Abstract
Natural killer group 2D (NKG2D), as an activating receptor, plays pivotal role in viral infectious diseases. Several single nucleotide polymorphisms (SNPs) in the NKG2D gene have characterized that the rs1049174G/C SNP of NKG2D is in the spotlight of notice because of its role in activating of human T cells. This study aimed to investigate rs1049174G/C genetic polymorphism in Chronic Hepatitis C (CHC) patients. The study compromised 107 CHC patients with genotype 1a and 1b. All recruited patients were under treatment with Peginterferon Alfa-2a/Ribavirin according to standard protocol. After completing treatment, 67 patients showed sustained virologic response (SVR) and the rest of patients did not respond to the treatment and considered as non-responder (NR). Genotyping of NKG2D rs1049174G/C SNP was performed using PCR–RFLP method in SVR and NR patients. The NKG2D rs1049174 genotypes frequency for GG, GC and CC were 45, 41 and 14 % respectively. Genotypes distribution were significantly different between SVR and NR groups (p = 0.005). So that the patients with the homozygous GG genotype demonstrated a higher response to Peginterferon Alfa-2a/Ribavirin therapy against HCV infection (OR = 6.0, 95 %CI 1.71–21.08, p = 0.005). In conclusion, the rs1049174 GG genotype of NKG2D receptor is an effective factor in successfully treatment of CHC patients by Peginterferon Alfa-2a/Ribavirin.
Keywords: Chronic hepatitis C, NKG2D, Peginterferon Alfa-2a/Ribavirin, rs1049174
Introduction
Chronic Hepatitis C virus (CHCV) infection is one of the major causes of chronic liver disease and has affected approximately 170–200 million people worldwide, comprising ~3 % of the global population [26]. About 80 % of patients with early acute HCV infection fail to clear the virus and will develop persistent infection which often cause chronic hepatitis, cirrhosis and hepatocellular carcinoma [23]. Despite this high burden of disease there is still no vaccine available for clinical use against HCV. In addition, there are several challenges associated with current and future treatment of chronic hepatitis C [3, 30]. A number of studies suggested that immune response has a crucial role in pathogenesis and outcome of hepatitis C virus infection [6, 22, 33, 39, 40]. A combined treatment including Peginterferon Alfa-2a/Ribavirin with or without protease inhibitor has become the standard course for the treatment of CHCV infection [10]. The efficacy of treatment depends on several factors such as baseline viral load, severity of liver disease, body weight, ethnicity and viral genotype with type 1 appearing to be less responsive to treatment than other types [44]. However, only about half of treated patients achieve a sustained virology response (SVR) in which serum HCV RNA become undetectable for at least 6 months after cessation of therapy. In this condition, completion of treatment course may not occur due to the cost and major side effects [25, 41]. Therefore, it is necessary to search for surrogate predictors as a guide to evaluate response to antiviral therapy.
NK cells include 30–50 % of lymphocytes population in the liver. They recognize and kill infected and tumorigenic cells and their function is controlled by net balance of inhibitory and activating receptor signals expressed on their surface. The activating receptors include NKG2D, CD94:NKG2C/E, NKp44, NKp30, NKp46, and CD16 (FC-γ-RIII) molecules [6]. NKG2D is one of the most important human NK cell activating receptor expressed on NKT cells, γδ cells, macrophages and a small number of CD4 + regulatory T cells and some CD8 + T lymphocytes. The ligands of NKG2D receptor are MHC class I chain-related A and B (MICA, MICB), and ULBP (UL-16-binding protein) which are expressed on tumor and virus infected cells. In early innate immune responses against tumor and virally infected cells, NKG2D signals can activate NK cells and CD8+ T cells by interaction with its ligands expressed on target cells [4, 17, 20, 32, 38, 45]. Molecular mechanisms used by viruses and tumor cells to escape from NKG2D recognition system highlight the importance of NKG2D receptor signaling against infection and tumorigenesis [1, 7, 29].
The NKG2D gene is located on the short (p) arm of human chromosome 12 in the region of natural killer complex (NKC). Studies show a single nucleotide polymorphism (SNP: rs1049174) located in 3′-untranslated regions (3′UTR) of the NKG2D gene that its haplotypes are associated with high activity of NK cells (HNK) or low activity (LNK). HNK phenotype (high NK cytotoxicity) is observed in rs1049174 G allele carriers, whereas the LNK (low NK cytotoxicity) is seen in C allele carriers [12, 13, 19].
Previous studies have shown an association between NKG2D polymorphism and cellular immune response against virus-infected and tumor cells [9, 24, 45]. Additionally, NKG2D and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) mediated signaling induce both apoptosis of fibrogenic hepatic satellite cell (HSC) and production of anti-fibrogenic cytokine, IFN-γ by NK cells, which can ameliorate liver fibrosis [14, 31]. The study by Chu et al. shows that decreased expression of NKG2D receptor on CD56+ CD3+ lymphocytes in HCV genotype 1 infected patients predicts poorer response to Peginterferon Alfa-2a/Ribavirin therapy compared to genotype 2 carriers (30). Therefore, in the current study we aimed to evaluate possible impact of NKG2D rs1049174 polymorphism on efficacy Peginterferon Alfa-2a/Ribavirin treatment in the patients with CHCV infection.
Materials and methods
Human subjects
The study was approved by ethics committee of Shahid Sadoughi University of Medical Sciences, Yazd, Iran and all patients gave informed consent to take part in the study. 107 Iranian patients with CHCV infection recruited of Shahid Sadoughi hospital and Bou-Ali Pathobiology laboratory during March 2010–June 2013 in Yazd, Iran. Clinical characteristics of the patients are summarized in Table 1.
Table 1.
Characteristics of recruited chronic hepatitis C patients
| Patients | SVR | NR patients | Pv |
|---|---|---|---|
| Patients number | 67 | 43 | – |
| Mean ± SD of age (year) | 45.8 ± 1.88 | 42.1 ± 2.22 | 0.239 |
| Sex (male/female) | 60/7 | 35/8 | – |
| HCV RNA (copy number/ml) before treatment | 8,454,434 ± 220,801 | 871,704 ± 292,996 | 0.154 |
| HCV genotype | 1a and 1b | 1a and 1b | |
| Length of treatment by Peginterferon/Ribavirin (weeks) | 48 | 48 | |
| ALT (IU/L) | 70 ± 68 | 66 ± 64 | 0.256 |
| ALP (IU/L) | 250 ± 124 | 264 ± 139 | 0.176 |
SVR Sustained virologic response, NR nonresponder, ALT alanine aminotransferase, ALP alkaline phosphatase
Patients with human immunodeficiency virus (HIV) or hepatitis B virus (HBV) coinfection, or having any evidence of clinically relevant liver disease such as apparent auto-immune hepatitis-primary biliary cirrhosis (AIH-PBC), hepatocellular carcinoma, sclerosing cholangitis, Wilson’s disease, α1-antitrypsin deficiency, former history of alcohol abuse or previous liver transplantation excluded of the study. Peripheral blood samples obtained from patients. Anti-HCV antibodies were assessed using a commercially available enzyme-linked immunosorbent assay kit (Diapro, Diaplus, Italy). Viral load assessment was performed by real-time polymerase chain reaction (Rotor-Gene 6000, Australia) method applying HCV RNA Quantification Kit (AJ Roboscreen, GmbH). Analytical sensitivity of the kit was 200 copies/ml and HCV-RNA-positive samples were genotyped using HCV genotyping kit (Sacace Biotechnologies Srl, Italy). Patients with HCV genotype 1 (1a, 1b) were selected.
All patients with detectable virus received a standard dose of Peginterferon α-2a (180 μg/week) subcutaneously plus weight-based ribavirin (1000 mg/days for weight <80 kg and 1200 mg/days for weight >80 kg). The patients were under treatment for 48 weeks as standard protocol for treatment of genotype 1 HCV infection. At least 6 months after treatment completion, 5 ml blood sample was taken from all patients of which 2 ml was used for DNA extraction, and the remaining 3 ml was centrifuged to separate serum. Serum viral load of all treated patients with genotype 1 were monitored using qPCR. According to presence or absence of HCV RNA in their blood, the patients with no detectable virus and detectable were considered as responder and non-responder respectively. NKG2D polymorphisms in the patients determined by qPCR.
DNA extraction and RFLP-PCR
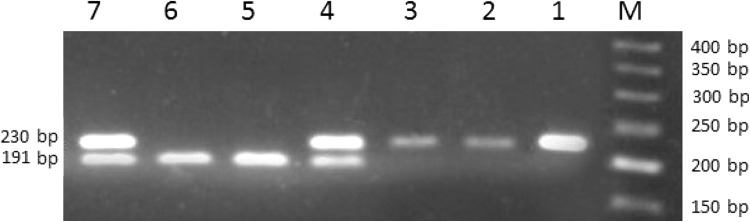
Genomic DNA was isolated from whole blood by a commercial DNA purification kit (Bioneer Co., Korea). NKG2D gene rs1049174 SNP was genotyped using polymerase chain reaction-restriction fragment length polymorphism (RFLP) method. Primers were designed by the Primer3 program. The forward and reverse primers were 5′-TTAAGGCTGGAGAATAATGC-3′ and 5′-TCAGTGAAGGAAGAGAAGG-3′, respectively, producing a 230 bp band. All PCR amplification reactions were performed in 25 μl volume, including 3 μl DNA (100 ng), 12.5 μl Taq Mastermix (Ampliqon, Denmark), 8.5 μl distilled water and 1 μl of each primer (forward and reverse). The PCR amplification was performed in the thermal cycler (Applied Biosystems, ABI, Foster City, CA, USA) with initial denaturation at 94 °C for 5-min, followed by 30 cycles of 30-s denaturation at 94 °C, 30-s annealing at 59 °C and 30-s elongation at 72 °C with the final extension at 72 °C for 5-min. Then, the 230 bp PCR product was digested with restriction enzyme Dde 1, according to kit instructions (New England Biolabs, Hitchin, UK). Digested fragments were separated by electrophoresis on a 3 % agarose gel, stained with green viewer. Gel images were visualized and captured by means of an E-gel imager (Life Technologies) instrument. Dde 1 digestion generated two fragments 191 and 39 bp (this band is not detectable in the agarose gel) for the C allele, whereas the undigested G allele was observed as a 230 bp fragment (Fig. 1).
Fig. 1.
RFLP genotyping for NKG2D rs1049174 polymorphism. Extracted DNA of patients and donors digested by Dde1 enzyme and the products ran on 3 % agarose gel. M: 50 bp DNA ladder, Samples 1, 2 and 3 correspond to GG; Samples of 4 and 7 show CC genotype and bands of 5 and 6 approve GC genotype
Statistical analysis
All statistical analyses were performed using the SPSS software for Windows, version 15.0 (SPSS Inc., Chicago, IL, USA). The association between genotypes and SVR were examined by computing the odds ratio (OR) and 95 % confidence intervals (95 % CI) from logistic regression analyses. p values below 0.05 were considered statistically significant. Hardy–Weinberg equilibrium (HWE) was tested with the χ 2 test for the SNP under consideration.
Results and discussion
Patients characteristics
Host immune system genetic diversity is one of the vital factor in inducing of effective immune response to HCV infection. Although genotype of the virus, treatment methods and different kinds drug are important too [27]. Genetic variation in IL28B is the most important pre-treatment predictor of achieving SVR with Peginterferon Alfa-2a/Ribavirin therapy in genotype 1 infected patients [15]. The HLA-ER/HLA-ER genotype has been shown to be protective against HCV genotypes 2 and 3 infection [36]. In the present study to exclude the genotype of HCV virus diversity as effective agent in immune system efficacy we only considered the patients who was infected by HCV genotype 1a and 1b. CHCV patients who has taken part in the present study were from central and south of east of Iran. All the patients who recruited to this study had not received any medication for HCV infection. According to HCV genotyping, the most patients showed genotype 1 and 3 and the prevalence of genotype of 1 and 3 were 73.3 and 26.7 % respectively. However, the patients with genotype 1 selected for this survey because this genotype is more resistant to usual treatments. On the other hand, concentration of ALT and ALP and serum albumin in the patients before starting treatment did not show significant differences (Table 1). The ALT and ALP enzymes in patient’s serum did not show significant relation with rs1049174 NKG2D polymorphism.
NKG2D polymorphism effects on Peginterferon Alfa-2a/Ribavirin treatment
In the infected patients with HCV genotype 1, rs1049174 determined according to methods section. Results are shown in Table 2. The frequency of GG genotype in the SVR patients were significantly higher than those in comparing to non-responder patients. The patients with the homozygous GG genotype showed a higher response to interferon therapy against HCV infection (OR = 6.0, 95 %CI 1.71–21.08, p = 0.005). Furthermore, carriers of G allele represented an increased level of response to HCV therapy compared to C allele carriers (OR = 2.46, 95 %CI 1.37–4.41, p = 0.003) as shown in Table 2.
Table 2.
NKG2D rs1049174 genotype and alleles frequencies in the patients infected by HCV genotype 1
| rs1049174 genotype | Number | SVR | Non-responder | Odds ratio (95 % CI) | Pv |
|---|---|---|---|---|---|
| CC | 15 (14 %) | 5 (7.5 %) | 10 (25 %) | Ref | – |
| CG | 44 (41 %) | 26 (38.8 %) | 18 (45 %) | 2.88 (0.84–9.88) | 0.134 |
| GG | 48 (45 %) | 36 (53.7 %) | 12 (30 %) | 6.00 (1.71–21.08) | 0.005 |
| Allele frequency | |||||
| C | 36 (26.9) | 38 (47.5) | Ref | – | |
| G | 98 (73.1) | 42 (52.5) | 2.46 (1.37–4.41) | 0.003 |
p values below 0.05 were considered significant
According to other studies, high load of HCV virus needs to longer treatment. Immune system efficacy certainly has affected HCV viral load. The NKG2D as an important activator receptor has substantial role in control of vial load. In the current study to investigate rs1049174 NKG2D polymorphism and the viral load of HCV, the patients before starting treatment divided into two groups. In group one HCV particles was higher than 800,000 copy number/ml and the group two included the patients with <800,000. NKG2D rs1049174 genotypes were compared in the two groups. Although C allele frequency in the SVR patients with <800,000 copy/ml was higher than the patients with more than 800,000, statistical analysis did not show significant difference (p = 0.056). The results shown in the Table 3.
Table 3.
HCV viral load in the chronic hepatitis C patients before treatment and NKG2D rs1049174 polymorphisms
| Patients | >800,000 virus copy number/ml | <800,000 virus copy number/ml | pv |
|---|---|---|---|
| Number of SVR patients | 17 | 50 | |
| Allele frequency in responder patients | |||
| C allele | 11 | 69 | 0.059 |
| G allele | 23 | 31 | 0.126 |
| Number of non-responder patients | |||
| Allele frequency in non-responder number | 12 | 28 | |
| C allele | 17 | 25 | 0.184 |
| G allele | 7 | 31 | 0.067 |
In the present study, we have found that patients who carried the G allele or GG genotype of NKG2D polymorphism rs1049174 were more likely to achieve SVR than patients with C allele or CC genotype. Our findings suggest that inheritance of NKG2D GG genotype/G allele has an important detrimental effect on SVR rates in CHC patients treated with Peginterferon Alfa-2a/Ribavirin. Several studies have demonstrated considerable role for NKG2D in protective function against virus infected or transformed cells [5, 37, 45]. The liver lymphocyte population is enriched with NK cells and these cells are one of the main cell populations, expressing NKG2D [11, 42]. The major NK cell functions include direct killing of target cells as well as cytokine production such as IFN-γ [11]. NKG2D ligation induces calcium flux, cytokine release, and cytotoxicity in NK cells. In addition, NKG2D can have costimulatory roles in CD8+ T cell activation, against viral infection [17, 43]. Earlier studies approved a connection between NKG2D haplotypes and natural cytotoxic activity of peripheral lymphocytes against cancer cells. In human NKG2D haplotype block-1(hb-1) has five tightly linked SNPs in hb-1. Furthermore, it has been shown that among these SNPs, G allele in the NKG2D polymorphism rs1049174, can be a good predictor of the high NK activity genotype, associated with cancer surveillance [12, 19]. However, there are few studies investigating NKG2D polymorphism and its impact on natural cytotoxic activity against viral infection. Juan Ma et al. [24], investigated a number of NK cell receptors gene polymorphism in Han Chinese chronic hepatitis B patients and proposed some NKG2D polymorphism and susceptibility to chronic hepatitis B virus infection. Additionally, there are many studies that have investigated immunological function of NKG2D in viral infection especially viral hepatitis. Amadei et al. [2] analyzed the expression of NKG2D on the surface of NK cells. In HCV infected patients compared with controls, NKG2D is more highly expressed in the acute phase of infection, but in chronic phase of infection there is conflicting evidence with regard to NKG2D expression, which has been described as being downregulated, upregulated or unchanged [8, 18, 21].
Immunomodulation patterns to helpful immune reaction in order to clearance of HCV infection or detrimental role of immune system in liver injury and fibrosis have not clear completely so far [28, 34, 35]. According to literature [16], NKG2D may also function as a costimulatory receptor on CD4+ regulatory T cells, suggesting that NKG2D signaling may induce dual tolerogenic and immunogenic response in hepatitis C infection. Nevertheless, it is unclear whether the NKG2D functions actually help to improve hepatitis C outcomes or may play pathological role in fibrosis and liver injury, however it is not surprising that NKG2D may play an important role in maintaining a balanced immune response that has significant influence on achieving SVR.
In conclusion, this study indicated a relation between NKG2D rs1049174 polymorphism and Peginterferon Alfa-2a/Ribavirin treatment method in central region of Iranian HCV patients. More studies in different populations and a larger number of subjects will be required to elucidate the association between this polymorphism and SVR. Because of great importance of NKG2D and other inhibitory and activating NK cell receptors and direct involvement of these receptors and due to the contradictory reports regarding the effect of immune response in liver disease severity and SVR, further genetic and biochemical studies of these receptors will provide further evidence for the clinical practice and the appropriate use of currently available therapy.
Acknowledgement
The authors appreciate all individuals who willingly participated in the current study. This research has been funded by Shahid Sadoughi University of Medical Sciences, under graduate thesis of AAS.
References
- 1.Alter G, Jost S, Rihn S, Reyor LL, Nolan BE, Ghebremichael M, et al. Reduced frequencies of NKp30 + NKp46 + , CD161 + , and NKG2D + NK cells in acute HCV infection may predict viral clearance. J Hepatol. 2011;55(2):278–288. doi: 10.1016/j.jhep.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amadei B, Urbani S, Cazaly A, Fisicaro P, Zerbini A, Ahmed P, et al. Activation of natural killer cells during acute infection with hepatitis C virus. Gastroenterology. 2010;138(4):1536–1545. doi: 10.1053/j.gastro.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansaldi F, Orsi A, Sticchi L, Bruzzone B, Icardi G. Hepatitis C virus in the new era: perspectives in epidemiology, prevention, diagnostics and predictors of response to therapy. World J Gastroenterol. 2014;20(29):9633–9652. doi: 10.3748/wjg.v20.i29.9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber A, Zhang T, Sentman CL. Immunotherapy with chimeric NKG2D receptors leads to long-term tumor-free survival and development of host antitumor immunity in murine ovarian cancer. J Immunol. 2008;180(1):72–78. doi: 10.4049/jimmunol.180.1.72. [DOI] [PubMed] [Google Scholar]
- 5.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235(1):267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheent K, Khakoo SI. Natural killer cells and hepatitis C: action and reaction. Gut. 2011;60(2):268–278. doi: 10.1136/gut.2010.212555. [DOI] [PubMed] [Google Scholar]
- 7.Coudert JD, Zimmer J, Tomasello E, Cebecauer M, Colonna M, Vivier E, et al. Altered NKG2D function in NK cells induced by chronic exposure to NKG2D ligand-expressing tumor cells. Blood. 2005;106(5):1711–1717. doi: 10.1182/blood-2005-03-0918. [DOI] [PubMed] [Google Scholar]
- 8.De Maria A, Fogli M, Mazza S, Basso M, Picciotto A, Costa P, et al. Increased natural cytotoxicity receptor expression and relevant IL-10 production in NK cells from chronically infected viremic HCV patients. Eur J Immunol. 2007;37(2):445–455. doi: 10.1002/eji.200635989. [DOI] [PubMed] [Google Scholar]
- 9.Espinoza JL, Takami A, Onizuka M, Sao H, Akiyama H, Miyamura K, et al. NKG2D gene polymorphism has a significant impact on transplant outcomes after HLA-fully-matched unrelated bone marrow transplantation for standard risk hematologic malignancies. Haematologica. 2009;94(10):1427–1434. doi: 10.3324/haematol.2009.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferenci P, Laferl H, Scherzer TM, Maieron A, Hofer H, Stauber R, et al. Peginterferon Alfa-2a/Ribavirin for 48 or 72 weeks in hepatitis C genotypes 1 and 4 patients with slow virologic response. Gastroenterology. 2010;138(2):503–512. doi: 10.1053/j.gastro.2009.10.058. [DOI] [PubMed] [Google Scholar]
- 11.Forel JM, Chiche L, Thomas G, Mancini J, Farnarier C, Cognet C, et al. Phenotype and functions of natural killer cells in critically-ill septic patients. PLoS ONE. 2012;7(12):e50446. doi: 10.1371/journal.pone.0050446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furue H, Kumimoto H, Matsuo K, Suzuki T, Hasegawa Y, Shinoda M, et al. Opposite impact of NKG2D genotype by lifestyle exposure to risk of aerodigestive tract cancer among Japanese. Int J Cancer. 2008;123(1):181–186. doi: 10.1002/ijc.23456. [DOI] [PubMed] [Google Scholar]
- 13.Furue H, Matsuo K, Kumimoto H, Hiraki A, Suzuki T, Yatabe Y, et al. Decreased risk of colorectal cancer with the high natural killer cell activity NKG2D genotype in Japanese. Carcinogenesis. 2008;29(2):316–320. doi: 10.1093/carcin/bgm260. [DOI] [PubMed] [Google Scholar]
- 14.Gao B, Radaeva S, Park O. Liver natural killer and natural killer T cells: immunobiology and emerging roles in liver diseases. J Leukoc Biol. 2009;86(3):513–528. doi: 10.1189/JLB.0309135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 16.Glassner A, Eisenhardt M, Kokordelis P, Kramer B, Wolter F, Nischalke HD, et al. Impaired CD4(+) T cell stimulation of NK cell anti-fibrotic activity may contribute to accelerated liver fibrosis progression in HIV/HCV patients. J Hepatol. 2013;59(3):427–433. doi: 10.1016/j.jhep.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 17.Groh V, Smythe K, Dai Z, Spies T. Fas-ligand-mediated paracrine T cell regulation by the receptor NKG2D in tumor immunity. Nat Immunol. 2006;7(7):755–762. doi: 10.1038/ni1350. [DOI] [PubMed] [Google Scholar]
- 18.Harrison RJ, Ettorre A, Little AM, Khakoo SI. Association of NKG2A with treatment for chronic hepatitis C virus infection. Clin Exp Immunol. 2010;161(2):306–314. doi: 10.1111/j.1365-2249.2010.04169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi T, Imai K, Morishita Y, Hayashi I, Kusunoki Y, Nakachi K. Identification of the NKG2D haplotypes associated with natural cytotoxic activity of peripheral blood lymphocytes and cancer immunosurveillance. Cancer Res. 2006;66(1):563–570. doi: 10.1158/0008-5472.CAN-05-2776. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson AM, Diefenbach A, McMahon CW, Xiong N, Carlyle JR, Raulet DH. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity. 2002;17(1):19–29. doi: 10.1016/S1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- 21.Jinushi M, Takehara T, Tatsumi T, Kanto T, Miyagi T, Suzuki T, et al. Negative regulation of NK cell activities by inhibitory receptor CD94/NKG2A leads to altered NK cell-induced modulation of dendritic cell functions in chronic hepatitis C virus infection. J Immunol. 2004;173(10):6072–6081. doi: 10.4049/jimmunol.173.10.6072. [DOI] [PubMed] [Google Scholar]
- 22.Keam SJ, Cvetkovic RS. Peginterferon-alpha-2a (40 kD) plus ribavirin: a review of its use in the management of chronic hepatitis C mono-infection. Drugs. 2008;68(9):1273–1317. doi: 10.2165/00003495-200868090-00006. [DOI] [PubMed] [Google Scholar]
- 23.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345(1):41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 24.Ma J, Guo X, Wu X, Li J, Zhu X, Li Z, et al. Association of NKG2D genetic polymorphism with susceptibility to chronic hepatitis B in a Han Chinese population. J Med Virol. 2010;82(9):1501–1507. doi: 10.1002/jmv.21855. [DOI] [PubMed] [Google Scholar]
- 25.Manns MP, Wedemeyer H, Cornberg M. Treating viral hepatitis C: efficacy, side effects, and complications. Gut. 2006;55(9):1350–1359. doi: 10.1136/gut.2005.076646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marinho RT, Vitor S, Velosa J. Benefits of curing hepatitis C infection. J Gastrointestin Liver Dis. 2014;23(1):85–90. [PubMed] [Google Scholar]
- 27.Matsuura K, Isogawa M, Tanaka Y. Host genetic variants influencing the clinical course of hepatitis B virus infection. J Med Virol. 2016;88(3):371–379. doi: 10.1002/jmv.24350. [DOI] [PubMed] [Google Scholar]
- 28.Oliviero B, Varchetta S, Paudice E, Michelone G, Zaramella M, Mavilio D, et al. Natural killer cell functional dichotomy in chronic hepatitis B and chronic hepatitis C virus infections. Gastroenterology. 2009;137(3):1151–1160. doi: 10.1053/j.gastro.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 29.Orange JS, Fassett MS, Koopman LA, Boyson JE, Strominger JL. Viral evasion of natural killer cells. Nat Immunol. 2002;3(11):1006–1012. doi: 10.1038/ni1102-1006. [DOI] [PubMed] [Google Scholar]
- 30.Ozaras R, Yemisen M, Balkan II. Current and future therapies for hepatitis C virus infection. N Engl J Med. 2013;369(7):679. doi: 10.1056/NEJMc1307589. [DOI] [PubMed] [Google Scholar]
- 31.Radaeva S, Sun R, Jaruga B, Nguyen VT, Tian Z, Gao B. Natural killer cells ameliorate liver fibrosis by killing activated stellate cells in NKG2D-dependent and tumor necrosis factor-related apoptosis-inducing ligand-dependent manners. Gastroenterology. 2006;130(2):435–452. doi: 10.1053/j.gastro.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 32.Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol. 2013;31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Investig. 2009;119(7):1745–1754. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat Med. 2013;19(7):859–868. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saez-Borderias A, Guma M, Angulo A, Bellosillo B, Pende D, Lopez-Botet M. Expression and function of NKG2D in CD4 + T cells specific for human cytomegalovirus. Eur J Immunol. 2006;36(12):3198–3206. doi: 10.1002/eji.200636682. [DOI] [PubMed] [Google Scholar]
- 36.Schulte D, Vogel M, Langhans B, Kramer B, Korner C, Nischalke HD, et al. The HLA-E(R)/HLA-E(R) genotype affects the natural course of hepatitis C virus (HCV) infection and is associated with HLA-E-restricted recognition of an HCV-derived peptide by interferon-gamma-secreting human CD8(+) T cells. J Infect Dis. 2009;200(9):1397–1401. doi: 10.1086/605889. [DOI] [PubMed] [Google Scholar]
- 37.Sene D, Levasseur F, Abel M, Lambert M, Camous X, Hernandez C, et al. Hepatitis C virus (HCV) evades NKG2D-dependent NK cell responses through NS5A-mediated imbalance of inflammatory cytokines. PLoS Pathog. 2010;6(11):e1001184. doi: 10.1371/journal.ppat.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. NKG2D function protects the host from tumor initiation. J Exp Med. 2005;202(5):583–588. doi: 10.1084/jem.20050994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stegmann KA, Bjorkstrom NK, Veber H, Ciesek S, Riese P, Wiegand J, et al. Interferon-alpha-induced TRAIL on natural killer cells is associated with control of hepatitis C virus infection. Gastroenterology. 2010;138(5):1885–1897. doi: 10.1053/j.gastro.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 40.Terilli RR, Cox AL. Immunity and hepatitis C: a review. Curr HIV/AIDS Rep. 2013;10(1):51–58. doi: 10.1007/s11904-012-0146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veldt BJ, Heathcote EJ, Wedemeyer H, Reichen J, Hofmann WP, Zeuzem S, et al. Sustained virologic response and clinical outcomes in patients with chronic hepatitis C and advanced fibrosis. Ann Intern Med. 2007;147(10):677–684. doi: 10.7326/0003-4819-147-10-200711200-00003. [DOI] [PubMed] [Google Scholar]
- 42.Viant C, Fenis A, Chicanne G, Payrastre B, Ugolini S, Vivier E. SHP-1-mediated inhibitory signals promote responsiveness and anti-tumour functions of natural killer cells. Nat Commun. 2014;5:5108. doi: 10.1038/ncomms6108. [DOI] [PubMed] [Google Scholar]
- 43.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 44.Witthoft T. Review of consensus interferon in the treatment of chronic hepatitis C. Biologics. 2008;2(4):635–643. doi: 10.2147/btt.s1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Basher F, Wu JD. NKG2D ligands in tumor immunity: two sides of a coin. Front Immunol. 2015;6:97. doi: 10.3389/fimmu.2015.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]