Abstract
Background
S2101 is one of the most potent LSD1 inhibitors, which can inhibit ovarian cancer cells viability. This study aimed to detect the mechanism behind the anticancer properties of S2101 in SKOV3 ovarian cells.
Material/Methods
Cell viability was tested by Cell Counting Kit-8 (CCK-8) assay. Cellular apoptosis and autophagy were evaluated by flow cytometric analysis using Annexin-V/PI staining methods and Green fluorescent protein (GFP)-fused-LC3 (GFP-LC3), respectively. Western blotting was performed for analyzing the Bax, Bcl-2, mTOR, p-mTOR, p62, LC3-I, LC3-II, AKT, and p-AKT protein expression.
Results
Our results show that the proportion of early apoptotic and late apoptotic cells increased significantly for cells treated with S2101 at a concentration of 100 μM for 48 h. Treatment of S2101 in SKOV3 cells resulted in upregulation of Bax and downregulation of Bcl-2 in a time-dependent manner, indicating that S2101 can induce apoptosis in SKOV3. There was a downward trend in the expression of p62 when the SKOV3cells were treated with 100 μm S2101 for 12 h, 24 h and 48 h. The conversion of LC3-I to LC3-II was increased significantly at 24 h and 48 h. Autophagy was induced by S2101 in SKOV3 cells, evidenced by an increase in punctuate localization of GFP-LC3 and a change in expression of autophagy-related proteins.
Conclusions
S2101 treatment decreased the levels of phosphorylated AKT and mTOR. S2101 inhibits SKOV3 cells viability and induces apoptosis and autophagy. The AKT/mTOR signaling pathway was found to be affected by S2101.
MeSH Keywords: Apoptosis, Autophagy, Ovarian Neoplasms
Background
Ovarian cancer is the most lethal gynecological malignancy and the second most common gynecologic cancer in the world, with a high incidence of metastasis and a high relapse rate [1]. It caused an estimated 14 270 deaths in 2014 in the USA. Because of the lack of effective diagnostics in early stages, the 5-year survival rate for ovarian cancer is only 27% [2]. New therapeutic strategies are urgently needed in the management of ovarian cancer [3].
Lysine-specific demethylase 1 (LSD1) is a nuclear enzymatic activity that demethylates mono- and di-methylated histone H3 at lysines 4 and 9, regulating the transcriptional actions of hormone-liganded nuclear receptors [4–7]. It can also demethylate non-histone lysine residues [8]. LSD1 has been found to be overexpressed in various cancers, including ovarian cancer [9,10], breast cancer [11], colon cancer [12], and gastric cancer [13]. LSD1 is flavin adenine dinucleotides-dependent (FAD) amine oxidase [9]. FDA-approved inhibitors of FAD amine oxidases such as mitochondrial-associated monoamine oxidase (MAO) A and B and polyamine oxidase (PAO) are non-selective inhibitors of LSD1 activity [14]. Recently, many selective LSD1 inhibitors have been developed, which can be grouped into 4 different classes based on their chemical structure. S2101 is known to be more selective than other LSD1 inhibitors (e.g., pargyline and TCP) in ovarian cancer cell lines, which can inhibit cell viability and it has potential value in therapeutic use [9]. However, the mechanism by which S2101 inhibits ovarian cancer cell proliferation remains unclear.
Autophagy is a lysosomal degradative process used to recycle obsolete cellular constituents and eliminate damaged organelles and protein aggregates [15]. When a chemotherapeutic drug induces oxidative stress and DNA damage, or when defective vascularization determines hypoxia and starvation, the upregulation of autophagy enables cancer cells to overcome the metabolic stress [16]. In brief, autophagy is a cytoprotective mechanism to protect normal cellular survival. When abnormality in the process of autophagy occurs, normal cellular functions become damaged, and the accumulated abnormalities will lead to systematic problems [17]. Abundant evidence shows that autophagy is associated with the genesis and development of cancers, and effective therapeutic strategies could induce or inhibit the process of autophagy in various cancers [18].
Based on previous studies, this study aimed to investigate the effects of S2101 on viability, apoptosis, and autophagy of SKVO3 ovarian cancer cells.
Material and Methods
Cell culture
The SKVO3 is a human ovarian cancer cell line that was donated by the Laboratory of Pharmacology, Harbin Medical University, China. Cells were cultured in RPMI 1640 medium (Millipore, Billerica, MA) containing 10% FBS, 100 U/mL penicillin, and 100 U/mL streptomycin (Millipore, Billerica, MA) in a humidified chamber at 5% CO2 and 37ºC.
Chemicals and antibodies
LSD1 inhibitor S2101 and autophagy inhibitor 3-methyladenine (3-MA) were purchased from the Millipore Company. The primary antibodies for Bax, Bcl-2, PARP, LC3-I/II, SQSTM1/p62, mTOR, phospho-mTOR, p70S6K, phospho-P70S6K, AKT, phospho-AKT, and β-actin were purchased from Cell Signaling Technology. FBS and RPMI 1640 medium were purchased from Millipore.
Cell viability assay
The cytotoxicity of S2101 was tested by Cell Counting Kit-8 (CCK-8) assay (Beyotime, Shanghai, China). SKOV3 cells were prepared and dispersed in 96-well cell culture plates at a cellular density of 1.0×104 cells/well. The cells in the exponential phase of growth were used. After incubating different concentrations of S2101 ranging from 0 to 200 μmol/L for 24 h and 48 h, 10 μL of CCK-8 solution in PBS was added to each well and incubated at 37°C for 2 h. Results were calculated as the absorbance of treated cells relative to untreated controls. Cell viability was assessed by a microplate reader at a wavelength of 450 nm.
Flow cytometric (FCM) analysis of apoptosis
Cellular apoptosis was quantified by flow cytometric analysis using Annexin-V/PI staining methods. Briefly, 48 h after 100 um S2101 incubation, both floating and attached cells were collected and washed twice with cold PBS. The cells were resuspended in prediluted binding buffer and propidium iodide (1 μg/ml) was then added. The samples were immediately detected by a FACSort flow cytometer (BD Biosciences, San Jose, CA, USA).
GFP-LC3
Green fluorescent protein (GFP)-fused-LC3 (GFP-LC3) was used to test autophagy. The SKOV3 cells were fused to 5 μg of GFP-LC3 plasmid using Lipofectamine 2000 (Invitrogen). The transfected cells were treated with 100 μm of S2101 for 48 h. The GFP-LC3 cells were visualized by Nikon fluorescence microscopy (magnification, ×100; 90i; Nikon Corporation, Tokyo, Japan). Cell numbers were counted to normalize the measurement, and the percentage of fluorescent cells was calculated.
Western blot analysis
After treatment, SKOV3 cells were harvested and lysed in radioimmunoprecipitation lysis buffer (Beyotime, Shanghai, China). The total proteins from each samples were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, and electro-transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). After blocking, the membranes were immunoblotted overnight at 4°C with primary antibodies, including anti-Bax, anti-Bcl-2, anti-PARP, anti-LC3-I/II, anti-SQSTM1/p62, anti-mTOR, anti-phospho-mTOR, anti-p70S6K, anti-phospho-P70S6K, anti-AKT, anti-phospho-AKT, and β-actin, followed by HRP-conjugated secondary antibodies at 37°C for 1 h. Signals were detected using an ECL system. The intensity was determined by densitometric analysis using Image J software.
Statistical analysis
All experiments were performed independently at least 3 times. Data are expressed as mean ± standard deviation (SD). The differences were analyzed using the 2-sided t test or ANOVA with SPSS 17.0 software. P<0.05 was considered statistically significant.
Results
S2101 inhibits growth of SKOV3 cells
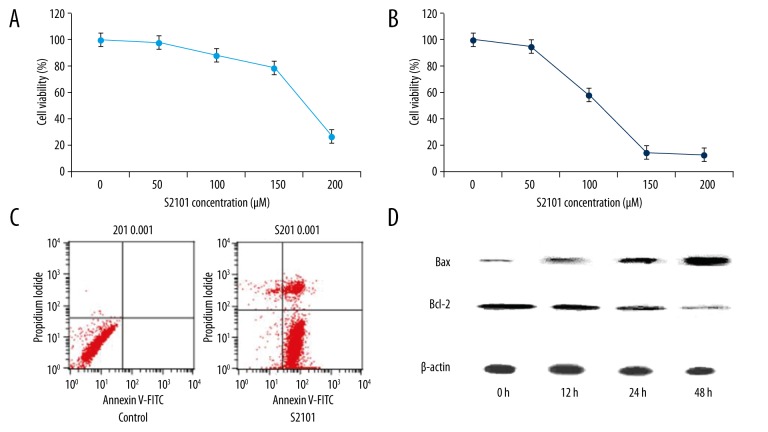
When SKVO3 cells were treated with S2101 at 0, 50, 100, 150, and 200 μM, the percentages of cellular viability over the control cells (100%) were 98.27%, 88.61%, 79.17%, and 27.17% for 24h treatment, respectively (Figure 1A); the percentages of cellular viability vs. the control cells (100%) were 94.83%, 58.23%, 14.24%, and 12.36% for 48 h treatment, respectively (Figure 1B). S2101 inhibits cell growth in dose-dependent and time-dependent manners, indicating an inhibitory effect of S2101 against excessive growth of SKOV3 cells.
Figure 1.
S2101 inhibits growth of SKOV3 cells. (A, B) The growth curve of SKOV3 cells treated with 0, 50, 100, 150 and 200 μm S2101 for 24 h and 48 h, respectively. (C) The flow cytometric analysis of apoptosis using Annexin-V-FITC/PI staining of SKOV3 cells treated with 100 μm S2101; (D) The detection of apoptosis-related protein Bax and Bcl-2 of SKOV3 cells treated with 100 μm S2101 by Western blot analysis.
Annexin-V/PI-stained flow cytometric analysis was used to determine whether the reduced cell viability was due to apoptosis. The proportion of both early apoptotic and late apoptotic cells increased significantly for those treated with S2101 at a concentration of 100 μM for 48 h as compared to control cells (Figure 1C). Moreover, apoptosis-related protein Bax and Bcl-2 were detected by Western blot analysis. Treatment of S2101 in SKOV3 cells resulted in upregulation of Bax and downregulation of Bcl-2 in a time-dependent manner, indicating that S2101 can induce apoptosis in SKOV3 (Figure 1D).
S2101 induces autophagy in SKOV3 cells
The expression of autophagy-related proteins was assessed by Western blot analysis to evaluate the effects of S2101 on autophagy in SKOV3 cells. Autophagosome formation involves the conjugation of cytosolic microtubule-associated protein light chain 3 (LC3-I) with phosphatidylethanolamine to form LC3-phosphatidylethanolamine (LC3-II) as an essential process [19,20]. The conversion of LC3-I to LC3-II is widely recognized as a marker protein of autophagy. P62 is also a special marker of autophagy.
Figure 2A shows that there was a downward trend in the expression of p62 when the SKOV3 cells were treated with 100 μm of S2101 for 12 h, 24 h, and 48 h. The conversion of LC3-I to LC3-II increased significantly at 24 h and 48 h. The percentage of cells with GFP-LC3 puncta was significantly increased along with the number and fluorescence intensity of SKOV3 cells treated with S2101 compared with the control group (Figure 2B).
Figure 2.
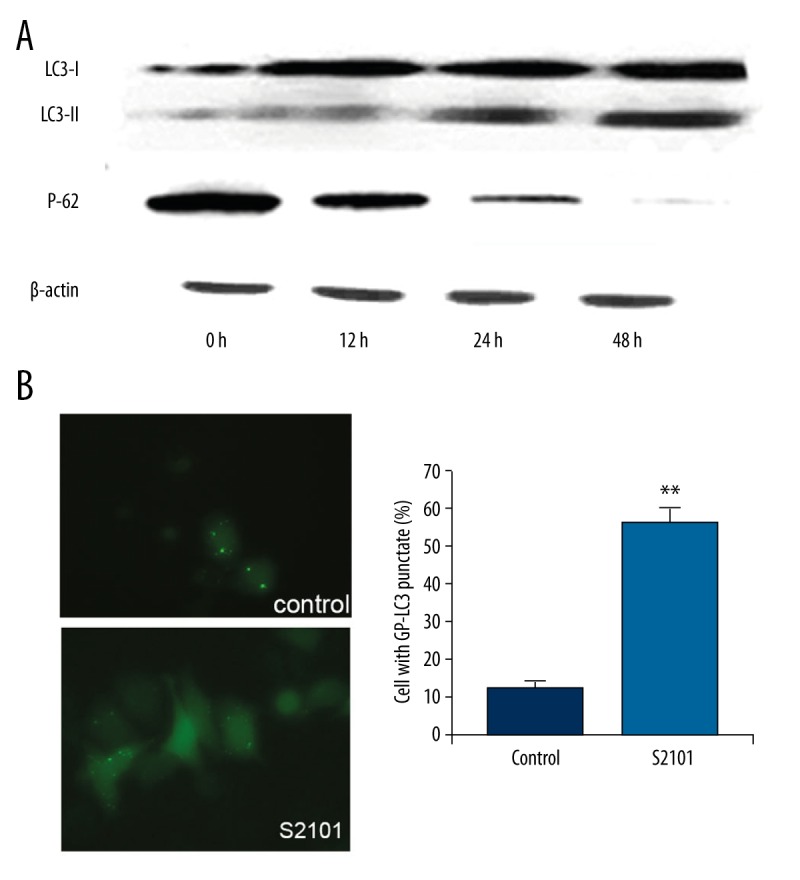
S2101 induces autophagy in SKOV3 cells. (A) The detection of autophagy-related protein LC3-I, LC3-II and P62 of SKOV3 cells treated with 100 μm S2101 by Western blot analysis. (B) The percentage of SKOV3 cells with GFP-LC3 puncta treated with 100 μm S2101.
SKOV3 cells viability can be inhibited by the block of autophagy
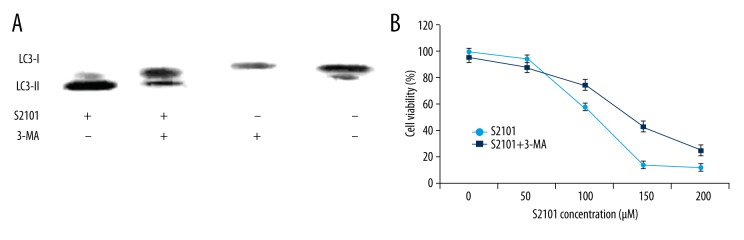
3-methyladenine (3-MA) was used to investigate the role of autophagy in S2101-induced growth suppression. As Figure 3A shows, pre-treatment with 3-MA resulted in reduced conversion of LC3-I to LC3-II as compared with the S2101 alone group.
Figure 3.
SKOV3 cell viability can be inhibited by blocking autophagy. (A) Pre-treatment with 3-MA resulted in reduced conversion of LC3-I to LC3-II in SKOV3 cells as compared with S2101 alone group. (B) S2101 treatment alone significantly inhibited cell proliferation compared with combination of S2101 and 3-MA
Autophagy may play a double-acting role in regulation of cell death, which can either promote or inhibit cell death. The relationship between apoptosis and autophagy in S2101-treated SKOV3 cells was detected. SKOV3 cells were pre-treated by 3-MA, followed by different concentrations of S2101 for 48 h. CCK-8 assay results showed that the group treated with S2101 alone significantly inhibited cell proliferation compared with S2101 and 3-MA (Figure 3B).
S2101 inhibits the AKT/mTOR/p70S6K signaling pathway
The AKT/mTOR/p70S6K pathway is considered to be related with autophagy. Autophagy is activated if the mTOR/p70S6K signature pathway is suppressed. Our results indicate that S2101 suppressed the phosphorylation of mTOR and p70S6K, but exerted little effect on total protein levels (Figure 4).
Figure 4.
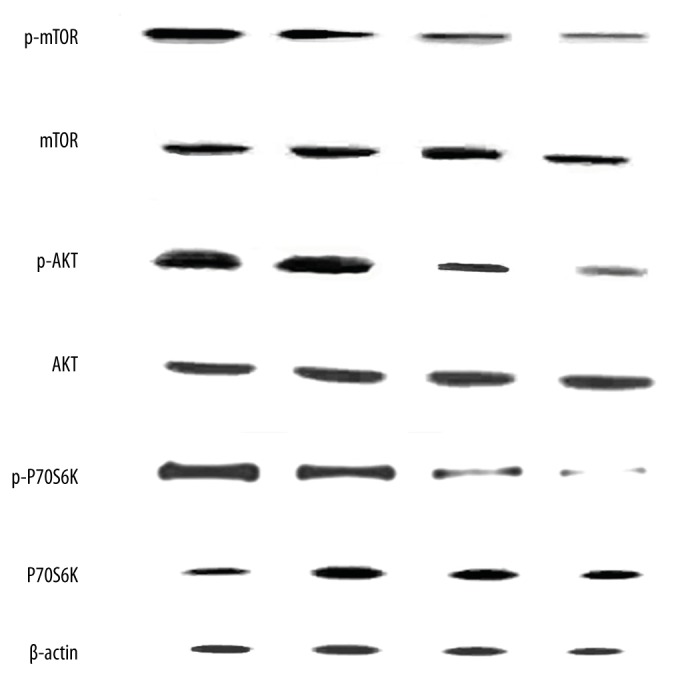
S2101 inhibits the AKT/mTOR/p70S6K signaling pathway.
AKT participated in the regulation of various signaling pathways, including cell proliferation, growth, survival, angiogenesis, and chemoresistance [21]. Our results show that the phosphorylation of AKT and the ratio of p-AKT/AKT were decreased after treatment with various doses of S2101 for 48 h.
Discussion
Ovarian cancer is one of most common cancers of women worldwide [1]. Although various treatments for ovarian cancer including surgery and chemotherapy have been developed, its long-term outcome remains poor [22,23]. Our results show that S2101 inhibited SKOV3 cells viability in a concentration- and time-dependent manner, and increased the apoptosis and autophagy in SKOV3, indicating that S2101, one of the most active inhibitors of LSD1, can kill ovarian cancer cells in vitro.
Autophagy is a necessary response to extracellular stimulus, allowing the elimination of toxic metabolites, intracellular pathogens, and damaged proteins and organelles, and providing energy and amino acids necessary for vital functions during metabolic stress [21–25]. The functions of normal human cells require the basal level of autophagy. However, abnormal autophagy may promote genesis and development of cancer. Emerging evidence shows that the induction of autophagy is a promising therapeutic strategy in cancer treatment. GFP-tagged LC3-expressing cells have been used as a specific marker to monitor autophagy [19,20]. Our results show that the punctate pattern of GFP-LC3 was increased in SKOV3 cells treated with S2101. Western blot analysis also showed a marked change in the expression of LC3-I/LC3-II and p62 in response to S2101 treatment.
Although autophagy and apoptosis are 2 different physiological processes, they are often interconnected through various crosstalk mechanisms [26]. The common methods to assess the relationship between autophagy and apoptosis rely on: i) the presence of autophagic features in apoptotic cells, and ii) rescue of cell apoptosis via changed level of autophagy [27]. To explore whether autophagy was involved in S2101-induced cell apoptosis, the pharmacological inhibition of autophagy (3-MA) was used. The results of CCK-8 assay showed that S2101 suppressed the growth of SKOV3 cells, which was partially reversed by pre-treatment with 3-MA, indicating that autophagy may co-occur with apoptosis in SKOV3 cells treated with S2101. S2101-induced autophagy may contribute to tumor cell death in part by enhancing apoptosis.
The mTOR signal pathway is involved in the induction of autophagy. Several studies have shown that the suppression of the mTOR pathway can induce autophagy in several human cancer cell lines, including lung cancer [28], liver cancer [29], ovarian cancer [30], and gastric cancer [31]. Our results showed the negative effect of S2101 on the mTOR/p70S6K pathway, indicating that inhibition of the mTOR pathway may be a mechanism of S2101-triggered autophagy in SKOV3 cells. As an upstream positive regulator of mTOR, AKT inactivates tuberous sclerosis complex 2 by phosphorylating it on 4 residues; the resulting compound is a negative regulator of mTOR, thereby activating mTOR [32]. The phosphorylation of AKT is one of the most important activators of mTOR. Our results suggest that the inhibition of AKT/mTOR/p70S6K signaling pathway, at least in part, participated in the induction of autophagy by treatment of S2101. Further, the AKT-mTOR axis may be involved in S2101-induced autophagic cell death as apoptosis was promoted, following by the induction of autophagy.
Conclusions
Our study provides evidence that S2101 inhibited the viability of human ovarian cells by inducing autophagy and apoptosis. In addition, the anticancer effect of 2101 was associated with suppression of the AKT/mTOR pathway. These results suggest that S2101 is an attractive therapeutic agent for developing alternative treatment protocols, and possibly for combining with other anticancer agents to overcome drug resistance and achieve better outcomes. In vivo studies using cancer animal models are currently underway in our lab.
Footnotes
Conflict of interest
The authors declare that there is no conflict of interest.
Source of support: Departmental sources
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Guarneri V, Piacentini F, Barbieri E, Conte PF. Achievements and unmet needs in the management of advanced ovarian cancer. Gynecol Oncol. 2010;117:152–58. doi: 10.1016/j.ygyno.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Ma L, Rao Q, et al. MiR-1271 inhibits ovarian cancer growth by targeting cyclin G1. Med Sci Monit. 2015;21:3152–58. doi: 10.12659/MSM.895562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metzger E, Imhof A, Patel D, et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464:792–96. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- 5.Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–39. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Bassets I, Kwon YS, Telese F, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–18. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nair SS, Nair BC, Cortez V, et al. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 2010;11:438–44. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi YJ, Matson C, Lan F, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–64. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 9.Konovalov S, Garcia-Bassets I. Analysis of the levels of lysine-specific demethylase 1 (LSD1) mRNA in human ovarian tumors and the effects of chemical LSD1 inhibitors in ovarian cancer cell lines. J Ovarian Res. 2013;6:75. doi: 10.1186/1757-2215-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Ge J, Lu Q, et al. Expression of Lysine-specific demethylase 1 in human epithelial ovarian cancer. J Ovarian Res. 2015;8:28. doi: 10.1186/s13048-015-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim S, Janzer A, Becker A, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–20. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 12.Hayami S, Kelly JD, Cho HS, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128:574–86. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 13.Magerl C, Ellinger J, Braunschweig T, et al. H3K4 dimethylation in hepatocellular carcinoma is rare compared with other hepatobiliary and gastrointestinal carcinomas and correlates with expression of the methylase Ash2 and the demethylase LSD1. Hum Pathol. 2010;41:181–89. doi: 10.1016/j.humpath.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Forneris F, Battaglioli E, Mattevi A, Binda C. New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin. FEBS J. 2009;276:4304–12. doi: 10.1111/j.1742-4658.2009.07142.x. [DOI] [PubMed] [Google Scholar]
- 15.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–44. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin G, Hill DK, Andrejeva G, et al. Dichloroacetate induces autophagy in colorectal cancer cells and tumours. Br J Cancer. 2014;111:375–85. doi: 10.1038/bjc.2014.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panda PK, Mukhopadhyay S, Das DN, et al. Mechanism of autophagic regulation in carcinogenesis and cancer therapeutics. Semin Cell Dev Biol. 2015;39:43–55. doi: 10.1016/j.semcdb.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimori T. Autophagy: A regulated bulk degradation process inside cells. Biochem Biophys Res Commun. 2004;313:453–58. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–18. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta. 2011;1813:1978–86. doi: 10.1016/j.bbamcr.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Codogno P. Shining light on autophagy. Nat Rev Mol Cell Biol. 2014;15:153. doi: 10.1038/nrm3751. [DOI] [PubMed] [Google Scholar]
- 23.Jung CH, Ro SH, Cao J, et al. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan J, Wang K, Xi M. MiR-494 inhibits epithelial ovarian cancer growth by targeting c-Myc. Med Sci Monit. 2016;22:617–24. doi: 10.12659/MSM.897288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Zhou ZW, Jin H, et al. Schisandrin B inhibits cell growth and induces cellular apoptosis and autophagy in mouse hepatocytes and macrophages: Implications for its hepatotoxicity. Drug Des Devel Ther. 2015;9:2001–27. doi: 10.2147/DDDT.S77071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behar M, Barken D, Werner SL, Hoffmann A. The dynamics of signaling as a pharmacological target. Cell. 2013;155:448–61. doi: 10.1016/j.cell.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 28.Wu SH, Hang LW, Yang JS, et al. Curcumin induces apoptosis in human non-small cell lung cancer NCI-H460 cells through ER stress and caspase cascade- and mitochondria-dependent pathways. Anticancer Res. 2010;30:2125–33. [PubMed] [Google Scholar]
- 29.Zhang DM, Liu JS, Deng LJ, et al. Arenobufagin, a natural bufadienolide from toad venom, induces apoptosis and autophagy in human hepatocellular carcinoma cells through inhibition of PI3K/Akt/mTOR pathway. Carcinogenesis. 2013;34:1331–42. doi: 10.1093/carcin/bgt060. [DOI] [PubMed] [Google Scholar]
- 30.He J, Yu JJ, Xu Q, et al. Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy. 2015;11:373–84. doi: 10.1080/15548627.2015.1009781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou L, Li Y, Song H, et al. Protective macroautophagy is involved in vitamin E succinate effects on human gastric carcinoma cell line SGC-7901 by inhibiting mTOR axis phosphorylation. PLoS One. 2015;10:e0132829. doi: 10.1371/journal.pone.0132829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–94. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]