected (Table 2) based on several factors, primarily choosing genes with interesting expression profiles in the whole retina samples, genes differentially expressed in neurons versus glia and/or genes of particular relevance to RGC plasticity. The results are shown in Figure 6a as fold changes relative to normalized AAV-GFP injected eyes; for comparison, expression levels for the same genes from the (unfixed) whole eye data base (Figure 1) are also shown (Figure 6b).
Table 2. qPCR primers designed and optimized in house for use on fixed tissue GCL samples.
| Gene Symbol | Refseq # | Product size | Temp | Forward primer | Reverse primer |
|---|---|---|---|---|---|
| Arg1 | NM_017134 | 93 bp | 60°C | ACG AAA CGG GAA GGT AAT CA | TGA TGC CCC AGA TGA CTT TT |
| Cfl1 | NM_017147 | 99 bp | 58°C | GGAGATTCTGGTAGGAGATGTGGG | CATAGAGAGCATAGCGGCAGTCC |
| Dpysl3 | NM_012934 | 128 bp | 60°C | GCGCATTAAAGCAAGGAGGA | GAGAGCCTCGAGTAGAGCCA |
| Egfr | NM_031507 | 290 bp | 60°C | GCACTCCTCCTCTAGACCCA | TCTCCTTGAGGGAACGCAAC |
| Gda | NM_031776 | 166 bp | 60°C | ATTTGGGCAGCGAGCATTTG | AAATCGCGGGGTCACTATGG |
| Gria2 | NM_017261 | 295 bp | 62°C | GTGAGGACTACCGCAGAAGG | TACTTCCCGAGTCCTTGGCT |
| Hras | NM_001098241.1 | 94 bp | 60°C | ACACCAAGTCCTTTGAAGACATCC | TTGTTGCCCACCAGCACCATTG |
| Klf7 | NM_001108800 | 119 bp | 60°C | AGACCCACACACATACCCACTGT | AGTTCACCTGGCTTCCTCCTTC |
| Lif | NM_022196 | 161 bp | 60°C | CCT CCC ATC ACC CCT GTA AA | ACG TTG TTG GGA AAT GGT TC |
| Mapk8 | NM_053829.1 | 94 bp | 60°C | GGGCTACAAGGAGAACGCTG | GGACGCATCTATCACCAGCA |
| Pten | NM_031606 | 141 bp | 60°C | TTGAAGACCATAACCCACCACAG | CACAAATCATTACACCAGTCCGTC |
| Ptpn11 | NM_013088 | 81 bp | 60°C | CAGACCTGGTGGAGCATTACAAG | TGTTGAGGGGCTGTTTGAGC |
| Rhoa | NM_057132 | 80 bp | 60°C | GGTTTATGTGCCCACGGTGTT | CCCATAAAGCCAACTCTACCTGC |
| Rock1 | NM_031098 | 159 bp | 60°C | CCTGATGAGCAACTACGATGTGC | TCGGCTAACTTCAAATGTCCAGAC |
| Sort1 | NM_031767 | 111 bp | 60°C | CAATAACACCTTCATTCGGACGG | AACACTCTTCCTCCTCGGCTTC |
| Stat1 | NM_032612 | 90 bp | 60°C | GAAAAGCAAGCGTAATCTCCAGG | TTCCTCTCCTCCTTCAGACAGTTG |
| PPIA | NM_017101 | 120 bp | 62°C | AGC ATA CAG GTC CTG GCA TC | TTC ACC TTC CCA AAG ACC AC |
| TBP | NM_001004198 | 183 bp | 60°C | GCC TGC GGC TGC TC GTT TTG | TGG GGA GGC CAA GCC CTG AG |
| YWHAZ | NM_013011 | 74 bp | 61°C | GAC GGA GCT GAG GGA CAT CTG C | GGC TGC GAA GCA TTG GGG ATC A |
| SDHA | NM_130428 | 134 bp | 60°C | TGG GGC GAC TCG TGG CTT TC | CCC CGC CTG CAC CTA CAA CC |
| HPRT | XM_003752155 | 186 bp | 60°C | ACCCTCAGTCCCAGCGTCGT | GGCCACAATGTGATGGCCTCCC |
bp, base pair; GCL, ganglion cell layer; qPCR, quantitative polymerase chain reaction.
Note that in the enriched GCL from AAV-BDNF-GFP and AAV-CNTF-GFP injected eyes, mRNA expression levels were generally increased relative to control, and the extent of these changes was in most cases much greater than that seen for whole retina. A clear exception was the Lif gene. Significant differences between AAV-BDNF-GFP and AAV-CNTF-GFP injected eyes were less frequently encountered in the enriched GCL data (asterisks, Figure 6a) compared with whole retina (Figure 6b). Relative to normalized AAV-GFP control levels, expression of only one gene (Klf7) was significantly increased in BDNF retinas, whereas 9 of the 16 genes (Arg1, Dpysl3, EgfR, Hras, Pten, Ptpn11, ROCK1, Sort1, and Stat1) were significantly altered after prolonged CNTF exposure. Occasionally, mRNA expression was increased in genes (e.g., Pten and Ptpn11) in the enriched GCL layer sample but expression of these genes was decreased in the whole retinal material.
Discussion
Previous studies in this laboratory applying AAV2 therapy to rats after optic nerve crush or in rats with a peripheral nerve grafted onto the cut optic nerve found that the efficacy of vector-mediated neurotrophic support of injured RGCs varied depending on the level of transduction in different parts of the retina.2,23,30 RGC survival and regeneration capability was consistently shown to be significantly higher in transduced compared with nontransduced areas. Differences were especially evident when using AAV2 encoding intracellular signaling molecules such as SOCS3 (ref. 29); intravitreal injection of AAV-SOCS3 and subsequent expression of SOCS3 protein almost completely suppressed ganglion cell regeneration in transduced retinal areas, but had little impact in nontransduced regions. In this study, in normal retinas, because our intravitreal AAV injections were made into lower temporal retina, we first compared differences in expression of endogenous retinal genes in retinal areas with high (temporal) versus low (nasal) transduction levels. Surprisingly, and in direct contrast to previous studies,2,23,29,30 very few differences in expression were seen, indicating a more global, retina-wide response to the ongoing cellular release of BDNF or CNTF.
After 1 year of a single intravitral application of either AAV-BDNF-GFP or AAV-CNTF-GFP, we saw many changes in endogenous retinal gene expression compared with AAV-GFP injected eyes, changes that in most cases also differed between BDNF and CNTF. We also observed several small but significant changes in gene expression between left (AAV-GFP injected) and right (noninjected) eyes. Expression levels of GFP were almost certainly higher in AAV-GFP injected retinas compared with post-IRES GFP expression,2,32 nonetheless, using similar AAV2 vectors to those used here, we previously showed that in transduced retinal areas the observed RGC transduction efficiencies for AAV-CNTF-GFP and AAV-BDNF-GFP were similar to those for AAV2-GFP (20 versus 25%).2 In any case, any hypothetical impact of differential levels of GFP on gene expression profiles would have been minimal in AAV-BDNF-GFP versus AAV-CNTF-GFP injected eyes, given that in both cases the GFP was post-IRES.
A greater number of genes was affected after prolonged CNTF exposure, usually involving a significant up-regulation. On the other hand, BDNF expression influenced fewer genes and the changes mostly involved down-regulation. Many of the gene expression changes were associated with cell signaling and transcription factors, as well as cell growth and differentiation. GO analysis provided confirmation of the wide-ranging nature of the gene changes, impacting on many biological processes including signal transduction, cell differentiation, response to stress, biosynthesis, metabolism, and cell death. These effects were more diverse than might be expected from known signaling effects of these two neurotrophic factors, and it seems likely that, after prolonged vector-mediated expression and secretion of neurotrophic factors, there were flow-on effects within and between different retinal cell types resulting in complex cascades of altered gene expression across the retina. The consequences of ongoing transgene expression after vector-based therapy can thus extend well beyond the immediate transduction site, and beyond the cell types initially targeted by a vector.
The arrays obtained from whole retina did not allow us to ascribe changes to signaling or biological processing to a particular cell type. Nonetheless, with regard to the cytokine and receptor cluster it is worth noting the significant and selective effects of AAV-CNTF-GFP on increasing expression of various growth factor and cytokine/chemokine receptor genes such as Cntfr, Cx3cr1, Egfr, and Ngfr. Expression levels of genes encoding other members of the cytokine family (LIF and Tgfb1) were also increased. AAV-BDNF on the other hand did not affect Ngfr gene expression but chronically reduced expression of the Egfr, Ntrk2, and Ntrk3 genes, the latter two genes encoding trkB and trkC receptors respectively. BDNF induced reduction in trkB mRNA expression has been reported previously in both normal and injured RGCs.33
Expression of a number of genes associated with cell growth and differentiation was increased, again mostly in retinas from AAV-CNTF-GFP injected eyes. Exceptions were the down-regulation of Limk2 and Raf1 in AAV-BDNF-GFP injected eyes. The products of many of the affected genes have been reported to be associated in some way with the cytoskeleton and/or Rho signaling (e.g., Cfl1, Gap43, Gda, Itgb1, Limk2, Pten, Rac1, and Rhoa). This is of particular interest because AAV-CNTF and AAV-BDNF have been shown to have different effects on axonal sprouting and long-distance regeneration,2,23 and we have previously shown that prolonged vector-mediated expression of BDNF or CNTF alters the dendritic architecture of RGCs.25
The group defined as “cell signaling and transcription factors” contained many genes with altered expression. After CNTF treatment, large increases in both Socs and Stat gene expression were seen with smaller but nonetheless significant increases in Jak1 and Jak2 mRNA levels. These are all genes known to be influenced by CNTF signaling through its cognate receptor, however even in AAV-BDNF-GFP exposed retinas there were significant (albeit smaller) increases in most Socs and Stat genes, presumably reflecting complex intracellular and intercellular effects arising from initial chronic BDNF production and release.
To look more closely at altered gene expression in RGCs, we developed a protocol in fixed retinal sections that allowed us to measure mRNA in only the inner part of the retina. This is still not exclusively an RGC population, due to the presence of displaced amacrines and some astrocytes in NFL, but the relative proportion of RGCs is considerably enriched in such samples. Overall, for the 16 genes tested, in both AAV-BDNF-GFP and AAV-CNTF-GFP injected eyes gene expression changes relative to AAV-GFP controls were much greater in enriched GCL samples compared with whole retina, perhaps to be expected given that the highest level of AAV2 transduction is found in this layer.26,27 Irrespective of the growth factor applied, gene expression was generally increased, although significant changes relative to normalized AAV-GFP controls were almost all confined to tissue from AAV-CNTF-GFP injected eyes.
Important differences in the expression of specific genes were seen in whole retina versus enriched GCL samples. For example, Lif expression was considerably less in the GCL analysis, presumably because Müller glia was absent. Of interest, 1 year after AAV-CNTF-GFP administration the Pten gene was down regulated in the whole retinal sample but was significantly upregulated in the enriched GCL sample. The protein PTEN (phosphatase and tensin homologue) via its actions in inhibiting PI3K/mTOR (mammalian target of rapamycin) pathway (and perhaps also Akt and GSK-3) has a repressive effect on RGC axonal growth, and in transgenic mice PTEN deletion promotes regenerative growth of RGC axons after optic nerve injury.34,35 The presence of AAV-CNTF-GFP in particular also resulted in increased expression of Rock1, a major activator of PTEN protein. EGFr expression was also increased, perhaps in GCL-resident astrocytes,36 as was the Dpysl3 gene, also known as the Crmp4 gene, a mediator of inhibitory growth signals.37 Given concomitant activation of growth stimulators (e.g., Arg1, Hras, and Stat1), the prolonged upregulation of a number of growth suppressor genes, including the Socs genes,29,38 in the enriched GCL sample suggests that cells are attempting to offset the continual growth-stimulating effects of chronic CNTF secretion. Another growth-promoting gene in RGCs (Kfl7)39 was upregulated in AAV-BDNF-GFP injected eyes, again an effect perhaps offset by a trend in these enriched RGC samples for increased Rock1 and Pten expression.
In conclusion, in addition to changes in dendritic architecture,25 we now show that ongoing AAV-mediated expression of BDNF or a secretable form of CNTF both result in widespread, long-lasting changes in endogenous gene expression in adult rat retinal tissue. Fold changes in the enriched GCL were greater than in whole retinal samples, and generally greater after AAV-CNTF exposure, strongly suggesting that cells were reacting against the persistent growth stimulus provided by the enhanced presence of transgenic neurotrophic proteins. With specific regard to retinal function, while we recently obtained evidence of recovery of some basic visual responses in AAV-CNTF injected rats with peripheral nerve bridges linking the transected optic nerve to visual midbrain,22 long-term gene therapy induced changes such as those described here and elsewhere25 almost certainly have an impact on retinal physiology. For example, others have shown that intraocular CNTF can adversely affect photoreceptor function,18,19,40,41 the effect apparently being dose-dependent,19,41 and CNTF induced upregulation of Socs3, although not as great when using vectors compared with recombinant protein,29,42 may also indirectly affect retinal vasculature.43
In the context of more general use of AAV in the clinic, long-term follow-ups indicate that recombinant AAV transduction and neurotrophic factor expression in the human nervous system is not, a priori, harmful.10,18 Nonetheless we argue that, in the absence of mechanisms for regulating transgene expression, sustained, complex and global alteration in endogenous mRNA expression after gene therapy in the CNS is a phenomenon that should be factored into any planned study and/or interpretation of functional neurological outcomes, particularly when using factors that require secretion to elicit their bioactivity.
Materials and Methods
Animals and intravitreal injections
A total of 36 female Wistar rats were used for these experiments (8–10 weeks old, Animal Resources Centre, Western Australia, Australia). For the whole eye experiment, 17 animals where used. Animals were allocated to three groups: AAV2-GFP (n = 6), AAV2-BDNF-GFP (n = 6), and AAV2-CNTF-GFP (n = 5). Nineteen animals were used for the GCL study: AAV2-GFP (n = 7), AAV2-BDNF-GFP (n = 8), and AAV2-CNTF-GFP (n = 4). All animal experimentation was approved by The University of Western Australia Animal Ethics Committee and conformed to the NHMRC guidelines. Animals were maintained on a 12 hour day/night cycle and supplied with food and water ad libitum throughout the experiment. Rats were anesthetized with a 1:1 mixture of ketamine (100mg/ml) and xylazine (20 ng/ml) (1–1.5 ml/kg bw, i.p.). Intravitreal AAV eye injections were carried out using a glass micropipette inserted through the sclera as described previously.26 In each rat, the vector injection was made into the lower temporal region of the left eye (4 µl volume). The GFP control AAV2 was generated from the pTRUF12 (GTC Virus Vector Core, NC) backbone and AAV2-BDNF and AAV2-CNTF vectors were generated from pTRUF12.1 plasmids (donated by Joost Verhaagen, Amsterdam, Netherlands) that contain a shorter promoter, previously described and tested in detail.2 The cytomegalovirus-chicken beta actin (CMV-CBA) promoter allows sustained transgene expression in transduced retinal cells.23,27 AAV2-CNTF-GFP (1 × 1012 GC/ml), AAV2-BDNF-GFP (8 × 1012 GC/ml), and AAV2-GFP (8 × 1012 GC/ml) were obtained from Gene Therapy Center Virus Vector Core Facility (University of North Carolina, Chapel Hill, NC).
Collection of whole retinal tissue
One year after the intravitreal injection of AAV2-GFP, AAV2-CNTF-GFP or AAV2-BDNF-GFP, animals were overdosed with an IP injection of sodium pentobarbitone (Lethabarb, Virbac, New South Wales, Australia; 200 μl/100 g). To ensure minimal RNase activity and optimal RNA quality, fast and clean techniques were used. Retinas were dissected out from left eye cups and were further divided into nasal and temporal halves. In some rats, retinas from the right (uninjected) eye were also collected. Retinas were then embedded in optimal cutting temperature (OCT) freezing media (Electron Microscopy Sciences, Hatfield, PA) snap frozen in liquid nitrogen and then stored at −80°C.
RNA isolation and cDNA synthesis of whole retinal tissue
Retinal tissues were washed in RNase free phosphate buffered saline (PBS) to remove OCT and immediately placed in RNA extraction buffer. Total RNA was isolated using the RNeasy Mini kit (Qiagen, Australia). Contaminating genomic DNA was eliminated by treatment with RNase-Free DNase Set (Qiagen). 250 ng of RNA was utilized for each reverse transcriptase reaction using the RT2 First strand kit (Qiagen) according to manufacturer protocol. The final number of usable RNA samples were AAV2-GFP (nasal n = 5, temporal n = 4), AAV2-BDNF-GFP (nasal n = 4 and temporal n = 5), and AAV2-CNTF-GFP (nasal n = 4 and temporal n = 5). RNA from retinal tissue for three uninjected right eyes was also isolated.
PCR array and statistical analysis of whole retinal tissue
A Custom SABiosciences 96 genes PCR array system (Custom RT Profiler array CAPR 10704R) (Qiagen) was used which included a rat genomic DNA contamination control, reverse transcription control, and a positive PCR control (Supplementary Table S1). The genes were categorized into four groups based on Qiagen functional gene groupings: (i) growth factors, cytokines, and receptors; (ii) cell growth and differentiation; (iii) survival and plasticity, and (iv) cell signaling and transcription factors. The PCR reactions were carried out using RT2 SYBR Green ROX FAST Mastermix (Qiagen) according to the manufacturer’s protocols. Arrays were run on a Corbett Rotor Gene 6000 instrument over the course of 2 weeks, using the recommended settings. RNA was considered DNA-free when 40 cycles of real-time PCR did not give an amplification signal for the genomic DNA. All genes were detected and successfully amplified with the exception of Ocm in sample “CNTF rat 5, nasal left eye”; while Lif and Ntrk1 in sample “GFP rat 3, temporal left eye”. Further outliers were identified using qBase+ (Biogazelle, Ghent, Belgium) and excluded from statistical analysis: “Bdnf in BDNF rat 1, nasal” and “BDNF rat 1 temporal”, and “Socs3 in CNTF rat 2, nasal”.
Genorm44 used to find genes with the highest stability (M), using the qBase+ software package (Biogazelle). For each comparison the highest six stable genes were selected. For comparison and confirmation of selected reference genes BestKeeper version 1 Microsoft Excel spreadsheet was also used.45 Subsequently data were normalized with the verified reference genes Tfrc, Gsk3b, Jak2, and Rpl13a.
The results from the samples and selected housekeepers were used in the Qiagen RT2 Profiler PCR Array Data Analysis Rotor-Gene Q Template v1.0 (September 10, 2010) Microsoft excel spreadsheet. This spreadsheet was used to generate a QC report on each array. Fold change (2−ΔΔCT) were normalized using Tfrc, Gsk3b, Jak2, and Rpl13a (with a Genorm M-value <0.03). The P-values calculated in the Qiagen RT2 Profiler PCR Array Data Analysis Rotor-Gene Q Template v1.0 were based on a Student’s t-test.
In further analysis the geometric mean of the expression for all genes was calculated relative to normalized expression data as outlined. Because of the significant involvement of the Jak2 gene in both BDNF and CNTF pathways, the Grb2 (Genorm M-value of 0.034) gene was used as an alternative normalization gene to compare expression changes. In comparison analysis the relative expression was calculated using the ddCt method.45 All statistical tests were performed using SPSS Statistics 19.0 (IBM, Armonk, NY). Left and right eye comparisons and nasal versus temporal comparisons were analyzed using t-tests. Changes between all three groups were assessed by Kruskal–Wallis (nonparametric) and differences between paired groups were assessed by Mann–Whitney U-tests (nonparametric) (significance levels *P < 0.05, **P < 0.01). Fold-changes of the genes were calculated as mean values of 2−∆∆CT or 1/2−∆∆CT relative to controls and were converted to log2 fold changes for use in graphs.46
After qBase software normalization using the same housekeeper genes as those used previously for nasal-temporal comparisons, data were analyzed using the nonparametric (ranked based) Kruskal–Wallis H-test for effect of treatment for each gene, followed by the nonparametric Mann–Whitney U-test for pairwise comparisons between treatments for significantly affected (P < 0.05) genes.
Perfusions and tissue collection of GCL and NFL
Nineteen animals were used for the enriched GCL study: AAV2-GFP (n = 7), AAV2-BDNF-GFP (n = 8), and AAV2-CNTF-GFP (n=4). One year after the animals received the gene therapy injection the animals were overdosed with an IP injection of Lethabarb (sodium pentobarbitone) and perfused with PBS (pH 7.4) containing 0.1% heparin followed by 4% paraformaldehyde in 0.1 mol/l phosphate buffer (pH 7.4). From this stage onwards techniques were set in place to minimize RNase activity. Retinas were dissected out from the eyecup and postfixed for 1 hour in 4% paraformaldehyde, then washed for 10 minutes in RNase free PBS. Tissue was snap frozen in OCT freezing media using liquid nitrogen and stored at −80°C for cryosectioning. To isolate the GCL and NFL, each retinal sample was cryosectioned at 16 µm on a cryostat (Leica, Germany). Sections were placed on Superfrost (Lomb Scientific, Australia) glass slides and stored at −80°C until needed. Slides were defrosted at room temperature for 15 minutes. To assist in visualization, sections were first washed in PBS and then in 95% EtOH before staining in a fresh 70:30 mixture of 2% Cresyl Violet in 95% EtOH or 0.5% Eosin Y in 95% EtOH, and finally rinsed in 95% EtOH and 100%EtOH. With the aid of a light microscope (Wild Heerbrugg, Switzerland) any remaining choroid and the outer layers of the eye were removed using a narrow blade, leaving essentially the inner RGC layer and innermost NFL. These layers were then scraped off the slides and placed in PDK solution (RNeasy FFPE kit, Qiagen) for RNA extraction.
RNA extraction and cDNA sample preparation of enriched retinal GCL
Total RNA was extracted from the remaining enriched GCL sample using the RNeasy FFPE kit (Qiagen), according to the manufacturer’s directions, which included treatment with recombinant DNase I (rDNase I; DNA-free; Ambion). The total RNA was eluted in 10 µl of RNase-free water and placed on ice before the RNA concentration and quality was assessed using the Nanodrop spectrophotometer (BioLab Group, Australia). RNA concentrations varied between 7 and 100 ng/ µl, with 260/280 ratio measurements between 1.8 and 2.10. Total RNA (up to 500 ng) was used to synthesize cDNA using the QuantiTect Reverse Transcription kit (Qiagen) according to the manufactures instructions and using random primers. The 20 µl reactions were purified using a PCR purification kit (MoBio) and the cDNA was eluted in 20 µl of sterile water. The resulting reactions were stored at −20°C until required for qPCR.
Quantitative RT-PCR and statistical analysis of enriched retinal GCL
Analyses of expression levels for gene transcripts were performed by qRT-PCR on the Rotorgene 6000 (Corbett Industries, Sydney, Australia) using IQ SYBR Green Supermix (BioRad). Primers for these target genes and two reference genes (Ppia and Hprt) were designed using Primer 3 software (MIT/Whitehead Institute, http://www-genome.wi.mit.edu) (Table 2). Each primer pair was positioned to span two exons thus ensuring no product was amplified from genomic DNA. Primers (Geneworks, Adelaide, South Australia) were diluted to 5 µmol/l working solutions. Each reaction was set up using 5 μl iQ SYBR 2X Mastermix (Bio-Rad, Gladesville, Australia) 1 μl of each forward and reverse primer, 1 μl cDNA template (20 ng cDNA per reaction) and 2 μl double deionized water (DDW).
Cycling conditions included an initial denaturation of 95°C for 10 minutes to activate the enzyme, followed by amplification of 45 cycles (denaturation at 95°C for 1 second, 15 seconds at the appropriate annealing temperature, and extension at 72°C for 5 seconds). All runs were repeated and values for each sample were averaged. Standard curves for each product were generated from gel-extracted (QIAEX II; Qiagen) PCR products using 10-fold serial dilutions and the Rotorgene 6000 software using the inbuilt second derivative maximum (SDM) equation. Specificity was ensured by single peak melting curves with single bands after gel electrophoresis and sequencing product used as standard. Cq values were exported from the Rotor-Gene software into Excel and analyzed. All samples were standardized against the two reference genes using the GeNorm algorithm as previously described44 and further analyzed using the ΔCt method.
Normalized data were analyzed for fold changes between groups by using SPSS Statistics 19.0 (IBM) software. A two tailed, unpaired student’s t-test was first used to compare the two different rAAV injection groups (AAV-CNTF and AAV-BDNF). Further statistics were performed to test for homogeneity of variance and data were statistically analyzed using a multigroup comparison Kruskal–Wallis test and then compared pair-wise using the nonparametric Mann–Whitney U-test (significance levels *P < 0.05, **P < 0.01).
Acknowledgments
This work was supported a grant from the WA Neurotrauma Research Program. We thank Marissa Penrose for assistance with preparation of the figures.
The authors declare no conflict of interest.
References
- Klein, RL, Muir, D, King, MA, Peel, AL, Zolotukhin, S, Möller, JC et al. (1999). Long-term actions of vector-derived nerve growth factor or brain-derived neurotrophic factor on choline acetyltransferase and Trk receptor levels in the adult rat basal forebrain. Neuroscience 90: 815–821. [DOI] [PubMed] [Google Scholar]
- Leaver, SG, Cui, Q, Plant, GW, Arulpragasam, A, Hisheh, S, Verhaagen, J et al. (2006). AAV-mediated expression of CNTF promotes long-term survival and regeneration of adult rat retinal ganglion cells. Gene Ther 13: 1328–1341. [DOI] [PubMed] [Google Scholar]
- Tuszynski, MH, Thal, L, Pay, M, Salmon, DP, U, HS, Bakay, R et al. (2005). A phase 1 clinical trial of nerve growth factor gene therapy for Alzheimer disease. Nat Med 11: 551–555. [DOI] [PubMed] [Google Scholar]
- Lipinski, DM, Thake, M and MacLaren, RE (2013). Clinical applications of retinal gene therapy. Prog Retin Eye Res 32: 22–47. [DOI] [PubMed] [Google Scholar]
- Bartus, RT, Weinberg, MS and Samulski, RJ (2014). Parkinson’s disease gene therapy: success by design meets failure by efficacy. Mol Ther 22: 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koilkonda, RD, Yu, H, Chou, TH, Feuer, WJ, Ruggeri, M, Porciatti, V et al. (2014). Safety and effects of the vector for the Leber hereditary optic neuropathy gene therapy clinical trial. JAMA Ophthalmol 132: 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire, CA, Ramirez, SH, Merkel, SF, Sena-Esteves, M and Breakefield, XO (2014). Gene therapy for the nervous system: challenges and new strategies. Neurotherapeutics 11: 817–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor, DM and Boulis, NM (2015). Gene therapy for neurodegenerative diseases. Trends Mol Med 21: 504–512. [DOI] [PubMed] [Google Scholar]
- Scarrott, JM, Herranz-Martín, S, Alrafiah, AR, Shaw, PJ and Azzouz, M (2015). Current developments in gene therapy for amyotrophic lateral sclerosis. Expert Opin Biol Ther 15: 935–947. [DOI] [PubMed] [Google Scholar]
- Marks, WJ Jr, Baumann, TL and Bartus, RT (2016). Long-term safety of patients with Parkinson’s disease receiving rAAV2-Neurturin (CERE-120) gene transfer. Hum Gene Ther 27: 522–527. [DOI] [PubMed] [Google Scholar]
- Trapani, I, Banfi, S, Simonelli, F, Surace, EM and Auricchio, A (2015). Gene therapy of inherited retinal degenerations: prospects and challenges. Hum Gene Ther 26: 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann, G, Ozduman, K and van den Pol, AN (2012). Oncolytic virus therapy for glioblastoma multiforme: concepts and candidates. Cancer J 18: 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Yepes, J, Zavala-Flores, L, Anandhan, A, Wang, F, Skotak, M, Chandra, N et al. (2014). Antioxidant gene therapy against neuronal cell death. Pharmacol Ther 142: 206–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, K, Nakajima, H, Guerrero, AR, Johnson, WE, Masri, WE and Baba, H (2014). Gene therapy strategies for the treatment of spinal cord injury. Ther Deliv 5: 591–607. [DOI] [PubMed] [Google Scholar]
- de Winter, F, Hoyng, S, Tannemaat, M, Eggers, R, Mason, M, Malessy, M et al. (2013). Gene therapy approaches to enhance regeneration of the injured peripheral nerve. Eur J Pharmacol 719: 145–152. [DOI] [PubMed] [Google Scholar]
- Godinho, MJ, Teh, L, Pollett, MA, Goodman, D, Hodgetts, SI, Sweetman, I et al. (2013). Immunohistochemical, ultrastructural, and functional analysis of axonal regeneration through peripheral nerve grafts containing Schwann cells expressing BDNF, CNTF or NT3. PLoS One 8: e69987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K, Klein, RL, Meyers, CA, King, MA, Hughes, JA, Millard, WJ et al. (2003). Long-term neuronal effects and disposition of ectopic preproNGF gene transfer into the rat septum. Hum Gene Ther 14: 1463–1472. [DOI] [PubMed] [Google Scholar]
- Zein, WM, Jeffrey, BG, Wiley, HE, Turriff, AE, Tumminia, SJ, Tao, W et al. (2014). CNGB3-achromatopsia clinical trial with CNTF: diminished rod pathway responses with no evidence of improvement in cone function. Invest Ophthalmol Vis Sci 55: 6301–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch, PK, MacLaren, RE, Durán, Y, Balaggan, KS, MacNeil, A, Schlichtenbrede, FC et al. (2006). In contrast to AAV-mediated Cntf expression, AAV-mediated Gdnf expression enhances gene replacement therapy in rodent models of retinal degeneration. Mol Ther 14: 700–709. [DOI] [PubMed] [Google Scholar]
- Leaver, SG, Cui, Q, Bernard, O and Harvey, AR (2006). Cooperative effects of bcl-2 and AAV-mediated expression of CNTF on retinal ganglion cell survival and axonal regeneration in adult transgenic mice. Eur J Neurosci 24: 3323–3332. [DOI] [PubMed] [Google Scholar]
- Harvey, AR, Ooi, JW and Rodger, J (2012). Neurotrophic factors and the regeneration of adult retinal ganglion cell axons. Int Rev Neurobiol 106: 1–33. [DOI] [PubMed] [Google Scholar]
- You, SW, Hellström, M, Pollett, MA, LeVaillant, C, Moses, C, Rigby, PJ et al. (2016). Large-scale reconstitution of a retina-to-brain pathway in adult rats using gene therapy and bridging grafts: An anatomical and behavioral analysis. Exp Neurol 279: 197–211. [DOI] [PubMed] [Google Scholar]
- Hellström, M and Harvey, AR (2011). Retinal ganglion cell gene therapy and visual system repair. Curr Gene Ther 11: 116–131. [DOI] [PubMed] [Google Scholar]
- Ruitenberg, MJ, Blits, B, Dijkhuizen, PA, te Beek, ET, Bakker, A, van Heerikhuize, JJ et al. (2004). Adeno-associated viral vector-mediated gene transfer of brain-derived neurotrophic factor reverses atrophy of rubrospinal neurons following both acute and chronic spinal cord injury. Neurobiol Dis 15: 394–406. [DOI] [PubMed] [Google Scholar]
- Rodger, J, Drummond, ES, Hellström, M, Robertson, D and Harvey, AR (2012). Long-term gene therapy causes transgene-specific changes in the morphology of regenerating retinal ganglion cells. PLoS One 7: e31061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, AR, Kamphuis, W, Eggers, R, Symons, NA, Blits, B, Niclou, S et al. (2002). Intravitreal injection of adeno-associated viral vectors results in the transduction of different types of retinal neurons in neonatal and adult rats: a comparison with lentiviral vectors. Mol Cell Neurosci 21: 141–157. [DOI] [PubMed] [Google Scholar]
- Hellström, M, Ruitenberg, MJ, Pollett, MA, Ehlert, EM, Twisk, J, Verhaagen, J et al. (2009). Cellular tropism and transduction properties of seven adeno-associated viral vector serotypes in adult retina after intravitreal injection. Gene Ther 16: 521–532. [DOI] [PubMed] [Google Scholar]
- Sharma, A, Pollett, MA, Plant, GW and Harvey, AR (2012). Changes in mRNA expression of class 3 semaphorins and their receptors in the adult rat retino-collicular system after unilateral optic nerve injury. Invest Ophthalmol Vis Sci 53: 8367–8377. [DOI] [PubMed] [Google Scholar]
- Hellström, M, Muhling, J, Ehlert, EM, Verhaagen, J, Pollett, MA, Hu, Y et al. (2011). Negative impact of rAAV2 mediated expression of SOCS3 on the regeneration of adult retinal ganglion cell axons. Mol Cell Neurosci 46: 507–515. [DOI] [PubMed] [Google Scholar]
- Hellström, M, Pollett, MA and Harvey, AR (2011). Post-injury delivery of rAAV2-CNTF combined with short-term pharmacotherapy is neuroprotective and promotes extensive axonal regeneration after optic nerve trauma. J Neurotrauma 28: 2475–2483. [DOI] [PubMed] [Google Scholar]
- Boyle, EI, Weng, S, Gollub, J, Jin, H, Botstein, D, Cherry, JM et al. (2004). GO::TermFinder–open source software for accessing Gene ntology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, H, Xu, Z, Ishii-Watabe, A, Uchida, E and Hayakawa, T (2000). IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol Ther 1: 376–382. [DOI] [PubMed] [Google Scholar]
- Chen, H and Weber, AJ (2004). Brain-derived neurotrophic factor reduces TrkB protein and mRNA in the normal retina and following optic nerve crush in adult rats. Brain Res 1011: 99–106. [DOI] [PubMed] [Google Scholar]
- Park, KK, Liu, K, Hu, Y, Smith, PD, Wang, C, Cai, B et al. (2008). Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322: 963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto, T, Yin, Y, Omura, K, Gilbert, HY, Kim, D, Cen, LP et al. (2010). Long-distance axon regeneration in the mature optic nerve: contributions of oncomodulin, cAMP, and pten gene deletion. J Neurosci 30: 15654–15663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, MR, Morrison, KC, Jacques, SJ, Leadbeater, WE, Gonzalez, AM, Berry, M et al. (2009). Off-target effects of epidermal growth factor receptor antagonists mediate retinal ganglion cell disinhibited axon growth. Brain 132(Pt 11): 3102–3121. [DOI] [PubMed] [Google Scholar]
- Nagai, J, Kitamura, Y, Owada, K, Yamashita, N, Takei, K, Goshima, Y et al. (2015). Crmp4 deletion promotes recovery from spinal cord injury by neuroprotection and limited scar formation. Sci Rep 5: 8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, PD, Sun, F, Park, KK, Cai, B, Wang, C, Kuwako, K et al. (2009). SOCS3 deletion promotes optic nerve regeneration in vivo. Neuron 64: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, DL, Apara, A and Goldberg, JL (2011). Krüppel-like transcription factors in the nervous system: novel players in neurite outgrowth and axon regeneration. Mol Cell Neurosci 47: 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, R, Song, Y, Kjellstrom, S, Tanikawa, A, Liu, Y, Li, Y et al. (2006). Regulation of rod phototransduction machinery by ciliary neurotrophic factor. J Neurosci 26: 13523–13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill, TJ, Prusky, GT, Douglas, RM, Yasumura, D, Matthes, MT, Nune, G et al. (2007). Intraocular CNTF reduces vision in normal rats in a dose-dependent manner. Invest Ophthalmol Vis Sci 48: 5756–5766. [DOI] [PubMed] [Google Scholar]
- Park, KK, Hu, Y, Muhling, J, Pollett, MA, Dallimore, EJ, Turnley, AM et al. (2009). Cytokine-induced SOCS expression is inhibited by cAMP analogue: impact on regeneration in injured retina. Mol Cell Neurosci 41: 313–324. [DOI] [PubMed] [Google Scholar]
- Sun, Y, Ju, M, Lin, Z, Fredrick, TW, Evans, LP, Tian, KT et al. (2015). SOCS3 in retinal neurons and glial cells suppresses VEGF signaling to prevent pathological neovascular growth. Sci Signal 8: ra94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J, De Preter, K, Pattyn, F, Poppe, B, Van Roy, N, De Paepe, A et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, MW (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans, J, Mortier, G, De Paepe, A, Speleman, F and Vandesompele, J (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.