Abstract
Solid tumors develop as three-dimensional tissue constructs. As tumors grow larger, spatial gradients of nutrients and oxygen and inadequate diffusive supply to cells distant from vasculature develops. Hypoxia initiates signaling and transcriptional alterations to promote survival of cancer cells and generation of cancer stem cells (CSCs) that have self-renewal and tumor-initiation capabilities. Both hypoxia and CSCs are associated with resistance to therapies and tumor relapse. Here, we conduct a systematic study to demonstrate that 3D cancer cell models, known as tumor spheroids, generated with a polymeric aqueous two-phase system (ATPS) technology capture these important biological processes. Similar to solid tumors, spheroids of triple negative breast cancer (TNBC) cells deposit major extracellular matrix proteins. Our molecular analysis establishes presence of hypoxic cells in the core region and expression of CSC gene and protein markers including CD24, CD133, and Nanog. Importantly, these spheroids resist treatment with chemotherapy drugs. We successfully target drug resistant spheroids through a combination treatment approach using a hypoxia-activated prodrug, TH-302, and a chemotherapy drug, doxorubicin. This study demonstrates that ATPS spheroids recapitulate important biological and functional properties of solid tumors and provide a unique model for studies in cancer research.
Keywords: spheroids, aqueous two-phase system, hypoxia, cancer stem cells, drug resistance
Graphical Abstract
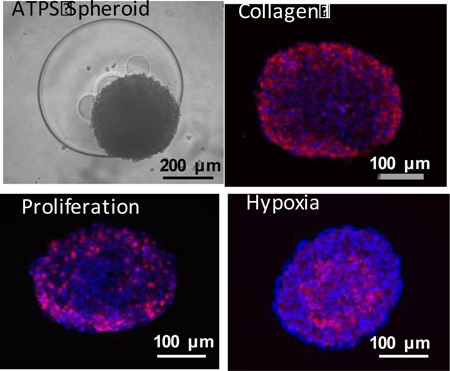
Three dimensional cancer spheroids formed with the aqueous two phase system technology (top, left) at a larger density reproduce key biological properties such as collagen I expression (top, right), density dependent proliferation of only outer cells (bottom, left), and inner core hypoxia (bottom, right) that correlate with drug resistance and cancer stem cell properties.
1. Introduction
During early stages of epithelial cancers, neoplastic cells invade through the basement membrane and proliferate abnormally as three-dimensional (3D) cellular masses supplied with nutrients through simple diffusion from surrounding vasculature.[1,2] The resulting solid tumors contain phenotypically and functionally heterogeneous cells and continue to grow until availability of oxygen and nutrients to cells distant from surrounding vasculature becomes limited.[1–3] Tumor hypoxia activates hypoxia-inducible factor (HIF) pathway by promoting protein stability of HIF-α subunits and their translocation to the nucleus. Binding of HIF transcription factors to hypoxia-responsive elements in target genes leads to activation of hundreds of genes that enable cells to adapt to low oxygen and survive in hypoxic environments.[4] Hypoxia promotes resistance to chemotherapy and radiation therapy through mechanisms such as altered cellular metabolism and genetic instability that drive formation of drug resistant subpopulations of cells.[5] More recent studies show that tumors often harbor cells with self-renewal and drug resistance capabilities, i.e., cancer stem cells (CSCs).[6,7] It is known that HIF-1 expression induces expression of genes, such as a gene encoding the stem cell factor that participate in stem cell maintenance.[8,9] Tumor hypoxia and presence of CSCs correlate with chemotherapy resistance, which is a main reason for failure of cancer treatment in patients with advanced, inoperable cancers.[10]
Widely-used monolayer (2D) cell cultures permit cancer cells unrestricted access to nutrients and oxygen, unlike solid tumors. As a result, critical elements such as mechanical and biochemical signaling and intercellular communications present in tumors are absent from standard 2D cell cultures.[11,12] To study solid tumors in vitro, it is essential to employ culture systems that preserve their biological properties. 3D cultures provide a unique tool to model tumor environments.[13] Cancer cell spheroids present a relevant model due to their 3D structure, complex intercellular network, and restricted diffusion of nutrients and oxygen.[14,15] Unfortunately, spheroid culture techniques face difficulties in production of consistently-sized spheroids in standard labware, ease of maintenance, drug treatment, analysis of cellular responses, and downstream molecular analysis. To facilitate routine use of spheroids, we have developed a high throughput spheroid printing microtechnology using a polymeric aqueous two-phase system (ATPS) that allows simple and quick spheroid formation in standard micro-well plates. Here, we utilize triple negative breast cancer (TNBC) as a model for biological and functional characterization and drug response analysis of ATPS spheroids. TNBC is the most lethal subtype of breast cancer. Treatment options for TNBC are limited, underscoring the need for new approaches to therapy.[16]
To generate spheroids with ATPS technology, a submicroliter drop of the denser polymeric aqueous phase mixed with cancer cells is robotically dispensed into wells of a non-adherent micro-well plate containing the immersion polymeric aqueous phase. The drop phase maintains cancer cells in close proximity and facilitates formation of a single spheroid upon incubation. The ATPS technology is adapted to standard 384-microwell plates where spheroids can easily be maintained or treated by direct addition of media or drug solutions, respectively. Additionally, drug responses can be conveniently evaluated using standard plate readers by direct addition of cellular viability reagents. We demonstrate that TNBC spheroids formed with the ATPS technology reproduce key biological properties and functionality of solid tumors including proliferation patterns, deposition of matrix proteins, hypoxia, expression of CSC markers, and drug resistance. Recapitulating these properties of tumors in vitro, coupled with the potential to incorporate other complexities of tumor microenvironment make the ATPS spheroid technology a unique 3D culture approach for generation of physiologically relevant tumor models for use in biological and drug screening studies.
2. Results and Discussion
2.1 ATPS Microprinting of Spheroids in 384-Microwell Plates
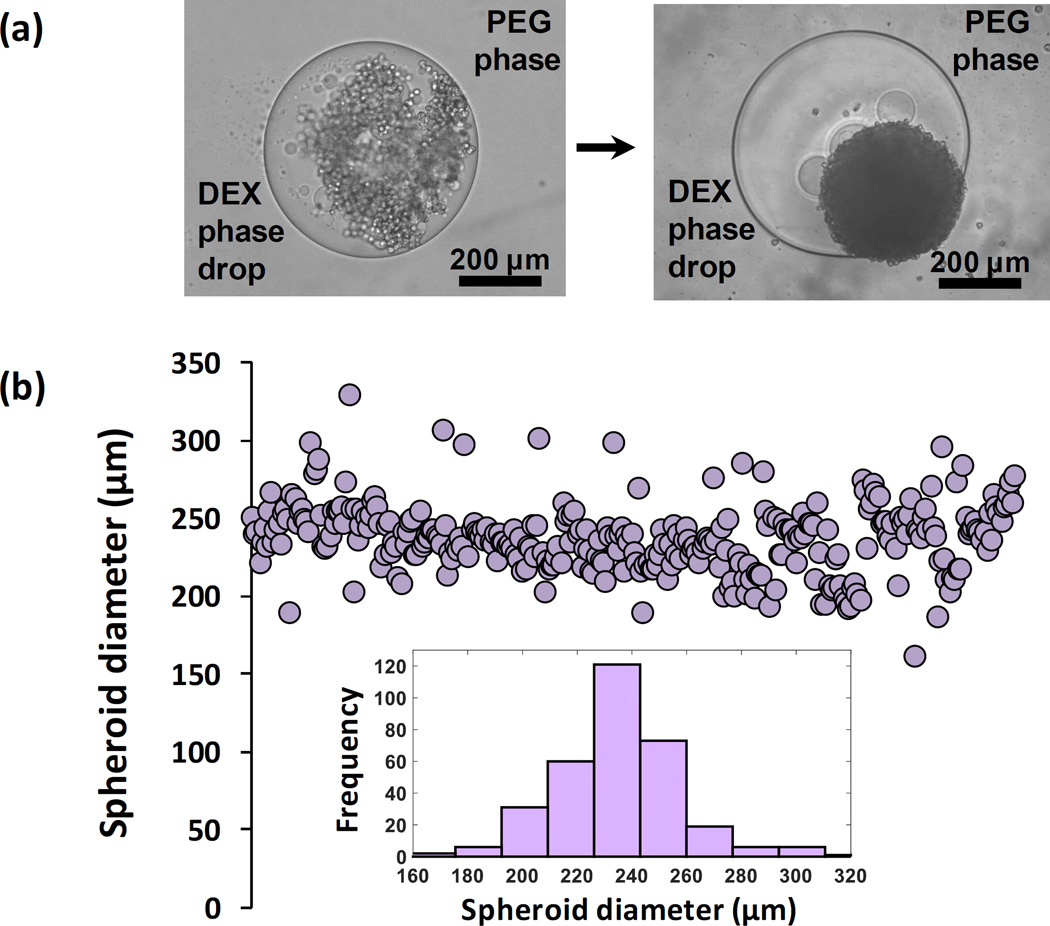
The ATPS spheroid technology forms a single MDA-MB-157 cell spheroid in each well of a standard 384-microwell plate within 24 hours of incubation (Figure 1a). The key to this efficient process is the immiscibility of a 0.3 µl aqueous DEX phase drop and the immersion PEG phase and favorable partition of cells to the drop phase due to an ultralow interfacial tension between the two aqueous phases.[17–19] Use of two highly aqueous phases ensures free diffusion of nutrients from the immersion phase to cells in the low-volume drop and removal of waste products of cells from the drop phase. Cancer cells restricted to the DEX phase drop spontaneously aggregate and form a spheroid. Printing MDA-MB-157 spheroids with a density of 1.5×104 cells results in an average diameter of 235 µm with a standard deviation of ~8% from the mean within each microplate (Figure 1b) that is reproducible across different plates. The variation of diameter for individual spheroids is typical of a normal distribution (Figure 1b, inset). The ATPS approach reproducibly generates consistently-sized spheroids at different cell densities (2.5×104, 5.0×104, and 1.0×105 shown in Figure SI-1). While this technique is compatible with manual pipetting, the use of liquid handling robotics reduces variability and labor and increases precision and efficiency of dispensing low-volume DEX phase drops into the aqueous PEG phase. Spheroid consistency and uniformity upon formation are critical to establish a similar metabolic activity baseline for all spheroids in a microplate and ensure that differences in metabolic activity are caused by treatment with drug compounds and not variations in size of spheroids. Once spheroids form, they are conveniently maintained by direct addition of fresh media to the wells. The addition reduces concentrations of polymers below minimum concentrations required for maintaining a two-phase system and converts the ATPS to a single media phase. Therefore, ATPS is solely used as a patterning medium to generate spheroids. Micropatterning with ATPS has additionally facilitated the generation of cancer cell niches for migration studies.[20,21]
Figure 1.
(a) Spontaneous spheroid formation using aqueous two phase system (ATPS) microtechnology; a compact MDA-MB-157 spheroid forms within 24 hrs of incubation (right). (b) This robotic approach produces spheroids of highly consistent diameter (b) with a normal distribution (inset).
2.2 Evaluation of Cell Viability of Spheroids
We used a standard PrestoBlue assay to quantify viability of ATPS spheroids since it is a one-step addition assay that eradicates the need for multiple wash steps or transfer of spheroids to special plates for viability analysis. PrestoBlue primarily has been used with 2D cultures, producing viability data similar to the conventional XTT assay.[22] Spheroids of four different cell densities (1.5×104, 2.5×104, 5.0×104, and 1.0×105) were formed in a 384-well plate and incubated with PrestoBlue. The media fluorescent intensity due to the reduction of resazurin to resorufin by metabolically active cells was measured for all spheroid densities every hour for 4 hours (Figure 2). Between incubation time points of 2 and 3 hours, the fluorescent signal became significantly different within each group, i.e., spheroids of the same cell density. With longer incubation time, the fluorescent signal differences became more pronounced. Based on a statistical analysis, it was validated that 3–4 hours of incubation was optimal to measure cell viability of spheroids. Slower diffusion of the resazurin component into spheroids is most likely the reason for the longer incubation time compared to 2D cultures. Incubations longer than 4 hours resulted in insignificant changes of the fluorescent signal within each group due to saturation. Thus, this test provided an optimal incubation time to evaluate cell viability of drug treated spheroids below. Our previous work also validated that the use of PrestoBlue allows us to sensitively detect increases in the fluorescent signal intensity from growing spheroids during long-term cultures.[23]
Figure 2.
Optimization of PrestoBlue assay to determine viability of cells in spheroid cultures. PrestoBlue is directly added to wells containing spheroids and metabolized by live cells. Based on the cellular metabolic activity, the solution emits a fluorescence signal detectable with standard plate readers. Incubation of spheroids with PrestoBlue for 3–4 hours optimally resolves cell viability.
2.3 Extracellular Matrix Deposition of Spheroids
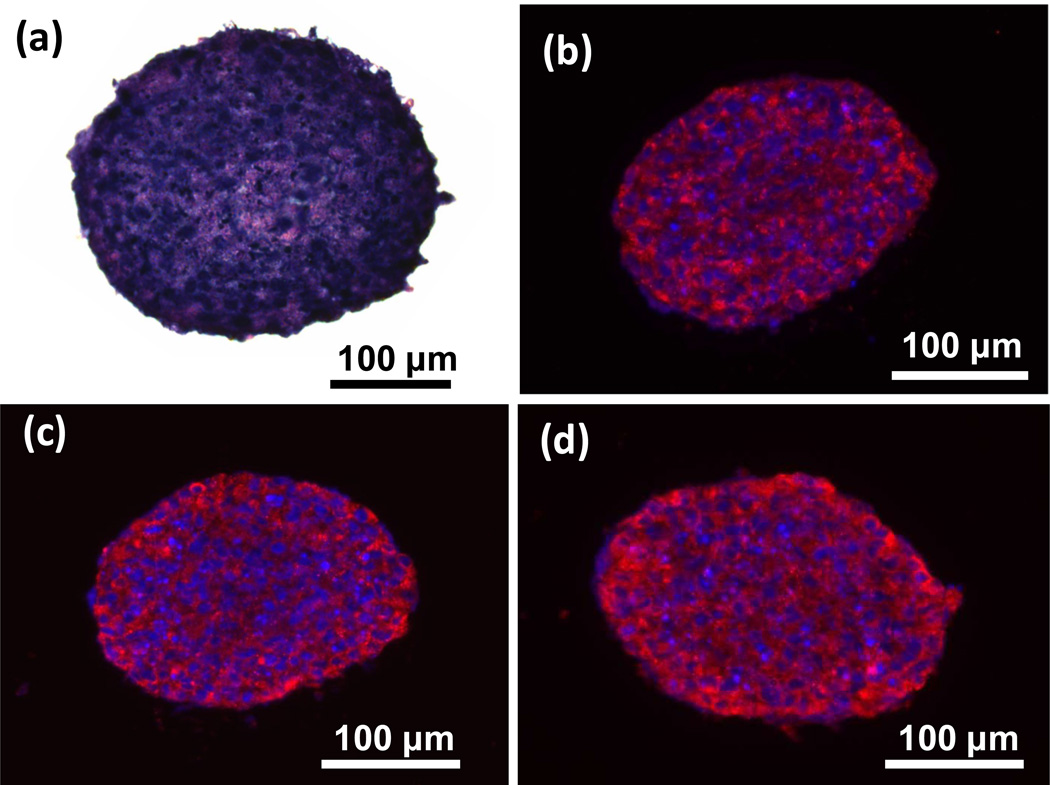
Cryosections of MDA-MB-157 spheroids were stained with H&E to examine their interior morphology. This staining showed a highly compact and dense cellular network without any gaps (Figure 3a), similar to a previous study that reports TNBC patient tumors to be highly cellularized and compact with a low fraction of extravascular and extracellular space.[24] Next, cryosections were immunostained for extracellular matrix (ECM) proteins type I collagen, laminin, and fibronectin. MDA-MB-157 spheroids of smaller cell density (1.5×104) showed expression of all three proteins, with greater abundance of collagen I and laminin (Figure 3b–d). Cell-ECM interactions play major roles in the tumor microenvironment.[25] Studies show that expression of both collagen I and fibronectin is elevated in breast cancer and associated with the tumor formation process.[25,26] Laminin is also highly expressed in invasive breast cancers and promotes cell migration, a process fundamental to metastasis.[26] The expression of ECM proteins is implicated in drug and radiation resistance of breast cancer cells.[26,27] Therefore, ATPS spheroids display critical cell-matrix interactions. Interestingly, larger density spheroids of MDA-MB-157 cells (1.0×105) showed lower expression of ECM proteins in the inner core cancer cells (Figure SI-2), suggesting reduced activity of these cells due to limited nutrients and oxygen.
Figure 3.
(a) H&E staining of a 1.5×104 cell density MDA-MB-157 spheroid shows a compact intercellular network. Blue and purple represent nuclei and cytoplasm, respectively. (b–d) Immunohistochemical analysis for ECM proteins (shown in red) show deposition of (b) collagen I, (c) fibronectin, and (d) laminin in ATPS spheroids. Blue represents nuclei staining with Hoechst.
2.4 Spheroid Size Dependent Proliferation and Hypoxia
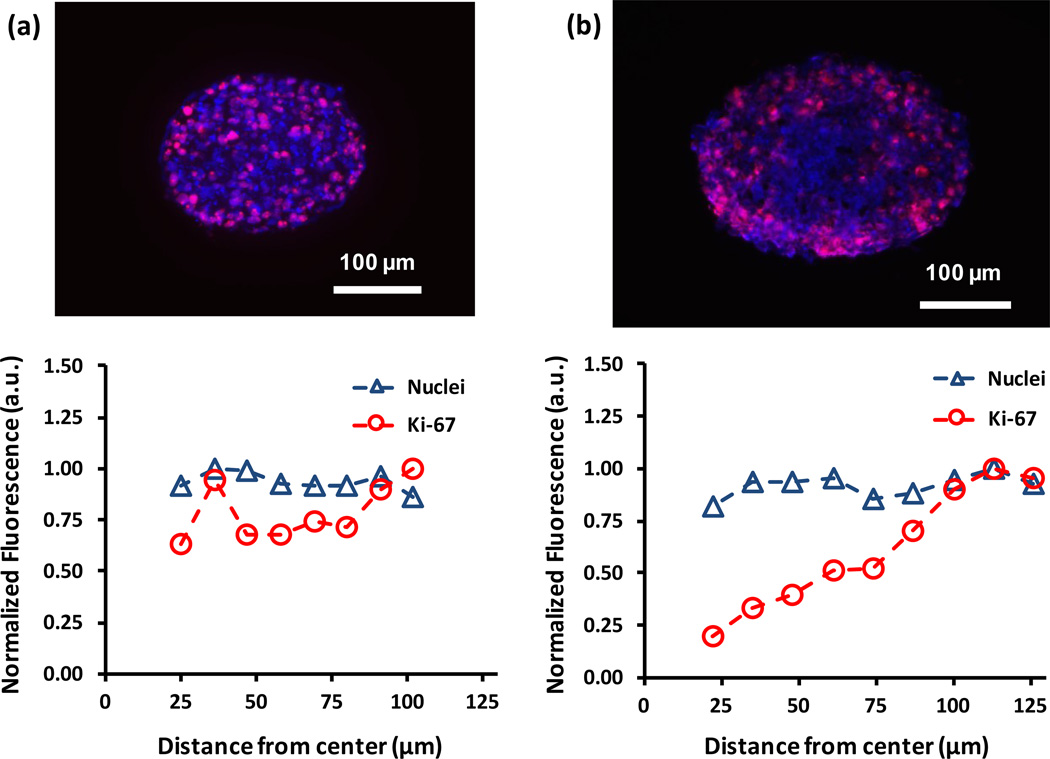
We immunostained cryosections of MDA-MB-157 spheroids formed with 1.5×104 and 1.0×105 cells for the cell proliferation marker Ki-67. This staining showed differential distribution of proliferative cells. In smaller spheroids, proliferative cells were distributed homogenously throughout the spheroid (Figure 4a). By comparison, proliferative cells in larger spheroids were mainly present toward the periphery of the spheroid (Figure 4b). Quantitative analysis of distribution of Ki-67+ cells confirmed this observation (Figure 4). Considering the average radius of large spheroids (220±22 µm), it is likely that diffusion limitations of nutrients and oxygen to cells residing in the core zone retards their proliferative activities. This is consistent with suggested free diffusion distance of 150–200 µm.[28] Smaller sized spheroids with a radius of 118±11 µm allow free diffusive molecular transport to support cellular proliferation in the central zone. Another potential cause for the lack of Ki-67+ cells in the core region of large spheroids is greater consumption of nutrients and oxygen by densely packed cells at the peripheral zone. Unlike several other cancer cells we have used, MDA-MB-157 cells are tightly packed within spheroids, evident from the H&E staining result (Figure 3a). Therefore, in addition to limited diffusive transport, close intercellular contacts and packing of cancer cells within spheroids and consumption of available oxygen and nutrients by cells in peripheral layers is a potential cause of lack of proliferative activity in the core region. Assuming a spherical shape for spheroids, 85% of cells in the large spheroids reside in a spherical shell defined by the radii of 1.5×104 and 1.0×105 cell spheroids, supporting the above explanation. Non-uniform distribution of proliferative cells is reminiscent of solid tumors with poor vascularization.[29]
Figure 4.
Immunostained cryosections of (a) 1.5×104 and (b) 1.0×105 cell density spheroids show the distribution of Ki-67 positive (pink) proliferative cells. Analysis of stained sections is used to compare the distribution of nuclei (blue) and proliferative cells throughout each section. The larger spheroid contains a non-uniform distribution of proliferative cells compared to the smaller spheroid.
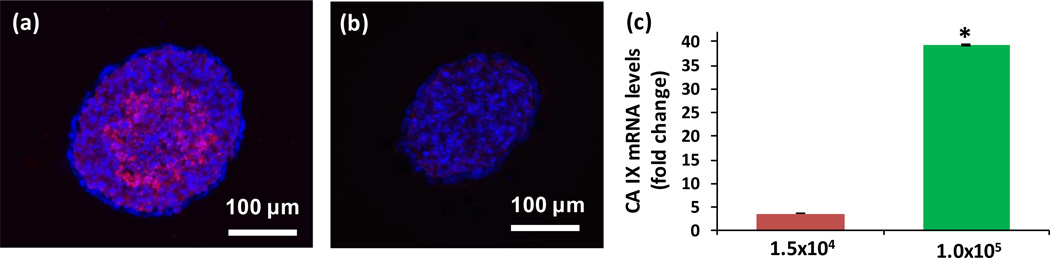
The disparity between oxygen demand and consumption of cancer cells in solid tumors generates hypoxia.[30] Tumor hypoxia has been implicated in processes including cancer cell survival, resistance to cell death, tumor angiogenesis, invasion, metastasis, radioresistance, and chemoresistance. Hypoxic regions occur more commonly in TNBC than other molecular subtypes of this disease.[28,31–33] Therefore, we investigated whether hypoxia could account for lack of cell proliferation in the core of the large TNBC spheroids. We used an exogenous 2-nitroimidazole probe, pimonidazole, that binds covalently to SH-containing molecules (thiols) in hypoxic tissues with less than 10 mmHg of oxygen partial pressure.[28,33] Cryosections of large TNBC spheroids stained positive in their core region (Figure 5a), whereas smaller spheroids lacked pimonidazole staining (Figure 5b). We further validated this result by measuring an endogenous transcriptional target of hypoxia-inducible factor-1 (HIF-1), carbonic anhydrase IX (CA IX). CA IX is a surrogate hypoxia marker, regulator of pH of the tumor microenvironment, and a prognostic factor found significantly more in basal-like breast tumors.[34,35] Our q-PCR analysis showed that large TNBC spheroids display a 39.2 fold change in CA IX gene expression compared to smaller spheroids with only a 3.6 fold change (Figure 5c). These results are consistent with cell proliferation patterns based on Ki-67 staining and explain the lack of proliferative activity. Considering slow cycling cells and activity of pro-survival pathways under hypoxic conditions,[36] the use of ATPS spheroids provides an opportunity to study hypoxia-mediated drug responses of cancer cells with a biologically relevant tumor model and without inducing systemic atmospheric oxygen deficiency in culture that is the current standard to generate hypoxia.
Figure 5.
Pimonidazole staining (pink) of cryosections of (a) 1.0×105 and (b) 1.5×104 cell density spheroids of MDA-MB-157 shows presence of a hypoxic core only in the larger spheroid. (c) q-PCR analysis of expression of the hypoxic marker CA IX in cells of both spheroids validates the results of immunostaining. mRNA levels are normalized with respect to a monolayer of MDA-MB-157 cells. Error bars represent the standard error of mean for three trials. Blue represents nuclei staining with Hoechst. (*p < 0.01)
2.5 Drug Resistance of Hypoxic Spheroids
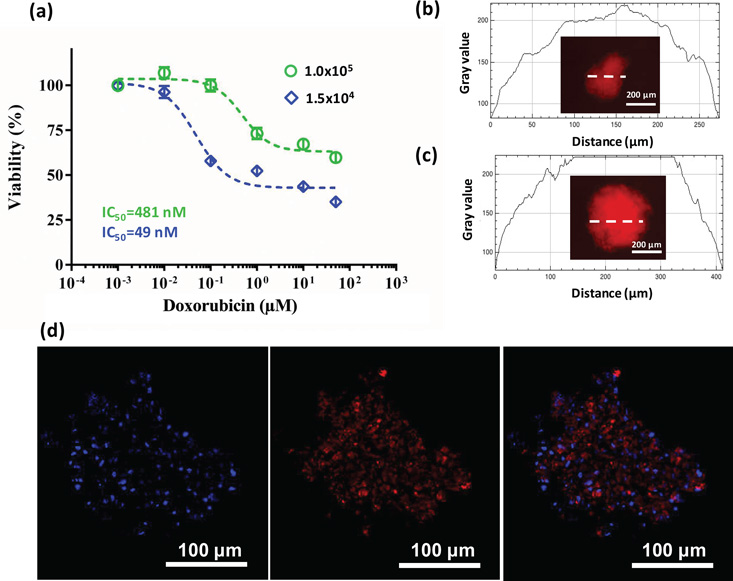
MDA-MB-157 spheroids of both 1.5×104 and 1.0×105 cell densities were treated with varying concentrations of doxorubicin, a standard chemotherapeutic drug currently used for TNBC treatment (Figure 6a).[37] Spheroids showed a sigmoidal response to doxorubicin treatment; however, the larger spheroids displayed drug resistance. With a ten-fold higher IC50 (481 nM), the resulting cell viability of larger spheroids was much higher. The largest difference was a percent viability of 42% that occurred at a drug concentration of 100 nM. Doxorubicin resistance of larger spheroids persisted at higher concentrations. First, we utilized the natural red fluorescence of doxorubicin to evaluate whether insufficient penetration of the drug into spheroids caused the observed resistance.[29] Fluorescent imaging of spheroids after 48 hrs of incubation with doxorubicin revealed a homogenous drug distribution within spheroids of both densities and a similar maximum signal intensity (Figure 6b,c) and presence of doxorubicin in the nuclei of cells (Figure 6d). This indicated that drug resistance of larger spheroids was not due to limited drug penetration.
Figure 6.
(a) Dose-response of 1.5×104 and 1.0×105 cell density MDA-MB-157 spheroids to doxorubicin treatment shows drug resistance of larger spheroids. Error bars represent the standard error of mean. (b), (c) Gray values of fluorescence intensity measurements along a sample line crossing spheroids of both densities show complete penetration of doxorubicin into spheroids after 48 hours of incubation. (d) Doxorubicin localization in the nuclei of 1.0×105 cell density spheroids after 48 hrs of treatment. Blue represents nuclei staining with Hoechst.
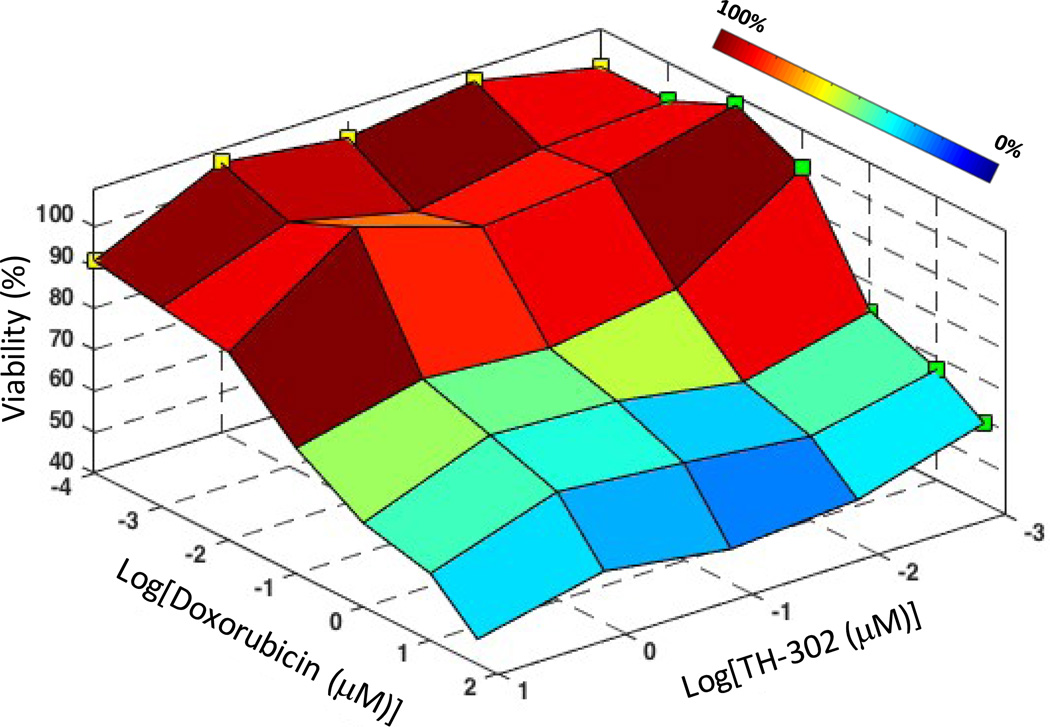
Next, we asked whether hypoxia in larger spheroids mediates drug resistance. Hypoxic cells may show resistance to anticancer drugs for several reasons, including reduced proliferation and metabolism due to insufficient oxygen and nutrients, lost sensitivity to p53-mediated apoptosis and consequent reduced sensitivity to certain drugs, and activation of genes involved in drug resistance including multidrug resistance 1 gene (MDR1) that encodes P-glycoprotein, acting as an efflux pump to reduce intracellular concentrations of some drugs.[10,28,33,38–43] We used a hypoxia activated prodrug, TH-302, that has progressed to clinical studies.[44] TH-302 is reduced under hypoxic conditions, releasing a DNA crosslinker bromo-isophosphoramide mustard.[45] This mechanism of TH-302 allows selective drug activation in hypoxic cells in a tumor and has shown hypoxia-induced cytotoxicity against 32 human cancer cell lines.[44] Larger spheroids were co-treated with doxorubicin and TH-302 over wide range of concentrations for each compound. We note that only the larger (hypoxic) spheroids were co-treated due to the hypoxia-dependent mechanism of TH-302. Consistent with previous studies, TH-302 alone at the concentrations used was ineffective.[44,46] However, a major reduction in drug resistance was observed at 100 nM doxorubicin with increasing TH-302 concentration (Figure 7), suggesting that these compounds synergistically target cancer cells. The greatest effect was ~30% reduced cell viability with combination treatment, i.e., 70.4% viability at 10 µM TH-302/100 nM doxorubicin compared to 99.9% viability with 100 nM doxorubicin treatment alone, at which the largest drug resistance was observed (see Figure 6). The 10 µM TH-302/100 nM doxorubicin combination of compounds was synergistic since the viability of co-treated larger spheroids (70.4%) was much lower than the viability data from treatment with each single compound (91.6% at 10 µM TH-302 and 99.9% at 100 nM doxorubicin). We further validated synergism between the two compounds by calculating a combination index (CI), which is a quantitative measure where values smaller than unity indicate synergism, using CompuSyn software.[47] The 10 µM TH-302/100 nM doxorubicin pair resulted in a CI value of 0.02, demonstrating highly synergistic effects of the two drugs.
Figure 7.
Surface plot of viability of 1.0×105 cell density spheroid of MDA-MB-157 cells co-treated with varying concentrations of doxorubicin and TH-302 shows synergistic enhancement in toxicity due to combination treatment. Color bar represents the cell viability range. Green and yellow squares represent cell viability of spheroids from treatment with doxorubicin only and TH-302 only, respectively.
This screening approach allows identifying an optimal pair of concentrations of the cytotoxic compound to target proliferating cells and hypoxia-activated drug to target hypoxic cells. A similar result was found with a combination of a chemopreventive agent (silibinin) and doxorubicin in which specific drug concentrations of 100 µM silibinin/25 nM doxorubicin produced a synergistic effect in breast cancer cells.[48] Our result is also consistent with a few other studies, including a phase II clinical trial, that demonstrated synergistic effects of TH-302 combined with chemotherapy and radiotherapy treatments against cancer cells.[45,49–51] The enhanced efficacy of doxorubicin at a low concentration of 100 nM by addition of 10 µM TH-302 is also advantageous from a practical standpoint to prevent/reduce cardiotoxic side effects.[52,53] Therefore, ATPS spheroids offer solid tumor models to identify effective drug combinations that reduce/overcome cancer cells resistance to chemotherapeutics.
We note that the combination of TH-302 and doxorubicin did not drop cell viability to that of 1.5×104 cell density spheroids, i.e., 52.2%, at 100 nM doxorubicin (diamonds in Figure 6a). This is potentially from the limited potency of the TH-302 compound and the involvement of additional mechanisms of resistance, such as the inoculum effect, which causes reduced cytotoxic activity of a chemotherapy drug as the cancer cell density increases.[54] The inoculum effect with doxorubicin results from insufficient number of drug molecules at high cell densities, causing a decreased drug accumulation in cancer cells.[54,55] For example, increasing the density of cancer cells resulted in a 27-fold decrease in the LD50 of doxorubicin against lymphoma cells[56] and 40 times lower growth-inhibitory capabilities (when comparing LD50 values) in leukemia cells.[55] Increasing doxorubicin concentrations substantially reduced this effect.[54] Our results also show that combination of 10 µM TH-302 and 50 µM doxorubicin reduces the cell viability to 45.9%, i.e., ~26% further decrease compared to the 10 µM TH-302/100 nM doxorubicin pair and suggesting that the inoculum effect plays a role in doxorubicin resistance of MDA-MB-157 spheroids. Nevertheless, this cell viability is still greater than that of the 1.5×104 cell density spheroids at a similar doxorubicin concentration (see Figure 6), indicating that doxorubicin resistance is caused, at least in part, by hypoxia.
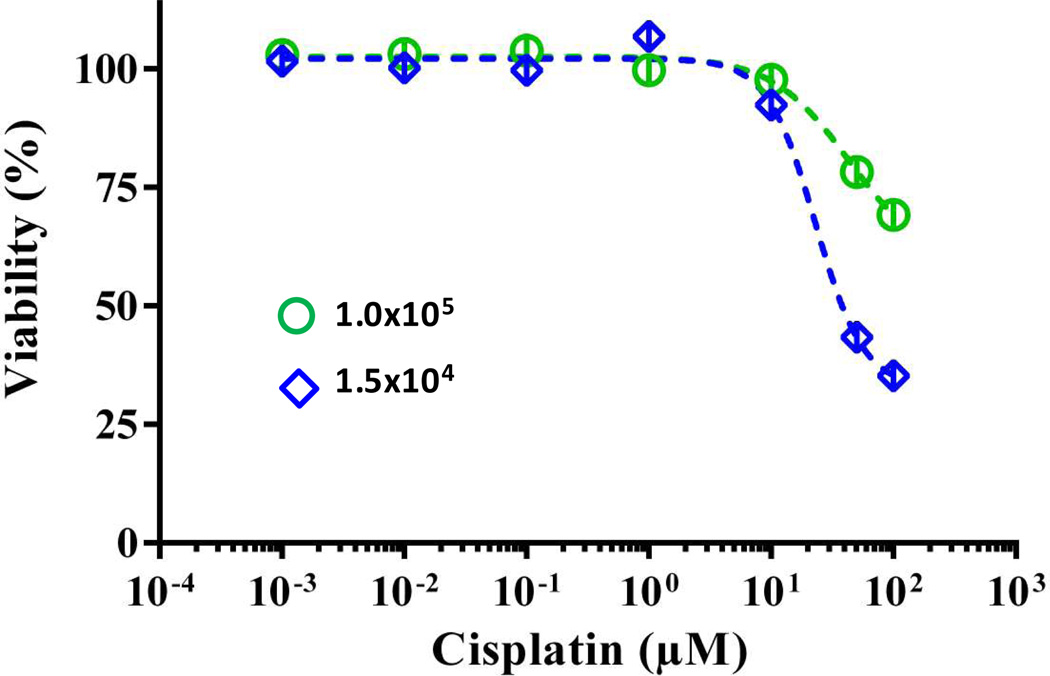
Previous studies showed that unlike with doxorubicin, the inoculum effect was not observed with cisplatin at various cell densities.[55,56] Therefore, we treated small and large spheroids of MDA-MB-157 cells with varying concentrations of cisplatin and determined cellular viability. Large spheroids still displayed resistance at effective concentrations of the drug and displayed ~35% greater viability than the small spheroids (Figure 8). Collectively, our results indicate that the hypoxia in large spheroids has a major influence on drug resistance of breast cancer cells in the absence of the inoculum effect, and that other phenomena such as the inoculum effect with certain chemotherapeutics and limited potency of TH-302 contribute to the observed doxorubicin resistance. A detail study is required to distinguish between such effects.
Figure 8.
Dose-response of MDA-MB-157 spheroids made with 1.0×105 and 1.5×104 cells to cisplatin treatment. Error bars represent the standard error of mean.
2.6 Cancer Stem Cell Markers in Hypoxic Spheroids
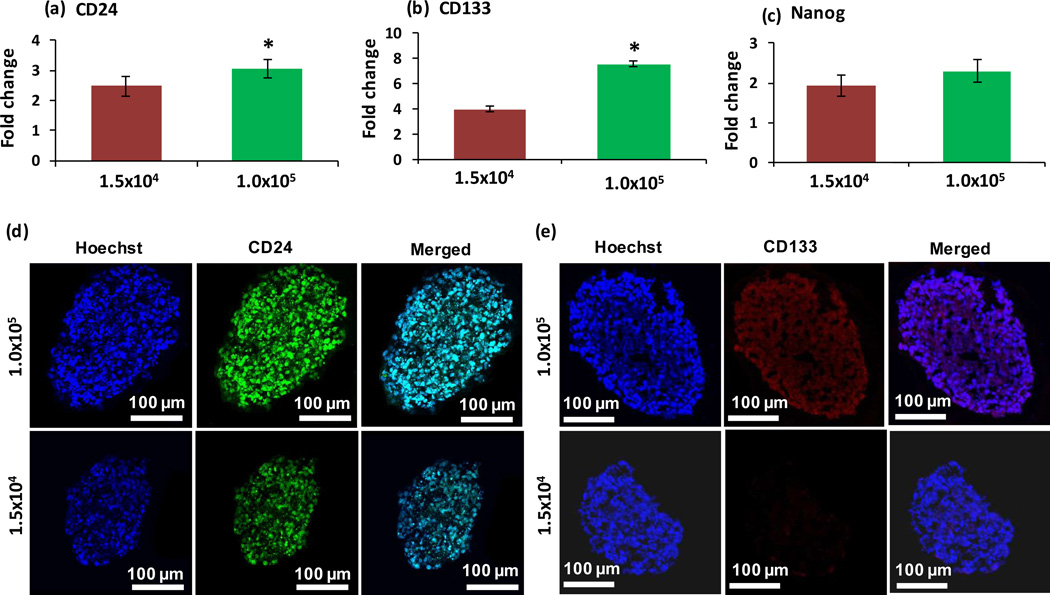
Hypoxia in solid tumors is associated with stem cell-like cancer cells with tumor initiation, recurrence, and metastasis capacity.[57–62] It has been shown that hypoxic environments house stem cells because of their preference for low oxidative DNA damage,[57] and that cancer stem cells (CSCs) rely on hypoxia induced factors for survival and self-renewal.[58] Chemotherapies typically kill the majority of cancer cells in tumors, but CSCs are able to evade therapy due to their resistance mechanisms.[63] The consequent survival of self-renewing CSCs causes recurrence of cancer.[63] To evaluate whether hypoxic tumor spheroids present stem cell markers and the utility of ATPS spheroids to study CSCs, we performed a gene expression study of breast CSC markers CD24, CD44, CD133 (prominin-1), ALDH1, and α6-integrin, and pluripotency markers Oct4, Sox2, and Nanog.[64] This analysis was done in parallel with 1.5×104 and 1.0×105 cell density spheroids of MDA-MB-157 cells, and resulting mRNA levels were normalized to that of a normoxic monolayer of cells for each marker gene. Among these markers, CD24, CD133, and Nanog showed greater expression in hypoxic, 1.0×105 cell density spheroids (Figure 9a–c). Other markers either showed similar expression in both spheroids or lower expression than the monolayer of cells (Figure SI-3). Increased expression of CD24 and CD133 in hypoxic spheroids was additionally confirmed at the protein level using immunostained cryosections (Figure 9d–e). Thus, our data suggests an association between hypoxia in the MDA-MB-157 TNBC spheroids and upregulated activity of these stem cell markers.
Figure 9.
q-PCR analysis of expression of (a) CD24, (b) CD133, and (c) Nanog in 1.5×104 and 1.0×105 cell density spheroids of MDA-MB-157 cells normalized against mRNA levels of a monolayer of cells. Expression levels are relative to -actin and hypoxanthine phosphoribosyltransferase (HPRT) and calculated using the ΔΔCt method. Fold change in mRNA expression represent 2−ΔΔCt. Error bars represent the standard error of mean for three trials. Largest cryosections of 1.5×104 and 1.0×105 cell density spheroids immunostained for cancer stem cell markers (d) CD24 (green) and (e) CD133 (red). Blue represents nuclei staining with Hoechst. (*p < 0.05)
Breast tumor cells with CD44high/CD24low surface markers expression were first proposed as breast CSCs with high tumorigenic capacity[65] and subsequently validated in other studies.[66–69] Nevertheless, growing evidence suggests heterogeneity in populations of CSCs and that breast CSCs cannot be stratified using this marker only.[70,71] Examining metastatic deposits of mammary tumors showed variable expression of CSC markers in different tumor samples and lack of CD44 expression in about one-third of tumor samples but expression of other CSC markers in these cells.[72] Elevated levels of CD24 alone correlated with advanced disease stages in several types of human epithelial cancers including breast cancer and increasing tumor grade and malignancy.[73,74] Recent studies also show that hypoxia promotes CD24 expression[75,76] and that CD44−/CD24+ phenotype predicts poor outcome.[67,77,78] Furthermore, expression of the transmembrane glycoprotein CD133 in primary breast tumors and breast cancer cell lines has been associated with self-renewal of cells,[79] tumor size and grade,[80] tumor angiogenesis,[81,82] invasiveness,[83] and metastasis.[84] Targeting CD133 prevents local tumor recurrence in mouse models of breast cancer.[85,86] Therefore, expression of CD24 and CD133, but not CD44, in MDA-MB-157 tumor spheroids in our study is consistent with these findings and agrees with identified variability in CSC markers among different lines of breast cancer cells.[87] Expression of embryonic stem cell pluripotency markers Sox2, Oct4, and Nanog has also been reported in poorly differentiated breast cancer cells and associated with poor clinical outcome,[88–90] suggesting that regulatory networks controlling the function of stem cells may also be active in breast tumors. Recent studies show that hypoxia leads to transcriptional activation of the gene encoding Nanog in breast CSCs and Nanog-expressing cells present tumor initiation capacity in NSG mice.[6,91,92] Overall, this study suggests association of hypoxia and CSCs in TNBC spheroids and that the ATPS technology provides a useful tumor model for future studies of CSCs and heterogeneity of these cells in distinct environments.
3. Conclusions
The aqueous two-phase system (ATPS) microprinting approach enabled convenient production of spheroids of well-defined size in standard 384-microwell plates and drug treatment and downstream biochemical analysis of cellular responses in the same plate. Resulting spheroids reproduced key properties of solid tumors including compact morphology and deposition of major ECM proteins. Increase in cell density of spheroids resulted in a preferential cell proliferation close to the periphery of spheroids and hypoxia in the core region, indicating consumption of extracellular oxygen by tightly packed layers of cells at the peripheral zone. Hypoxic spheroids showed resistance to a standard chemotherapy drug (doxorubicin) over a wide concentration range. Imaging of drug diffusion into spheroids eliminated widely-proposed diffusion limitations as the cause of resistance and emphasized a major role for hypoxia. A combination treatment of hypoxic spheroids with doxorubicin and a hypoxia-activated prodrug (TH-302) significantly reduced drug resistance, suggesting a potential strategy against multidrug resistance in tumors. Hypoxia in tumor spheroids was associated with the expression of several genes associated with breast cancer stem cells. Altogether, these findings demonstrated the utility of aqueous two-phase tumor spheroids in cancer research to develop biomimetic tumor models that exhibit key biological and functional properties of solid tumors such as hypoxia and drug resistance, and present a tool to study cancer stem cells.
4. Experimental Section
4.1 Aqueous Two-Phase System (ATPS) Preparation
Polyethylene glycol (PEG, Sigma), Mw: 35,000 Da, and dextran (DEX, Pharmacosmos), Mw: 500,000 Da, were used to form an ATPS due to their biocompatibility.[93] PEG and DEX were prepared in complete cell culture media at 5% (w/v) and 12.8% (w/v), respectively. The polymer solutions were vortexed and then kept in a 37°C water bath for 1 hour to ensure complete dissolution. The PEG phase solution was filtered with a 0.2 µm filter to remove any impurities. Both polymer solutions were then stored at 4°C until use.
4.2 Cell Culture
Prior to spheroid formation, MDA-MB-157 triple negative breast cancer (TNBC) cells were grown in T175 flasks at 37°C and 5% CO2 to a confluent monolayer. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Sigma) supplemented with 10% fetal bovine serum (FBS, Sigma), 1% glutamine (Life Technologies), and 1% antibiotic (Life Technologies). Once confluent, cells were rinsed with PBS and dissociated by incubating with 5 ml of trypsin for ~7 min. The cells were collected and neutralized with 10 ml of complete medium, and centrifuged down for 5 min at 1000 rpm. The supernatant was removed and the cells were resuspended in 1 ml of medium for counting with a hemocytometer.
4.3 Spheroid Formation Using ATPS
A 384-well plate (destination plate) for spheroid formation was prepared by adding 30 µl of the immersion PEG phase to each well. The DEX phase was mixed at an equal volume with the MDA-MB-157 cell suspension at a density of 3.0×104 or 2.0×105 cells per 0.3 µl. The addition of the DEX phase solution to these cell suspensions diluted DEX concentration to 6.4% (w/v) and adjusted the cell densities to 1.5×104 and 1.0×105 cells per 0.3 µl. Each well from a column of a 384-well plate (source plate) was filled with ~20 µl of the aqueous DEX phase containing cells. A liquid handling robot (Bravo SRT, Agilent) was used to mix the content of the wells of the source plate and then aspirate 0.3 µl from each well.[94] Then the solution was dispensed as a single drop into each well of one column of the PEG phase-containing destination plate. This process was repeated for all columns of the destination plate, which was then placed in an incubator. Within 24 hours of incubation, a single spheroid of MDA-MB-157 cells formed in each well.
4.4 Evaluation of Viability of Spheroids
A standard cellular metabolic activity detection assay (PrestoBlue) was utilized to determine the viability of spheroids. Metabolically active and viable cells reduce the resazurin compound in PrestoBlue, producing fluorescence detectable with a standard plate reader. Since the PrestoBlue assay had only been optimized by the manufacturer for monolayer cultures, the required incubation time was adjusted for 3D spheroid cultures. This was determined by forming MDA-MB-157 spheroids of 1.5×104, 2.5×104, 5.0×104, and 1.0×105 cell density, adding the PrestoBlue reagent at 10% of total well volume, and measuring their fluorescent signal at different time points. The different cell density spheroids were generated using the ATPS approach with a cell culture media renewal at 24 hrs and maintaining cultures for an additional 24 hrs prior to viability evaluation. Based on this optimization study, we selected an incubation period that produced a significant difference in the fluorescent signal among different sizes of spheroids.
4.5 Histological Examination of Spheroids
To perform histological examination, a previously established spheroid cryosectioning protocol was adapted.[95] First, 3-day old MDA-MB-157 spheroids were gently pipetted from wells into 200 µl microcentrifuge tubes. Supernatant media transferred with spheroids was removed from tubes. Spheroids were fixed with 100 µl of 4% formaldehyde for 10 minutes at room temperature. Following fixation, the spheroids were gently rinsed with 100 µl of PBS three times for 5 minutes. Spheroids were then incubated with 100 µl of 30% (w/v) sucrose solution at 4°C until they sank to the bottom of the microcentrifuge tubes. This ensured water removal from spheroids and prevented crystal formation upon freezing. Next, an equal volume (100 µl) of a tissue freezing medium (Triangle Biomedical Sciences) was added to each tube and incubated at 4°C overnight. The next day, cryomolds were prepared by flash-freezing with dry ice a thin layer of the tissue freezing medium in biopsy-sized cryomolds. Spheroids were then pipetted from the microcentrifuge tubes and dispensed onto the layer of the tissue freezing medium in the cryomold. The frozen layer of the medium was slightly melted at room temperature prior to mixing with the newly added medium containing the spheroid sample. It was important in this process to avoid bubble formation in the medium that would cause hole formation when cryosectioning. Once the tissue embedding medium was homogenous, a fresh second layer was added to the mold to immerse the spheroid sample, which then was flash frozen on dry ice to form frozen molds. Frozen molds containing spheroid samples were stored at −80°C until use. A cryostat was used to section the samples to 10 µm-thick slices. The slices were transferred onto Superfrost Plus microscopic slides (Fisher) upon sectioning. Sections were stained with hematoxylin and eosin (H&E) to examine spheroid morphology and immunostained following standard procedures for specific markers such as a cell proliferation protein marker (Ki-67, Cell Signaling), hypoxia (pimonidazole, HypoxyProbe), extracellular matrix proteins type I collagen (Abcam), laminin (Sigma), and fibronectin (Sigma), and cancer stem cell markers CD24 (Abcam) and CD133 (Novus Biologicals). Expression of these proteins was detected using a fluorescent secondary antibody, Cy3 conjugated goat anti-rabbit (Ki-67, hypoxia, ECM), FITC conjugated donkey anti-mouse (CD24), or Alexa Fluor 594 conjugated donkey anti-mouse (CD133), obtained from Jackson ImmunoResearch. Nuclei were stained with Hoechst (Life Technologies). Fluorescent images were captured using an inverted fluorescent microscope (Axio Observer, Zeiss) equipped with a high resolution camera (AxioCam MRm, Zeiss) or using a confocal microscope (Fluoview FV1000, Olympus). ImageJ (NIH) was used for image processing and analysis of Ki-67 expression in spheroid sections. Each spheroid section in captured images was manually divided into elliptical rings; starting from the border toward the center of each section, each ring was 10% smaller in diameter than its preceding one. The number and size of rings used for analysis was based on the size of spheroid sections. The total fluorescence of each ring was measured and normalized to its area using the integrated density and area measurements in ImageJ.
4.6 Preparation of Drug Compounds and Testing
A 50 mM stock solution of doxorubicin (Sellekchem) was prepared in DMSO and stored at −80°C according to the manufacturer’s instructions. TH-302 (Santa Cruz Biotechnology) was dissolved in DMSO at a concentration of 44.5 mM and stored at −20°C according to the manufacturer’s instructions. The desired drug concentrations for experiments were prepared by serial dilution of each stock solution in the complete cell culture medium. Since the drug solutions were directly added to wells containing half of the desired total volume, drug concentrations were prepared at 2× the desired concentration (for single drug experiments) or 4× the desired concentration (for combination drug experiments). Cisplatin (Spectrum Chemicals) was dissolved in ultrapure sterile water at a stock concentration of 2 mg/ml, which was then serially diluted to 2× working concentrations for the drug test against MDA-MB-157 TNBC spheroids.
After MDA-MB-157 spheroid formation in ATPS at 24 hours of culture (day 1), 30 µl of cell culture medium was directly added to the wells already containing the PEG phase solution. This addition of the medium disrupted the initial concentrations of PEG and DEX, causing a single phase solution of medium with small residues of polymers. After 72 hours of spheroid culture (day 3), 30 µl of medium was removed from each well using a multichannel pipette. Then, 30 µl of drug solutions at varying concentrations (prepared at 2× the desired concentrations) was added to wells for single drug testing with doxorubicin or cisplatin. For combination drug testing with doxorubicin and TH-302, 15 µl of each drug solution at varying concentrations (prepared at 4× the desired concentrations) was added to wells. Wells containing control (non-treated) spheroids received 30 µl of fresh medium. Spheroids were incubated for 48 hours, and their viability was evaluated using the PrestoBlue assay.
4.7 q-PCR Analysis with Spheroids
MDA-MB-157 cells cultured as a monolayer and spheroids of 1.5×104 and 1.0×105 cells were lysed on the third day of culture using a TRK lysis buffer (Omega Biotek) and homogenized by passing through homogenizer mini columns (Omega Biotek). Total RNA was isolated from the samples using an RNA isolation kit (Omega Biotek). DNA was removed using RNase-free DNase (Omega Biotek). Purity and concentration of isolated RNA was assessed using OD 260/280 spectrophotometry (Synergy H1M, Biotek instruments). cDNA was synthesized from 1 µg of total RNA using random hexamer primers (Roche). Real time q-PCR was performed in a LightCycler 480 instrument II using a SYBR Green Master Mix (Roche). Briefly, 50 ng of cDNA was combined with the primer and the SYBR green Master Mix to a final volume of 15 µl. The reactions were pre-incubated at 95°C for 5 min followed by 45 cycles of amplification, i.e., at 95°C for 10 sec, at 60°C for 10 sec, and at 72°C for 10 sec. Specific primer sequences for all the genes investigated are listed in Table SI-1. Expression levels of mRNA for different gene markers of CSCs and hypoxia were calculated relative to β-actin and hypoxanthine phosphoribosyltransferase (HPRT) using the ΔΔCt method. The fold change in mRNA expression was determined according to the 2−ΔΔCt method. Statistical analysis was performed between the larger and smaller spheroid fold changes using a Student’s t-test in Microsoft Excel software.
Supplementary Material
Acknowledgments
The work is supported by National Institutes of Health (R21CA182333), Ohio Third Frontier (TECG20140954), and National Science Foundation (1264562). We thank Ms. Ashley E. Mohrman for helping with confocal microscopy.
Contributor Information
Stephanie L. Ham, Department of Biomedical Engineering, The University of Akron, Akron, OH 44325, United States
Ramila Joshi, Department of Biomedical Engineering, The University of Akron, Akron, OH 44325, United States.
Gary D. Luker, Department of Radiology, Microbiology and Immunology, and Biomedical Engineering, University of Michigan, Ann Arbor, MI 48109, United States
Hossein Tavana, Department of Biomedical Engineering, The University of Akron, Akron, OH 44325, United States.
References
- 1.Xu X, Farach-Carson MC, Jia X. Biotechnol. Adv. 2014;32:1256. doi: 10.1016/j.biotechadv.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fidler IJ. Nat. Rev. Cancer. 2003;3:1. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 3.Visvader JE. Nature. 2011;469:314. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 4.Trédan O, Galmarini CM, Patel K, Tannock IF. J Natl. Cancer Inst. 2007:1441. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 5.Teicher BA. Cancer Metastasis Rev. 1994;13:139. doi: 10.1007/BF00689633. [DOI] [PubMed] [Google Scholar]
- 6.Lu H, Samanta D, Xiang L, Zhang H, Hu H, Chen I, Bullen JW, Semenza GL. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E4600. doi: 10.1073/pnas.1513433112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnero A, Lleonart M. Insid. Cell. 2016;1:96. [Google Scholar]
- 8.Semenza GL. Trends Pharmacol. Sci. 2012;33:207. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenza GL. Cell. 2012;148:399. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohwer N, Cramer T. Drug Resist. Updat. 2011;14:191. doi: 10.1016/j.drup.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Pampaloni F, Reynaud EG, Stelzer EHK. Nat. Rev. Mol. Cell Biol. 2007;8:839. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Sarvestani SK, Moeinzadeh S, He X, Jabbari E. Tissue Eng. Part A. 2013;19:669. doi: 10.1089/ten.tea.2012.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee GH, Lee JS, Wang X, Lee SH. Adv. Healthc. Mater. 2016;5:56. doi: 10.1002/adhm.201500107. [DOI] [PubMed] [Google Scholar]
- 14.Ham SL, Joshi R, Thakuri PS, Tavana H. Exp. Biol. Med. (Maywood) 2016;241:939. doi: 10.1177/1535370216643772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwal R, Jurney P, Raythatha M, Singh V, Sreenivasan SV, Shi L, Roy K. Adv. Healthc. Mater. 2015;4:2269. doi: 10.1002/adhm.201500441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guarneri V, Dieci MV, Conte P. Drugs. 2013;73:1257. doi: 10.1007/s40265-013-0091-6. [DOI] [PubMed] [Google Scholar]
- 17.Atefi E, Mann JA, Tavana H. Langmuir. 2014;30:9691. doi: 10.1021/la500930x. [DOI] [PubMed] [Google Scholar]
- 18.Atefi E, Joshi R, Mann JA, Tavana H. ACS Appl. Mater. Interfaces. 2015;7:21305. doi: 10.1021/acsami.5b05757. [DOI] [PubMed] [Google Scholar]
- 19.Tavana H, Mosadegh B, Takayama S. Adv. Mater. 2010;22:2628. doi: 10.1002/adma.200904271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemmo S, Nasrollahi S, Tavana H. Biotechnol. J. 2014;9:426. doi: 10.1002/biot.201300227. [DOI] [PubMed] [Google Scholar]
- 21.Ham SL, Nasrollahi S, Shah KN, Soltisz A, Paruchuri S, Yun YH, Luker GD, Bishayee A, Tavana H. Integr. Biol. 2015;7:792. doi: 10.1039/c5ib00121h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lall N, Henley-Smith CJ, De Canha MN, Oosthuizen CB, Berrington D. Int. J. Microbiol. 2013;2013 doi: 10.1155/2013/420601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemmo S, Atefi E, Luker GD, Tavana H. Cell. Mol. Bioeng. 2014;7:344. doi: 10.1007/s12195-014-0349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boisserie-Lacroix M, Mac Grogan G, Debled M, Ferron S, Asad-Syed M, Brouste V, Mathoulin-Pelissier S, Hurtevent-Labrot G. Diagn. Interv. Imaging. 2012;93:183. doi: 10.1016/j.diii.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 25.van Dijk M, Göransson Sa, Strömblad S. Exp. Cell Res. 2013;319:1663. doi: 10.1016/j.yexcr.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Oskarsson T. Breast. 2013;22(Suppl 2):S66. doi: 10.1016/j.breast.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 27.Cordes N, Blaese MA, Plasswilm L, Rodemann HP, Van Beuningen D. Int J Radiat Biol. 2003;79:709. doi: 10.1080/09553000310001610240. [DOI] [PubMed] [Google Scholar]
- 28.Wilson W, Hay M. Nat. Rev. Cancer. 2011;11:393. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 29.Minchinton AI, Tannock IF. Nat. Rev. Cancer. 2006;6:583. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 30.Shannon AM, Bouchier-Hayes DJ, Condron CM, Toomey D. Cancer Treat. Rev. 2003;29:297. doi: 10.1016/s0305-7372(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 31.Ward C, Langdon SP, Mullen P, Harris AL, Harrison DJ, Supuran CT, Kunkler IH. Cancer Treat. Rev. 2013;39:171. doi: 10.1016/j.ctrv.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Hunter FW, Hsu H-L, Su J, Pullen SM, Wilson WR, Wang J. Mol. Cancer Ther. 2014:2501. doi: 10.1158/1535-7163.MCT-14-0476. [DOI] [PubMed] [Google Scholar]
- 33.Brown J, Wilson W. Nat. Rev. Cancer. 2004;4:437. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- 34.Aomatsu N, Yashiro M, Kashiwagi S, Kawajiri H, Takashima T, Ohsawa M, Wakasa K, Hirakawa K. BMC Cancer. 2014;14:400. doi: 10.1186/1471-2407-14-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan EY, Yan M, Campo L, Han C, Takano E, Turley H, Candiloro I, Pezzella F, Gatter KC, Millar EKA, O’Toole SA, McNeil CM, Crea P, Segara D, Sutherland RL, Harris AL, Fox SB. Br. J. Cancer. 2009;100:405. doi: 10.1038/sj.bjc.6604844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Guerrab A, Zegrour R, Nemlin CC, Vigier F, Cayre A, Penault-Llorca F, Rossignol F, Bignon YJ. PLoS One. 2011;6:1. doi: 10.1371/journal.pone.0025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClendon AK, Dean JL, Rivadeneira DB, Yu JE, Reed CA, Gao E, Farber JL, Force T, Koch WJ, Knudsen ES. Cell Cycle. 2012;11:2747. doi: 10.4161/cc.21127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, Johnson RS. Mol. Cell. Biol. 2003;23:359. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan R, Paré GC, Frederiksen LJ, Semenza GL, Graham CH. Mol. Cancer Ther. 2008;7:1961. doi: 10.1158/1535-7163.MCT-08-0198. [DOI] [PubMed] [Google Scholar]
- 40.Achison M, Hupp TR. Oncogene. 2003;22:3431. doi: 10.1038/sj.onc.1206434. [DOI] [PubMed] [Google Scholar]
- 41.Erler JT, Cawthorne CJ, Williams KJ, Koritzinsky M, Wouters BG, Wilson C, Miller C, Demonacos C, Stratford IJ, Dive C. Mol. Cell. Biol. 2004;24:2875. doi: 10.1128/MCB.24.7.2875-2889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie J, Xiao Y, Zhu X, Ning Z, Xu H, Wu H. Med. Oncol. 2016;33:42. doi: 10.1007/s12032-016-0755-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Nat. Rev. Cancer. 2013;13:714. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 44.Meng F, Evans JW, Bhupathi D, Banica M, Lan L, Lorente G, Duan JX, Cai X, Mowday AM, Guise CP, Maroz A, Anderson RF, Patterson AV, Stachelek GC, Glazer PM, Matteucci MD, Hart CP. Mol Cancer Ther. 2012;11:740. doi: 10.1158/1535-7163.MCT-11-0634. [DOI] [PubMed] [Google Scholar]
- 45.Sun JD, Liu Q, Ahluwalia D, Li W, Meng F, Wang Y, Bhupathi D, Ruprell AS, Hart CP. Cancer Biol. Ther. 2015;16:438. doi: 10.1080/15384047.2014.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meng F, Bhupathi D, Sun JD, Liu Q, Ahluwalia D, Matteucci MD, Hart CP. BMC Cancer. 2015;15:422. doi: 10.1186/s12885-015-1387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou T. Pharmacol. Rev. 2006;58:621. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 48.Tyagi A, Agarwal C, Chan D, Agarwal R. Oncol. Rep. 2004;11:493. [PubMed] [Google Scholar]
- 49.Sun JD, Ahluwalia D, Liu Q, Li W, Wang Y, Meng F, Bhupathi D, Matteucci MD, Hart CP. Am. J. Cancer Res. 2015;5:2139. [PMC free article] [PubMed] [Google Scholar]
- 50.Peeters SGJA, Zegers CML, Biemans R, Lieuwes NG, Van Stiphout RGPM, Yaromina A, Sun JD, Hart CP, Windhorst AD, Van Elmpt W, Dubois LJ, Lambin P. Clin. Cancer Res. 2015;21:2984. doi: 10.1158/1078-0432.CCR-15-0018. [DOI] [PubMed] [Google Scholar]
- 51.Chawla SP, Cranmer LD, Van Tine BA, Reed DR, Okuno SH, Butrynski JE, Adkins DR, Hendifar AE, Kroll S, Ganjoo KN. J Clin. Oncol. 2014;32:3299. doi: 10.1200/JCO.2013.54.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arola OJ, Saraste A, Pulkki K, Kallajoki M, Parvinen M, Voipio-Pulkki L-M. Cancer Res. 2000;60:1789. [PubMed] [Google Scholar]
- 53.Zhang Y-W, Shi J, Li Y-J, Wei L. Arch. Immunol. Ther. Exp. (Warsz) 2009;57:435. doi: 10.1007/s00005-009-0051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobayashi H, Takemura Y, Ohnuma T. Cancer Chemother. Pharmacol. 1992;31:6. doi: 10.1007/BF00695987. [DOI] [PubMed] [Google Scholar]
- 55.Takemura Y, Kobayashi H, Miyachi H, Hayashil K, Sekiguchil S, Ohnuma T. Cancer Chemother. Pharmacol. 1991;27:417. doi: 10.1007/BF00685154. [DOI] [PubMed] [Google Scholar]
- 56.Ohnuma T, Arkin H, Holland JF. Br. J. Cancer. 1986;54:415. doi: 10.1038/bjc.1986.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keith B, Simon MC. Cell. 2007;129:465. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heddleston J, Li Z, Lathia J, Bao S, Hjelmeland A, Rich J. Br. J. Cancer. 2010;102:789. doi: 10.1038/sj.bjc.6605551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muz B, de la Puente P, Azab F, Luderer M, Azab AK. Blood Cancer J. 2014;4:e262. doi: 10.1038/bcj.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao H, Liu N, Lin MC, Zheng J. Cancer Lett. 2016 doi: 10.1016/j.canlet.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 61.Dhawan A, Madani Tonekaboni SA, Taube JH, Hu S, Sphyris N, Mani SA, Kohandel M. Sci. Rep. 2016;6:18074. doi: 10.1038/srep18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subarsky P, Hill R. Clin. Exp. Metastasis. 2003;20:237. doi: 10.1023/a:1022939318102. [DOI] [PubMed] [Google Scholar]
- 63.Vinogradov S, Wei X. Nanomedicine (Lond) 2012;7:597. doi: 10.2217/nnm.12.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Medema JP. Nat. Cell Biol. 2013;15:338. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 65.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3983. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marotta LLC, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-qimron N, Kim JJ, Choudhury Sa, Maruyama R, Wu Z, Gönen M, Mulvey La, Bessarabova MO, Huh SJ, Silver SJ. J Clin. Invest. 2011;121:2723. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmed MaH, Aleskandarany Ma, Rakha Ea, Moustafa RZa, Benhasouna A, Nolan C, Green AR, Ilyas M, Ellis IO. Breast Cancer Res. Treat. 2012;133:979. doi: 10.1007/s10549-011-1865-8. [DOI] [PubMed] [Google Scholar]
- 68.Lin Y, Zhong Y, Guan H, Zhang X, Sun Q. J Exp. Clin. Cancer Res. 2012;31:59. doi: 10.1186/1756-9966-31-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma F, Li H, Wang H, Shi X, Fan Y, Ding X, Lin C, Zhan Q, Qian H, Xu B. Cancer Lett. 2014;353:153. doi: 10.1016/j.canlet.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 70.Pattabiraman DR, Weinberg RA. Nat. Rev. Drug Discov. 2014;13:497. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huebschman ML, Lane NL, Liu H, Sarode VR, Devlin JL, Frenkel EP. Breast Cancer Targets Ther. 2015;7:231. doi: 10.2147/BCTT.S87041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu YJ, Shen D, Yin X, Gavine P, Zhang T, Su X, Zhan P, Xu Y, Lv J, Qian J, Liu C, Sun Y, Qian Z, Zhang J, Gu Y, Ni X. Br. J. Cancer. 2014:1. doi: 10.1038/bjc.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kristiansen G, Sammar M, Altevogt P. J Mol Histol. 2004;35:255. doi: 10.1023/b:hijo.0000032357.16261.c5. [DOI] [PubMed] [Google Scholar]
- 74.Wang Z, Shi Q, Wang Z, Gu Y, Shen Y, Sun M, Deng M, Zhang H, Fang J, Zhang S, Xie F. Pathol. Res. Pract. 2011;207:505. doi: 10.1016/j.prp.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 75.Thomas S, Harding MA, Smith SC, Overdevest JB, Nitz MD, Frierson HF, Tomlins SA, Kristiansen G, Theodorescu D. Cancer Res. 2012;72:5600. doi: 10.1158/0008-5472.CAN-11-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Imaizumi A, Onishi H, Yamasaki A, Kawamoto M, Umebayashi M, Morisaki T, Hasumi K. Anticancer Res. 2016;36:653. [PubMed] [Google Scholar]
- 77.Kristiansen G, Winzer K-J, Mayordomo E, Bellach J, Schluns K, Denkert C, Dahl E, Pilarsky C, Altevogt P, Guski H, Dietel M. Clin. Cancer Res. 2003;9:4906. [PubMed] [Google Scholar]
- 78.Mylona E, Giannopoulou I, Fasomytakis E, Nomikos A, Magkou C, Bakarakos P, Nakopoulou L. Hum. Pathol. 2008;39:1096. doi: 10.1016/j.humpath.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 79.Sansone P, Ceccarelli C, Berishaj M, Chang Q, Rajasekhar VK, Perna F, Bowman RL, Vidone M, Daly L, Nnoli J, Santini D, Taffurelli M, Shih NNC, Feldman M, Mao JJ, Colameco C, Chen J, DeMichele A, Fabbri N, Healey JH, Cricca M, Gasparre G, Lyden D, Bonafé M, Bromberg J. Nat. Commun. 2016;7:10442. doi: 10.1038/ncomms10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kapucuoğlu N, Bozkurt K, Başpınar S, Koçer M, Eroğlu H, Akdeniz R, Akçil M. Pathol. Res. Pract. 2015;211:740. doi: 10.1016/j.prp.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 81.Zhang D, Sun B, Zhao X, Ma Y, Ji R, Gu Q, Dong X, Li J, Liu F, Jia X, Leng X, Zhang C, Sun R, Chi J. Mol. Cancer. 2014;13:207. doi: 10.1186/1476-4598-13-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lv X, Wang Y, Song Y, Pang X, Li H. Oncol. Lett. 2016:1750. doi: 10.3892/ol.2016.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim DG, Kim KH, Seo YJ, Yang H, Marcusson EG, Son E, Lee K, Sa JK, Lee HW, Nam D-H. Oncotarget. 2016 doi: 10.18632/oncotarget.8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bock C, Kuhn C, Ditsch N, Krebold R, Heublein S, Mayr D, Doisneau-Sixou S, Jeschke U. J Cancer Res. Clin. Oncol. 2014;140:1873. doi: 10.1007/s00432-014-1750-z. [DOI] [PubMed] [Google Scholar]
- 85.Ieni A, Giuffrè G, Adamo V, Tuccari G. Anticancer Res. 2011;31:1315. [PubMed] [Google Scholar]
- 86.Swaminathan SK, Roger E, Toti U, Niu L, Ohlfest JR, Panyam J. J Control. Release. 2013;171:280. doi: 10.1016/j.jconrel.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 87.Stuelten CH, Mertins SD, Busch JI, Gowens M, Scudiero DA, Burkett MW, Hite KM, Alley M, Hollingshead M, Shoemaker RH, Niederhuber JE. Stem Cells. 2010;28:649. doi: 10.1002/stem.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ben-porath I, Thomson MW, Carey VJ, Ge R, George W, Regev A, Weinberg Ra. Nat Genet. 2008;40:499. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi M, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R, Martin A. Oncogene. 2012;31:1354. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 90.Piva M, Domenici G, Iriondo O, Miriam R, Zabalza I, Kypta R, Comaills V, Barredo I, Jose aL, Vivanco MM. EMBO Mol. Med. 2014;6:66. doi: 10.1002/emmm.201303411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CMA, Hubaud A, Stadler B, Choi M, Bar M, Tewari M, Liu A, Vessella R, Rostomily R, Born D, Horwitz M, Ware C, Blau CA, Cleary MA, Rich JN, Ruohola-Baker H. Cancer Res. 2011;71:4640. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Proc. Natl. Acad. Sci. 2016:201602883. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tavana H, Jovic A, Mosadegh B, Lee QY, Liu X, Luker KE, Luker GD, Weiss SJ, Takayama S. Nat. Mater. 2009;8:736. doi: 10.1038/nmat2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ham SL, Atefi E, Fyffe D, Tavana H. J Vis. Exp. 2015:1. doi: 10.3791/52754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Timmins N, Nielsen L. Methods Mol. Med. 2007;140 doi: 10.1007/978-1-59745-443-8_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.