SUMMARY
Generating Tier 2 HIV neutralizing antibody (nAb) responses by immunization remains a challenging problem, and the immunological barriers to induction of such responses with Env immunogens remain unclear. Here, some rhesus monkeys developed autologous Tier 2 nAbs upon HIV Env trimer immunization (SOSIP.v5.2), while others did not. This was not because HIV Env trimers were immunologically silent, as all monkeys made similar ELISA-binding antibody responses; the key difference was nAb versus non-nAb responses. We explored immunological barriers to HIV nAb responses by combining a suite of techniques, including longitudinal lymph node fine needle aspirates. Unexpectedly, nAb development best correlated with booster immunization GC B cell magnitude and Tfh characteristics of the Env-specific CD4 T cells. Notably, these factors distinguished between successful and unsuccessful antibody responses, as GC B cell frequencies and stoichiometry to GC Tfh cells correlated with nAb development but did not correlate with total Env Ab binding titers.
Graphical Abstract
INTRODUCTION
The efficacy of the vast majority of human vaccines is associated with antibody responses (Plotkin, 2010). We still know critically little about the immunological processes important for the generation of neutralizing antibodies and how to maximize such responses during immunization (Burton and Hangartner, 2016; Corti and Lanzavecchia, 2013; Crotty, 2014). Induction of protective antibodies by vaccination is dependent both on features of the immune system and the immunogen. While CD4 T cells and B cell responses are essential, the specific characteristics of those responses that are responsible for successful development of vaccine-elicited neutralizing antibodies remain poorly characterized for any vaccine. The immunogen must properly mimic the relevant pathogen structure. Minimal, if any, neutralizing antibody responses to meaningful clinical isolate HIV strains (Tier 2 or Tier 3) have been detected in human HIV vaccine trials (Mascola and Montefiori, 2010; Sliepen and Sanders, 2016). The immunogens in those vaccine trials were HIV Env gp120 monomers, not trimers. Rapid progress has been made in recent years in the engineering of HIV vaccine immunogens. The construction of the BG505 SOSIP.664 trimer now permits presentation of a recombinant, well-ordered HIV-1 Env trimer to the immune system (Julien et al., 2013; Lyumkis et al., 2013; Sanders et al., 2013). The structure of SOSIP Env trimer is an accurate reflection of the native membrane-expressed HIV-1 Env trimer (Lee et al., 2016).
Non-human primates are an important animal model for testing candidate HIV vaccines due to their evolutionary relatedness to humans (Hessell and Haigwood, 2015). While NHP HIV protein immunization studies have been done for over 20 years, significant Tier 2 neutralizing antibody responses in immunized non-human primates have only been reported in two studies, published recently (Hessell et al., 2016; Sanders et al., 2015). In a first study of four rhesus macaques (RM) immunized with BG505 SOSIP.664, some animals developed low levels of autologous Tier 2 neutralizing antibodies, whereas in the same study BG505 gp120 immunized macaques did not (Sanders et al., 2015). High titers of non-neutralizing antibody responses, such as those targeting the V3 loop tip, were also generated in BG505 SOSIP.664 immunized animals. Competition between Ab responses to 'difficult' HIV neutralizing epitopes and 'easy' non-neutralizing Env epitopes may limit generation of neutralizing antibodies (Hu et al., 2015; Sattentau, 2014).
HIV bnAbs are highly somatically mutated and the mutations are necessary for neutralization breadth (Burton and Mascola, 2015; Haynes et al., 2016; Klein et al., 2013). Mutations occur in GC B cells during affinity maturation, whereby GC B cells undergo repeated rounds of somatic hypermutation of their antigen-binding B cell receptor (BCR) and selection by GC T follicular helper (GC Tfh) CD4 T cells (Crotty, 2014; Victora and Nussenzweig, 2012).
Germinal centers occur in lymphoid tissues, such as lymph nodes, but not peripheral blood. Consequently, direct studies of germinal centers have not been possible in HIV bnAb+ individuals. Nevertheless, markers connected to GC activity have been associated with the generation of HIV bnAbs. A subset of circulating memory Tfh cells correlated with the generation of HIV bnAbs (Locci et al., 2013). In addition, plasma concentrations of the chemokine CXCL13 correlate with lymph node GC activity (Havenar-Daughton et al., 2016a) and were elevated in bnAb+ individuals (Cohen et al., 2014; Havenar-Daughton et al., 2016a). The importance of GC responses and Tfh cells for the generation of HIV nAbs is also supported by studies of SIV-infected macaques (Hong et al., 2014; Petrovas et al., 2012; Yamamoto et al., 2015).
Therefore, data suggest that generation of neutralizing antibody responses against HIV will likely require an immunization regimen that optimizes the induction of GC Tfh cells and GCs. However, for candidate HIV vaccines these concepts are only inferences. Indeed, a recent large NHP vaccine study with an HIV gp140 immunogen found no association between SHM and HIV Ab responses (Francica et al., 2015). Little is known about the immunological factors determining whether or not neutralizing antibody responses develop in response to Env trimer immunization.
RESULTS
Generation of autologous HIV neutralizing antibodies in NHPs immunized with BG505 SOSIP.v5.2
HIV bnAbs consistently protect NHPs against SHIV infection in passive transfer experiments when the circulating neutralizing Ab (nAb) titers are ≥ ~1:100 (Gautam et al., 2016; Moldt et al., 2012; Shingai et al., 2014) with detectable protection even at titers of 1:20 (Gautam et al., 2016). Immunization of NHPs with a well-formed HIV Env trimer BG505 SOSIP.664 has not consistently elicited autologous Tier 2 nAb of substantial magnitude (≥ 1:50 titer). We immunized a cohort of RMs with a modified BG505 N332 SOSIP designed to improve the stability of the protein (BG505 SOSIP.v5.2 [ATP and RWS, manuscript in prep]). In addition, we utilized a modified immunization regimen that included subcutaneous bilateral immunizations, which may engage more B and CD4 T cells (Figure 1A). Two different adjuvants were tested in parallel, with groups of 8 (PLGA/R848+MPL, ‘PLGA’) and 4 (Iscomatrix) animals. Two weeks after the final immunization, serum was tested for autologous Tier 2 nAb responses to BG505 T332N pseudovirus. The majority of immunized animals (9 of 12; 75%) generated detectable nAb responses (Figure 1B, S1; Table S1). No significant difference was observed between the two adjuvants. Therefore, all animals from both adjuvant+BG505 SOSIP groups were pooled together for all subsequent analyses. Seven of 12 animals developed peak titers of ≥ 1:50. Four animals generated BG505 nAb titers >1:200 (defined here as top neutralizers). These are the best autologous Tier 2 nAb responses yet reported for HIV Env protein immunized NHPs. Based on the results of nAb passive transfer studies, we infer that nAb titers may be sufficient for protection against an autologous Tier 2 virus, but this remains to be tested. The animals in this study were not challenged, as the study was designed as an immunogenicity study and necropsy tissues were required for full characterization of the anti-Env vaccine elicited responses.
Figure 1. Generation of autologous neutralizing antibodies by immunization with BG505 SOSIP.v5.2.
A. Schematic timeline of the BG505 SOSIP.v5.2 immunization study. Nx = necropsy.
B. BG505 (N332) nAb titers, wk 20. Average from two independent assays. Different adjuvants are indicated by blue and red.
C. BG505 Env ELISA titers. Vertical lines = immunization dates.
D. BG505 neutralization titers (wk 20) compared to BG505 trimer binding IgG titers (week 20).
E. BG505 neutralization titers compared to Tier 1 neutralization titers of SF162 (left) or MN.3 (right).
F. Ratio of BG505 trimer binding titers to V3 peptide titers 2 wks post 3rd or 4th immunization.
See also Figure S1 and Table S1.
Env Binding Antibodies
Substantial BG505 SOSIP-binding IgG endpoint titers were detected by ELISA for all BG505 SOSIP.v5.2 immunized animals (Figure 1C and S1B). Thus, the difference between RMs that did or did not develop HIV neutralizing antibodies in response to BG505 SOSIP.v5.2 protein immunization was not that the immunogen was inherently immunological silent in some animals. Therefore, the immunological process limiting for nAb responses to Env trimers was unknown. We then examined this knowledge gap.
Longitudinal analyses revealed that the BG505 SOSIP—binding IgG responses in plasma after the priming immunization were minimal (Figure 1C). This is a common feature of protein immunizations in large animals and humans (Evans et al., 2001; McElrath et al., 2000; West, 1989) and has been previously observed for BG505 SOSIP (Sanders et al., 2015). BG505 SOSIP—binding IgG in plasma increased substantially after the 2nd immunization, and after the 3rd and 4th immunizations (Figure 1C). Peak titers did not differ statistically between the two adjuvant groups (Figure S1B).
BG505 nAb titers did not correlate with BG505 SOSIP—binding IgG after BG505.v5.2 immunization (Figure 1D). Additionally, Tier 2 nAbs were not detected to heterologous Clade A strains (see Methods). The observation that BG505 SOSIP—binding IgG titers were not predictive of autologous BG505 nAb titers indicated that the BG505 SOSIP—binding IgG was composed of antibody responses to a mixture of neutralizing and non-neutralizing epitopes in ratios that are variable from one animal to another. Non-neutralizing Abs can bind the exposed base of the trimer, the V3 loop tip, CD4i sites, or gp120 breakdown products. Anti-V3 antibody responses can be an immunodominant yet irrelevant response to Env immunization or HIV infection (Mascola et al., 1996; Mascola and Montefiori, 2010; McGuire et al., 2014). One aspect of BG505 SOSIP.v5.2 protein design was the modification of the BG505 SOSIP.664 V3 region to reduce exposure of the V3 loop in vitro (de Taeye et al., 2015). A BG505 V3 peptide ELISA was performed to detect V3 loop—specific IgG in BG505 SOSIP.v5.2—immunized RMs. The BG505 SOSIP.v5.2 modifications did not eliminate the immunization-induced antibody response to the BG505 V3 loop (Figure S1C) suggesting that the V3 loop becomes exposed in vivo during natural protein breakdown processes (Hu et al., 2015). As the V3 loop is a major target of irrelevant Tier 1 nAb responses (Mascola et al., 1996; Morner et al., 2009; Sanders et al., 2013), Tier 1 nAb responses were measured. Autologous BG505 Tier 2 nAb titers in BG505 SOSIP.v5.2— immunized RMs did not correlate with Tier 1 nAb titers to MN.3 and SF162 pseudoviruses (Figure 1E), indicating that HIV Env trimer-immunization elicited neutralizing antibodies against Tier 1 and Tier 2 viruses are unrelated. The V3 loop Ab response correlated with the Tier 1 MN.3 nAb response, consistent with the V3 loop being in an open confirmation on Tier 1 HIV strain pseudoviruses (r = 0.64, p = 0.03; Figure S1D). Interestingly, the V3 loop responses plateaued after the 2nd immunization while total BG505 SOSIP IgG continued to increase, resulting in a higher ratio of SOSIP ELISA to V3 loop ELISA specific Abs after the 4th versus 3rd immunization (Figure 1F). In summary, serology showed that a range of autologous nAb responses were generated by BG505 SOSIP.v5.2 immunization, including animals with high autologous Tier 2 nAb titers and animals with undetectable autologous nAb, without correlation to the overall magnitude of the anti-Env IgG response. Therefore, what was the immunological basis for the qualitatively different response by nAb responders and non-responders? We considered that understanding GC and Tfh cell responses within the draining lymph nodes (LN) may be critical to assess the immunological processes responsible for generating immunization elicited HIV Tier 2 neutralizing antibodies.
Fine needle aspirates quantitatively monitor germinal center activity in draining lymph nodes
GC B cells and GC Tfh cells are not present in blood and therefore can only be studied with lymphoid tissue samples. Whole LN excision has been rarely performed in human vaccine studies. Whole LN excisional biopsies can be performed as part of vaccine studies in NHPs, however removal of a draining LN has numerous drawbacks and can only be done once, thus precluding longitudinal assessment of the vaccine-induced GC responses. This inability to directly quantify GC activity (GC B cells and GC Tfh cells) in response to an immunization has remained a vexing ‘black box’ problem for vaccine immunology of monkeys and humans. Therefore, we used fine needle aspirates (FNAs) of the draining LN to sample GC B cells and GC Tfh cells. The LN FNA technique is widely used in clinical oncology for non-quantitative diagnostic purposes as a simple outpatient procedure (Diamantis et al., 2009). Importantly, longitudinal FNAs could theoretically be used to monitor GC cells within the same LN of the same animal after each immunization. However, as LN FNAs have only rarely been used for vaccine studies, several issues needed to be addressed: 1) Can FNAs provide enough cells to gain immunologically useful information? 2) Do repeated FNAs interfere with the GC or antibody responses? 3) Are the GC B cell and GC Tfh cell populations obtained by FNA sampling representative of those in the whole LN? Each of these questions impacted the potential value of LN FNA sampling.
To address these issues, a pilot LN FNA study was conducted in a group of uninfected and SHIV-infected RMs at necropsy (Figure S2A). The frequencies of both GC B cells (p < 0.0001; Figure S2B) and GC Tfh cells (p < 0.0001; Figure S2C) correlated with the respective cell frequencies in the matched whole LNs. These results supported the potential utility of LN FNAs in NHP vaccine studies. LN FNAs were then performed on 8 of the 12 BG505 SOSIP.v5.2 immunized RMs 3 weeks after each immunization, with the remaining 4 animals left without LN FNAs as a control. The average cell yield per LN FNA was ~1×106 (Figure 2A). The vast majority of FNAs provided >1×105 cells (95%; 91 of 96), sufficient for flow cytometric analysis of GC Tfh cells and GC B cells (Figure 2B). One issue to address was whether the LN FNA procedure might impact immune responses. While we considered this unlikely, as only ~106 cells are removed and LN FNAs are well tolerated in humans, it was nevertheless important to investigate. After the final immunization, whole lymph node biopsies were taken from animals that did or did not undergo serial LN FNAs (Figure 2C). Neither the frequency of GC B cells nor GC Tfh cells was affected by the serial LN FNAs (Figure 2D and 2E). Furthermore, antibody titers did not differ between the animals that did or did not undergo serial LN FNAs (Figure 2F).
Figure 2. FNAs monitor GC activity in draining lymph nodes.
A. Cell recovery from LN FNAs (n = 24).
B. Flow cytometry from LN FNA samples gated on GC B cells among CD20+ B cells (left) or GC Tfh cells among CD4+ T cells (right).
C. Experimental group design. Bx = whole LN biopsy.
D. GC B cell frequency in whole LN biopsies, 3 wk post 4th immunization (n = 12 per group). LN were either previously sampled by FNA (group 1) or not (group 2).
E. GC Tfh cell frequency in whole LN biopsies, 3 wks post 4th immunization (n = 12 per group). LN were either previously sampled by FNA (group 1) or not (group 2).
F. BG505 SOSIP.v5.2 ELISA titers at wk 0 and wk 14 for animals that had undergone previous LN FNAs (group 1) or not (group 2).
G. Percentage of cells sampled per LN by FNA (n = 24).
H. GC B cell frequency determined from FNA or whole LN biopsy (n = 24). Each point represents an individual LN sampled by FNA and then by whole LN biopsy.
I. GC Tfh cell frequency, as in panel H.
See also Figure S2.
Finally, we asked if LN FNA samples were quantitatively representative of the LN GC B cell and GC Tfh cell frequencies. Twenty-four pairs of FNAs and whole LN biopsies from BG505 SOSIP.v5.2 immunized animals were analyzed (Figure 2C, Group 3). A LN FNA was performed and then the same LN was immediately removed for analysis. On average, FNA samples represented 3.1% of the cellularity of whole LNs (Figure 2G). The frequency of GC B cells among total CD20+ B cells in the LN FNAs correlated strongly with the GC B cell frequency in the whole LN biopsy (r =0.79, p < 0.0001; Figure 2H). The frequency of GC Tfh cells also correlated strongly (r =0.66, p < 0.0005; Figure 2I). In sum, FNAs of draining LNs after vaccination provided sufficient cells for immunological analysis, was non-disruptive to the antibody and GC responses, and properly quantified GC B and GC Tfh cell populations within a LN.
Massive germinal center responses to priming protein immunization
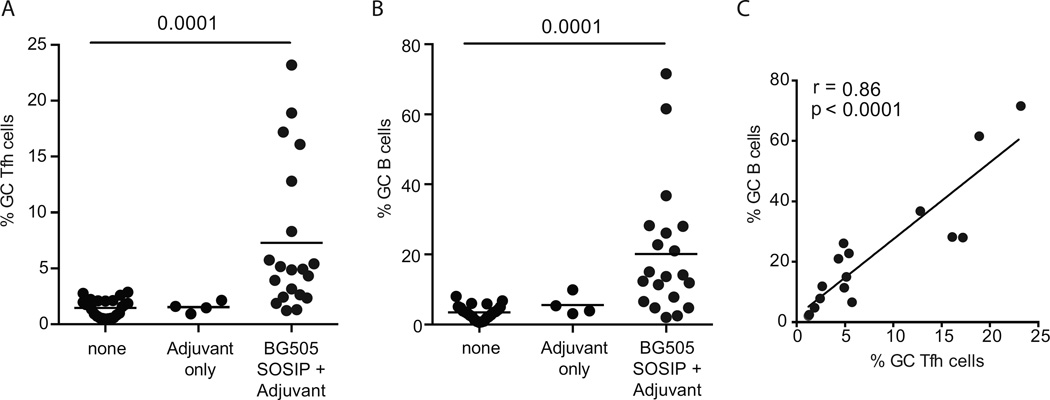
A priming immunization with a protein antigen is generally considered a small immunological event in humans and large animal models (Evans et al., 2001; McElrath et al., 2000; West, 1989), unlike in mice. Antibody titers in plasma are minimally elevated after an initial immunization (Figure 1C) (Sanders et al., 2015). In addition, CD4 T cell responses in the blood after protein immunizations are generally undetectable or barely detectable (McElrath et al., 2000; Verschoor et al., 1999). However, very little is directly known about the magnitude of the CD4 T cell and B cell responses at the primary site of the immune response to a protein immunization in monkeys or humans. Thus, GC responses were examined in the RMs immunized once with BG505 SOSIP.v5.2. GC Tfh cells were dramatically increased in protein + adjuvant immunization groups 21 days after the priming immunization (unimmunized vs adjvant+BG505 SOSIP.v5.2, p = 0.0001) with mean GC Tfh cell frequencies of 7.3% of total CD4 T cells, respectively (Figure 3A and S3A). The frequencies of GC B cells were vastly increased in protein + adjuvant immunization groups (unimmunized vs adjuvant+BG505 SOSIP.v5.2, p = 0.0001; Figure 3B). On average, GC B cell frequencies of 20.2% were observed in draining LNs after a single immunization. That is vastly higher than protein immunized mice [3% or lower (Hu et al., 2015; Kuraoka et al., 2016)]. The frequency of GC B and GC Tfh cells in immunized RMs strongly correlated in the primary immune response (r = 0.86, p < 0.0001; Figure 3C), consistent with the interdependence of GC B cells and GC Tfh cells (Crotty, 2011; Johnston et al., 2009; Victora et al., 2010). The ‘adjuvant only’ control group did not differ from the unimmunized group (Figure 3A and 3B), confirming that the GC responses to the SOSIP immunization were antigen specific. In summary, strikingly high frequencies of GC B and GC Tfh cells are induced in draining LNs of monkeys after a single protein + adjuvant immunization, discordant from the primary serological response to the immunogen.
Figure 3. Priming protein immunization generates massive GC responses.
Analysis of GC LN cells from three groups of RMs: 1) unimmunized (‘none’); 2) injected with only PLGA(MPL+R848) (‘adjuvant only’); 3) immunized with either BG505 SOSIP.v5.2 + PLGA(MPL+R848) or BG505 SOSIP.v5.2 + Iscomatrix (‘adjuvant + BG505 SOSIP’).
A. GC Tfh cells, as % of CD4+ T cells.
B. GC B cells, as % of CD20+ B cells.
C. Correlation of GC B cell and GC Tfh frequencies in draining LNs after the 1st immunization.
See also Figure S3.
Longitudinal monitoring of germinal center responses in individual LN during an immunization series
Multiple immunizations will likely be required to successfully generate HIV bnAbs via a vaccine (Burton and Hangartner, 2016; Haynes et al., 2016; Klein et al., 2013). Therefore, booster immunizations need to generate germinal centers. In mice, booster immunizations of protein + adjuvant predominantly induce strong plasmablast responses, but can induce some 2° GC responses (Dogan et al., 2009; Pape et al., 2011). The extent to which secondary GCs occur after immunizations in monkeys or humans and how the 2° GC response compares to the 1° GC response in the same individual is unknown.
Six weeks after the priming immunization, the first booster immunization was given (Figure 1A). As a group, draining LNs from immunized RMs had statistically higher GC B cell frequencies than unimmunized RMs (p = 0.03; Figure 4A), while there was a trend for increased GC Tfh cells (p = 0.1; Figure 4B). Since LN FNA sampling was obtained from the same draining LN sites after each immunization, GC recall responses could be directly compared to the priming responses. Mean GC B cell frequencies after the 2nd immunization were only 60% that observed to the 1st immunization (13.7% vs. 22.4%; paired p = 0.006; Figure 4C). Similarly, mean GC Tfh cell frequencies were only 43% the amount observed to the 1st immunization (3.4% vs. 8.0%, paired p = 0.004; Figure 4D). Thus, the 2nd immunization generated markedly fewer GC Tfh and GC B cells than the primary immunization.
Figure 4. Longitudinal monitoring of GC responses.
A. Frequency of GC B cells in draining LNs 3 wks post 2nd immunization (wk 9) compared to matched LNs from unimmunized RMs (‘none’).
B. Frequency of GC Tfh cells, as per A.
C. Frequency of GC B cells, as per A, with Wilcoxon matched pairs rank test.
D. Frequency of GC Tfh cells, as per B, with Wilcoxon matched pairs rank test.
E. GC B cell frequency (draining LNs at wk 3, average of Left and Right inguinal LN) compared to BG505 binding titers.
F. GC Tfh cell frequency, as per E.
G. BG505 Env-specific IgG+ plasmablasts in blood in RMs that did (2nd Imm) or did not receive the 2nd immunization.
H. Longitudinal peak GC B cell frequencies in left (L) and right (R) draining inguinal LN from individual RMs, 3 wks after each of 4 immunizations. Blue indicates animals receiving PGLA adjuvant; red indicates animals receiving Iscomatrix adjuvant.
I. Frequency of GC B cells (Left) and GC Tfh cells (Right) in draining LNs 3 wks post 3rd immunization (week 15) compared to matched LN from unimmunized RMs.
J. As per I, 3 wks post 4th immunization (week 21).
K. GC B cells in LNs plotted by time since most recent immunization.
See also Figure S3.
After the 2nd immunization, anti-Env plasma antibody titers increased, by a factor of 17 on average (Figure 1C). The frequency of GC B cells and GC Tfh cells after the 1st immunization exhibited strong trends with the BG505 SOSIP binding titers 2 weeks after the 2nd immunization (GC B cells: r = 0.62, p = 0.06; GC Tfh cells: r = 0.62, p = 0.06; Figure 4E and 4F). In addition, Env-specific plasmablasts were present in the blood 5 days after the 2nd immunization, but not in unimmunized animals (p = 0.001, Figure 4G). Taken together, robust GC activity induced by the priming immunization led to strong plasmablast and Env antibody responses upon the next immunization.
Diverse patterns of GC activity were observed for individual animals (Figure 4H and Figure S3D), but overall GC responses after all booster immunizations tended to be lower than the primary response. The frequency of GC B and GC Tfh cells correlated at each individual time point and over all time points (All time points: r = 0.83, p < 0.0001; Figure S3C). Examination of the GC responses of the RMs to the 3rd and 4th immunizations revealed that mean frequencies of GC B and GC Tfh cells in the draining LN were significantly higher than the unimmunized control group at both time points (Figure 4I and 4J).
To investigate why GC responses generated by booster immunization may be lower in magnitude than those generated by the primary immunization, GC responses were examined 6 weeks after the final immunization. This was chosen to match the interval between previous immunizations. Substantially elevated frequencies of GC B cells and GC Tfh cells were still present in the draining LNs 6 weeks after the final immunization (GC B cells, unimmunized vs immunized, 3.5% vs 10.5%, p <0.0001; Figure 4K. GC Tfh cells, 1.5% vs 3.0%, p <0.0001; Figure S3E). Additional LN FNAs revealed GCs persisting for up to 120 days after immunization (Figure 4K). Therefore, given the magnitude of the GC responses observed ~42 days after the final immunization, and the magnitude of the GC responses observed at 21 days after each immunization, it is highly likely that the booster immunizations at 42 day intervals were given in the face of substantial ongoing GC responses initiated by the previous immunization. That may have negative consequences.
Env-specific Tfh cells in LN
The intrinsic biology of GC Tfh cells dictates these cells be stingy cytokine producers (Dan et al., 2016; Havenar-Daughton et al., 2016b). This explains why it has been very difficult or impossible to detect antigen-specific GC Tfh cells by intracellular cytokine staining in most contexts (Hessell et al., 2016), resulting in the impression that antigen-specific GC Tfh cells were quite rare. We have developed a cytokine-independent methodology, referred to as the activation induced marker (AIM) method (Dan et al., 2016; Havenar-Daughton et al., 2016b), for detecting antigen-specific GC Tfh cells. AIM shows acceptable background in adjuvant-only immunized macaques and retains fidelity of the GC Tfh cell phenotype after stimulation. Strikingly, a median of 6.4% of GC Tfh cells were Env-specific 21 days after the final Env trimer immunization (Range: 1.3 to 39.3%; Figure 5A and 5B). Env-specific non-Tfh cell responses were also present (Figure 5C and 5D). Together, these data demonstrate surprisingly robust antigen-specific GC Tfh and non-Tfh CD4 T cell responses to a protein immunization.
Figure 5. Env-specific Tfh responses in LN.
A. Env-specific GC Tfh cells (CXCR5hi PD-1hi) in draining LNs by AIM assay. CD25+OX40+ cells are gated of GC Tfh cells after stimulation (BG505 Env or SEB) or left unstimulated (—). Representative flow cytometry plots are shown.
B. Env-specific GC Tfh in draining LN 3 wks post 2nd, 3rd, and 4th immunizations. Dotted line indicates positive signal threshold.
C. Total Env-specific CD4 T cells in draining LNs by the AIM assay. CD25+OX40+ cells are gated from total CD4+ T cells after stimulation (BG505 Env or SEB) or left unstimulated (—). Representative flow cytometry plots are shown.
D. Total Env-specific CD4 T cells in the draining LN 3 wks after the 2nd, 3rd, and 4th immunizations. Dotted line indicates positive signal threshold.
GC B cells and autologous nAbs
Directly probing the GCs at the primary site of the immune response allowed for interrogation of specific immunological factors potentially predictive of the generation of HIV nAbs. The strongest immunological feature associated with the development of BG505 nAbs was the frequency of GC B cells after booster immunizations (Figure 6A–B). Correlation between nAbs and the average frequency of GC B cells in response to the 3rd and 4th immunizations was observed for the 8 serial LN FNA animals, of borderline statistical significance (r = 0.71, p = 0.06, Figure 6A). A wider analysis, including all animals for which LN samples were available, demonstrated a strong association between GC B cell activity after the final (4th) immunization and the development of nAbs (p = 0.007; Figure 6B). The median frequency of LN GC B cells in top neutralizers was almost 3 fold higher than in non/low neutralizers (median GC B cells, Top: 20.3% vs Non/low: 7.6%; Figure 6B). Tier 2 nAb development was also associated with GC B cell frequencies after the 2nd and 3rd immunizations (Figure 6B). In contrast, neither BG505 SOSIP IgG ELISA binding titers (p = 0.88, Figure S4A) nor Tier 1 nAbs (p = 0.58, Figure S4B) correlated with GC B cell activity to the later immunizations, highlighting the specificity of the association between the ability of the immune system of select animals to re-elicit strong GC responses to booster immunizations and the development of a strong autologous Tier 2 nAb response. In conclusion, GC B cell frequencies in draining LNs after the booster immunizations were strongly associated with successful development of autologous Tier 2 neutralizing antibodies.
Figure 6. GC B cell relationships to autologous nAbs and longitudinal sequencing analysis of clonal lineages and affinity maturation identified in LN FNAs of BG505 SOSIP.v5.2 immunized RMs.
A. GC B cells post 3rd and 4th immunizations compared against BG505 neutralization titer. Each point represents the average of four GC B cell measurements (2 draining LNs at 2 time points) for an individual animal. Longitudinal GC B cell data was only acquired from the eight RM undergoing FNA analysis. Neutralizing titers are log transformed values.
B. GC B cell frequencies in draining LNs after each immunization for Top neutralizers (red) and non/low neutralizers (black). Points represent individual LNs. Dotted line indicates the mean frequency of GC B cells in unimmunized RM LNs.
C. The top 50 largest clonal lineages from two representative BG505 SOSIP.v5.2 immunized RMs. LN BCR sequences from Top neutralizer RJk15 (left) and non-neutralizer RUj15 (right). Each BCR lineage is a separate horizontal bar. Frequency of the lineage sequences found in each LN at each time point are demarcated. Colors correspond to specific LNs at specific time points. Blue = left inguinal LN. Burgundy = right inguinal LN. Colors progress from light to dark over time (3 wks to 21wks). The color key is shared with panel D. Boxes to the right of each chart tabulate recurrent lineages in multiple LNs (gray), lineages present in both left and right LNs (dark gray), and lineages with ‘winner’ (increasing frequencies over subsequent immunizations; orange), ‘extreme winner’ (greater than 90% of all sequence was found after the final immunization; yellow) and ‘loser’ (decreasing frequencies over subsequent immunizations; purple) distributions. For shared representation between R-ILN and L-ILN a cutoff of 1% was used.
D. A BG505 HIV Env trimer-specific LN B cell BCR sequence lineage tree from top neutralizer animal, ROp15. The tree is rooted to the most related germline sequence, positioned in the upper left. 884 sequences (centroids, each shown as a branch) are shown. The closest branches to the sequences of the BG505 SOSIP.v5.2 binding B cell found in the blood/spleen are indicated by asterisks.
See also Figure S4 and S5.
Given the large magnitude of the GC responses in the LNs of top neutralizers, we reasoned that GC B cell clonal lineages responding to HIV Env may be individually identifiable by NGS as clusters of closely related BCR sequences. We sequenced IgM and IgG BCRs from LN FNA samples using NGS (Figure 6C–D and Figure S5). Large to very large lineages of 50–30,000 independent sequences from the LNs of the Env-immunized animals were readily organizable into phylogenic trees. We focused analysis on the top 50 largest lineages per immunized animal (Figure S5B, Figure 6C). Phylogenetic trees were densely populated with closely related sequences (Fig 6D), many differing by only a single nucleotide, consistent with expectations for large active GCs, and also likely including some plasma cell and memory B cell sequences. Mutation frequencies per lineage increased over time during the course of the immunization (p < 0.04, Figure S6D–E). Three of the top 50 largest lineages from one of the top neutralizers, ROp15, were confirmed to be BG505 trimer-specific, by cross-referencing the NGS lineages against an initial set of 109 BCR sequences of BG505 SOSIP.v5.2 trimer binding B cells isolated from peripheral blood or spleen at necropsy (Figure 6D, Figure S5C and S5F). Based on the large magnitude of the GC responses in the draining LNs of immunized animals above that of unimmunized animals (Figure 6B) and animals receiving adjuvant alone, the majority of B cell lineages observed in the LN FNA NGS are likely Env-specific, but that awaits future studies with additional sorts of blood Env-binding B cells.
Analysis of the phylogeny of the largest BG505 SOSIP-specific lineage (884 independent sequences, see Methods) from ROp15 revealed a number of interesting characteristics. The lineage emerged early in the immune response (detectable after the 1st immunization) and BCR sequences close to the unmutated common ancestor (UCA) were identified (Figure 6D, Figure S5C and S5F). The lineage contained interspersed IgM and IgG switched B cells (data not shown). The lineage was detected after each of the four immunizations. The lineage had evidence of extensive hypermutation and affinity maturation over time. Additionally, the lineage was found in both left and right draining lymph nodes (Figure 6D, Figure S5C and S5F). Expanding analysis to the top 50 largest lineages per animal, these patterns were consistent. LN FNA BCR NGS revealed unmutated common ancestor clones for the majority of the lineages (Figure S5G). Furthermore, the majority (60–80%) of lineages were observed in multiple LN FNA samples (Figure 6C). A substantial fraction of the lineages were found in both the left and right draining LNs (26–66%, Figure 6C). In sum, LN FNA BCR NGS analysis identified large clonal lineages which could be traced longitudinally over the course of the immunization regimen. For BG505 SOSIP-specific sequences from a top neutralizer, the lineages were detected at the earliest time point, were detectable at all subsequent time points, were detectable in both left and right lymph nodes, and exhibited increased mutations over time.
Tfh cell quality is associated with autologous nAbs
Tfh cells are not only required for GC reactions to occur but are also limiting (Johnston et al., 2009; Victora et al., 2010). After the 4th immunization, which had the strongest association between GC B cells and nAbs (Figure 6B), the correlation between GC B cell and GC Tfh frequencies was modest (r = 0.53, p =0.05; Figure S3A) when compared to the tight relationship between GC B cells and GC Tfh cell frequencies after the 1st immunization (r = 0.86, p < 0.0001; Figure 3C). This led us to consider whether the quality of the Tfh cells differed. One way in which Tfh quality can be conceptualized is the number of GC B cells supported per GC Tfh cell. The GC B:GC Tfh cell stoichiometry was more than doubled in top neutralizer RMs compared to non/low neutralizer RMs (GC B cell to GC Tfh ratio: top, 6.8 vs non/low, 3.1, p = 0.005, Figure 7A–B and S6A. GC B cell to GC Tfh ratio: top vs low, p = 0.049; top vs non, p = 0.029, Figure S6B). The increased GC B:GC Tfh cell ratio suggested that top neutralizers had larger germinal center responses because their Tfh cells were capable of supporting larger GC B cell populations.
Figure 7. GC Tfh cell quality is associated with autologous nAb.
A. GC B cell:GC Tfh cell ratio in top neutralizers (red) versus non/low neutralizers (black) [after the 4th immunization]. Each point represents an individual LN. The best-fit linear correlations were constrained to pass through the origin. The slopes (m) for each line were found to be significantly different (F test, p = 0.0076).
B. Quantitation of GC B cell to GC Tfh cell ratio after the fourth immunization from RM neutralizer groups. Points represent individual LNs, bars indicates medians.
C. Representative flow cytometry plots of AIM+ (CD25+ Ox-40+) Env-specific CD4 T cells in PBMCs, one week after the 4th immunization.
D. Quantitation of Env-specific CD4 T cell responses in PBMCs of immunized RMs. Cells from unimmunized RMs were used as controls.
E. RNA-seq analysis of Env-specific CD4 T cells, comparing gene expression of top neutralizer and non-neutralizer RMs. Four Tfh-associated genes are highlighted (CD40LG, IL21, ICOS, and IL12Rb1).
F. RNA-seq counts of genes of interest from Env-specific CD4 T cells. Top, and Non/Low neutralizing groups are shown. Four Tfh-associated genes (CD40LG, IL21, ICOS, and IL-12Rb1) and a control gene (YWHAZ) are shown. Bars indicate median values. Average counts are plotted for samples where the RNAseq was repeated. One-way Mann-Whitney p values are shown.
See also Figure S6 and Table S2.
LN FNA GC Tfh cell numbers were too limiting for additional analysis. Therefore, we examined whether Env-specific CD4 T cells were detectable in peripheral blood. Substantial frequencies of Env-specific CD4 T cells were detected in blood at both 1 and 3 weeks after the 1st immunization (control vs week 1, p = 0.0017; control vs week 3, p = 0.0017; Figure 7C and 7D). Env-specific CD4 T cells responses were also robust in blood after the final immunization, at week 19 (control vs week 19, median: 0.16% vs 0.92%, p = 0.0018; Figure 7D) and week 24 (control vs week 24, median: 0.16% vs 2.4%, p = 0.0003; Figure 7D). Elevated frequencies of Env-specific CXCR5+ CD4 T cells were also detected in the blood at the same time points (p < 0.003, Figure S6C and S6D). To further assess the quality of the CD4 T cell responses, 200–5000 AIM+ Env-specific CD4 T cells were sorted from PBMCs after the 4th immunization and analyzed by RNA-seq (Figure S6E, S6F, and Table S2). Extensive gene expression differences were observed between Env-specific CD4 T cells in high neutralizers and non-neutralizers (1012 upregulated, 1354 downregulated > 2-fold; Figure 7E). Strikingly, Tfh-associated genes were more highly expressed in top vs non-neutralizers (Figure 7E). These included the two most critical proteins for Tfh help to B cells, IL-21 and CD40L. IL21 was one of the most differentially expressed genes between top neutralizers and non-neutralizers (8-fold increase, Fig S6G) with a wide range of expression among low neutralizers. In addition, two prominent proteins in human Tfh cell development, ICOS and IL-12Rβ (Crotty, 2014; Ma et al., 2012; 2009; Schmitt et al., 2013) were also more highly expressed in top neutralizers than non-neutralizers (Figure S6G). Significantly higher CD40LG, ICOS, and IL12Rb1 gene expression was observed when top neutralizers were compared to the group of non/low-neutralizers (CD40LG, p = 0.036; ICOS, p = 0.017; IL12Rb1, p = 0.018, Figure 7F). Thus, generation of autologous Tier 2 nAbs was associated with Tfh cell quality as measured by GC B cell to GC Tfh stoichiometry and Env-specific CD4 T cell gene expression data. Together, the correlations between autologous Tier 2 nAbs, GC B cell frequency, and Tfh cell quality identifies GC responses as a central limiting process for immunization-induced generation of autologous Tier 2 nAbs.
DISCUSSION
Here we demonstrate generation of significant titers of autologous HIV Tier 2 nAbs in the majority of BG505 SOSIP.v5.2 immunized RMs. While the BG505 nAbs results are the best reported to date, the observation of nAb responders and non-responders highlights the difficulties facing HIV vaccine development. The T and B cell responses of these animals were intensively interrogated using a suite of innovative techniques, including longitudinal LN FNAs to quantify GC Tfh cells and GC B cells, longitudinal GC BCR sequencing, antigen-specific GC Tfh quantitation by AIM, antigen-specific peripheral blood Tfh quantitation by AIM, and RNA-seq of Env-specific CD4 T cells. GC B cell frequencies in the draining LN in response to the final immunization was the most predictive factor for development of autologous nAbs, with the top neutralizers having three fold more responding GC B cells than animals that only made non-neutralizing Ab responses. Furthermore, the results indicate Tfh cell quality is a key factor for development of autologous nAbs. Thus, GCs are limiting for nAb development in response to immunization with a candidate HIV vaccine immunogen. These results suggest that direct probing of GCs should be considered as a central immune correlate in HIV vaccine trials.
HIV neutralizing epitopes of circulating Tier 2 HIV viruses are difficult epitopes for B cell response to recognize (Burton and Hangartner, 2016; Hu et al., 2015; McGuire et al., 2014; Sanders et al., 2015). For example, immunization with gp120, which contains the high mannose patch, does not elicit N332 supersite-specific bnAbs. In HIV infection, autologous nAbs generally take many months to develop. nAb responses to SOSIP immunization require three or more immunizations to develop (de Taeye et al., 2015; Sanders et al., 2015), if at all (Hu et al., 2015). In contrast, non-neutralizing Ab epitopes, such as the V3 loop, are easy epitopes for B cells to recognize and are robustly elicited by any HIV Env construct. In the case of HIV-1 Env, difficult neutralizing epitopes and easy non-neutralizing epitopes are on the same immunogen molecule, and therefore B cells recognizing each type of epitope will compete for the same protein available. Based on data here and in the literature, we posit that non-neutralizing B cell responses such as V3 specificities dominate the early response. This presents a daunting challenge for Env-specific B cells with neutralization potential (i.e., targeting Env epitopes for which neutralization is possible if the antibody has sufficient affinity), as they are at a significant competitive disadvantage to the Env-specific B cells targeting non-neutralizing epitopes. Thus, in this scenario, the factor that distinguishes animals that develop nAbs from animals that fail to develop nAbs may be the capacity of their immune system to elicit and maintain Env-specific B cells targeting neutralizing epitopes early on in the immune response, even though those B cells targeting neutralizing epitopes are less competitive than high affinity (or high frequency) B cells specific to easy non-neutralizing epitopes.
How can B cells with neutralizing specificities but initial low affinity for the immunogen best be elicited and maintained in the GC response? Three possibilities are: 1) increasing Tfh help, 2) increasing the number of B cells exposed to the immunogen, 3) favoring presentation of neutralizing epitopes over non-neutralizing epitopes on the immunogen. CD4 T cell help can be a limiting factor for successful progression of a low affinity B cell into GCs in the face of competition from high affinity B cells in the context of nitrophenol hapten immunization in mice (Schwickert et al., 2011). Thus, by extension, early availability of Tfh cells may be required for successful recruitment of low affinity HIV Env-specific B cells with nAb potential. Data presented here indicate a role for Tfh cells in the development of vaccine-elicited HIV nAbs. Therefore, methods of inducing additional Tfh cells could be useful (Locci et al., 2016), or enhancing the quality of the Tfh cells. Second, if B cells with neutralizing specificities are rare, engaging larger numbers of B cells could increase the likelihood of stimulating a nAb B cell lineage. Diverse GC responses were observed between the right and left draining LN of the same animal after bilateral immunizations implying a stochastic element to the response and highlighting the potential value immunizing at two sites to increase the number of responding B and T cells. A third mechanism by which B cells with neutralizing specificities could be favored may be minimizing presentation of non-neutralizing epitopes. To this end, designs to stabilize Env trimers (de Taeye et al., 2015; Guenaga et al., 2016) or protect the immunogen via slow release mechanisms (Hu et al., 2015; Tam et al., 2016) are of interest. One potential metric for assessing the relative neutralizing and non-neutralizing Ab responses is the BG505-specific:V3-specific IgG ratio. However, this ratio did not correlate with neutralization, likely due to the presence of other non-neutralizing epitopes present on the trimer (Hu et al., 2015). A shift in the ratio of BG505-specific:V3-specific IgG when immunizing with BG505.v5.2 was observed, suggesting that BG505 SOSIP.v5.2 partially limits V3 loop immunogenicity, which may shift the immunodominance profile of the B cell response.
GC B cell sequencing data indicate that memory B cells actively re-circulated after each immunization and reseeded new GCs. The phylogeny structures suggest that the seeding of new GCs by memory B cells was likely to come not from the most mutated branches of the lineage, but instead from moderately mutated subclones. This is consistent with cross sectional data from mouse immunizations with model antigens (Kaji et al., 2012), suggesting a certain stochasticism to both the GC response (Tas et al., 2016) and the recall of memory B cell clones for reseeding new GCs, and perhaps a bias toward recall of less mutated memory B cells. Intriguingly, for a top neutralizer animal, the largest lineages contained more BCR sequences from later time points, consistent with initially subdominant responses (i.e. lineages with neutralizing potential) becoming more dominant in top neutralizer animals over time.
LN FNAs provided a powerful technique for the identification and quantitation of GC B and GC Tfh cells and for longitudinal GC analysis in each animal. FNA LN in humans were previously shown to be non-disruptive to the LN architecture (Prasad et al., 1996). We expanded on the recent experimental use of LN FNAs in NHPs which identified GC Tfh cells at steady state (Xu et al., 2013) and LN FNA analysis in humans where total CD4 T cells responses were measured after immunization (Tatovic et al., 2015). We demonstrated that GC B cells are identifiable in LN FNAs, GC Tfh cells and GC B cells are quantitatively represented in LN FNAs, vaccine-specific GC Tfh cells could be quantitated in LN FNAs, and clonal expansion and sequence evolution of antigen-specific B cell responses can be tracked longitudinally in fine detail in LN FNAs.
Unexpectedly, robust GC B and GC Tfh cell populations were induced in the draining LN of NHPs after a single protein + adjuvant immunization, even while minimal circulating Ab was generated. Also unexpectedly, the 2nd immunization yielded significantly lower frequencies of GC cells than the 1st immunization. Given the magnitude and longevity of the GC responses observed, the short interval between immunizations appeared to have contributed to impairing the GC response to the booster immunization. Why poor GC boosting occurred at the shorter interval is unknown. One possibility is that new GC reactions may quench existing germinal centers (McHeyzer-Williams et al., 2015), prematurely terminating SHM and causing some cost to the initiation of new germinal centers.
Somatic hypermutation increased in the major GC B cell lineages upon each successive immunization in this study. A recent gp140 HIV Env immunization study did not find an association between somatic hypermutation and different adjuvant groups (Francica et al., 2015). However, no Tier 2 nAbs were elicited in any animals in that study, only non-neutralizing antibodies. Thus, we speculate that only ‘easy’ non-neutralizing/ Tier 1A B cell responses were elicited by that immunogen, which rapidly plateaued in affinity. In contrast, here development of autologous Tier 2 nAbs was observed in the majority of the animals immunized with BG505.v5.2, and major GC B cell lineages did evolve extensively during the course of the immunizations.
Understanding the GC response by directly accessing and interrogating GC cells may be particularly important in upcoming human Phase I clinical trials that will test immunogens that are not expected in and of themselves to immediately generate HIV bnAbs, but are designed to be important initial steps on a path to bnAb development (Jardine et al., 2016). Our findings indicate that GC Tfh cell and GC B cell magnitude and quality are likely to be central for success of those approaches and should be examined directly or by surrogate indicators. These findings and approaches reported here are also likely to be valuable for vaccine development to other high priority pathogens.
EXPERIMENTAL PROCEDURES
A detailed description of the experimental methods, procedures, flow cytometry panels (Table S3) and associated references are available in the Supplementary Materials.
Statistics
The Mann-Whitney test was used for evaluating differences among groups. The Wilcoxon test was used to evaluate differences between samples obtained at different time points from the same animal, unless otherwise noted. The Spearman correlation test was used for correlative analysis unless otherwise noted. Prism 6.0 (GraphPad) was used for all statistical analyses.
Supplementary Material
Acknowledgments
We thank Francois Villinger for assistance with FNA pilot studies (University of Louisiana at Lafayette). We thank the Flow Cytometry Core at the La Jolla Institute for Allergy and Immunology for expert cell sorting assistance. We thank Vijay Pandurangan (LJI), Jason Greenbaum, and Alex Fu of LJI bioinformatics for assistance with the RNA-seq analysis. This work was supported by the Scripps CHAVI-ID (SC, GS, DB, BP, RA) and National Primate Research Center funding (P51 RR000165/OD011132 to the Yerkes National Primate Research Center), with additional grants from NIAID (Emory Center for AIDS Research, NIH Grant # P30-AI-504, GS. HIVRAD, RWS, JPM), the Gates Foundation (CAVD BP, SC), and the European Research Council (ERC-StG-2011–280829-SHEV, RWS).
Footnotes
Accession Numbers
RNA-seq data is available in the Gene Expression Omnibus (GEO) database under accession number.
AUTHOR CONTRIBUTIONS
CHD, GS, and SC designed the experiments. CHD, DGC, MP, SMR, JSW, KK, SR, KML, AURM, SG, AG, and SPK performed the experiments. ATP, PW, SWT, MJG, and RWS contributed critical reagents. CHD and SC wrote the paper. CHD, DGC, MP, BB, SMR, ML, BP, JPM, RA, DRB, RWS, GS, and SC analyzed the data and edited the paper.
The authors declare no competing interests.
REFERENCES
- Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu. Rev. Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen K, Altfeld M, Alter G, Stamatatos L. Early preservation of CXCR5+ PD-1+ helper T cells and B cell activation predict the breadth of neutralizing antibody responses in chronic HIV-1 infection. J. Virol. 2014;88:13310–13321. doi: 10.1128/JVI.02186-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annu. Rev. Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Lindestam Arlehamn CS, Weiskopf D, da Silva Antunes R, Havenar-Daughton C, Reiss SM, Brigger M, Bothwell M, Sette A, Crotty S. A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4+ T Cells in Blood. The Journal of Immunology. 2016;197:983–993. doi: 10.4049/jimmunol.1600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye SW, Ozorowski G, Torrents de la Peña A, Guttman M, Julien J-P, van den Kerkhof TLGM, Burger JA, Pritchard LK, Pugach P, Yasmeen A, et al. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamantis A, Magiorkinis E, Koutselini H. Fine-needle aspiration (FNA) biopsy: historical aspects. Folia Histochem. Cytobiol. 2009;47:191–197. doi: 10.2478/v10042-009-0027-x. [DOI] [PubMed] [Google Scholar]
- Dogan I, Bertocci B, Vilmont V, Delbos F, Mégret J, Storck S, Reynaud C-A, Weill J-C. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- Evans TG, McElrath MJ, Matthews T, Montefiori D, Weinhold K, Wolff M, Keefer MC, Kallas EG, Corey L, Gorse GJ, et al. QS-21 promotes an adjuvant effect allowing for reduced antigen dose during HIV-1 envelope subunit immunization in humans. Vaccine. 2001;19:2080–2091. doi: 10.1016/s0264-410x(00)00415-1. [DOI] [PubMed] [Google Scholar]
- Francica JR, Sheng Z, Zhang Z, Nishimura Y, Shingai M, Ramesh A, Keele BF, Schmidt SD, Flynn BJ, Darko S, et al. Analysis of immunoglobulin transcripts and hypermutation following SHIV(AD8) infection and protein-plus-adjuvant immunization. Nature Communications. 2015;6:6565. doi: 10.1038/ncomms7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam R, Nishimura Y, Pegu A, Nason MC, Klein F, Gazumyan A, Golijanin J, Buckler-White A, Sadjadpour R, Wang K, et al. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 2016 doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. Structure-Guided Redesign Increases the Propensity of HIV Env To Generate Highly Stable Soluble Trimers. J. Virol. 2016;90:2806–2817. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Lindqvist M, Heit A, Wu JE, Reiss SM, Kendric K, Belanger S, Kasturi SP, Landais E, Akondy RS, et al. CXCL13 is a plasma biomarker of germinal center activity. Proceedings of the National Academy of Sciences. 2016a;113:2702–2707. doi: 10.1073/pnas.1520112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Reiss SM, Carnathan DG, Wu JE, Kendric K, Torrents de la Peña A, Kasturi SP, Dan JM, Bothwell M, Sanders RW, et al. Cytokine-Independent Detection of Antigen-Specific Germinal Center T Follicular Helper Cells in Immunized Nonhuman Primates Using a Live Cell Activation-Induced Marker Technique. The Journal of Immunology. 2016b;197:994–1002. doi: 10.4049/jimmunol.1600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Shaw GM, Korber B, Kelsoe G, Sodroski J, Hahn BH, Borrow P, McMichael AJ. HIV-Host Interactions: Implications for Vaccine Design. Cell Host and Microbe. 2016;19:292–303. doi: 10.1016/j.chom.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell AJ, Haigwood NL. Animal models in HIV-1 protection and therapy. Curr Opin HIV AIDS. 2015;10:170–176. doi: 10.1097/COH.0000000000000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell AJ, Malherbe DC, Pissani F, McBurney S, Krebs SJ, Gomes M, Pandey S, Sutton WF, Burwitz BJ, Gray M, et al. Achieving Potent Autologous Neutralizing Antibody Responses against Tier 2 HIV-1 Viruses by Strategic Selection of Envelope Immunogens. The Journal of Immunology. 2016;196:3064–3078. doi: 10.4049/jimmunol.1500527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JJ, Amancha PK, Rogers KA, Courtney CL, Havenar-Daughton C, Crotty S, Ansari AA, Villinger F. Early lymphoid responses and germinal center formation correlate with lower viral load set points and better prognosis of simian immunodeficiency virus infection. The Journal of Immunology. 2014;193:797–806. doi: 10.4049/jimmunol.1400749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, de Taeye SW, Sok D, Ozorowski G, Deresa I, et al. Murine Antibody Responses to Cleaved Soluble HIV-1 Envelope Trimers Are Highly Restricted in Specificity. J. Virol. 2015;89:10383–10398. doi: 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereño-Orbea J, Kalyuzhniy O, Deresa I, et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse P-J, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji T, Ishige A, Hikida M, Taka J, Hijikata A, Kubo M, Nagashima T, Takahashi Y, Kurosaki T, Okada M, et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. Journal of Experimental Medicine. 2012;209:2079–2097. doi: 10.1084/jem.20120127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, Harrison SC, Kepler TB, Kelsoe G. Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity. 2016;44:542–552. doi: 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M, Wu JE, Arumemi F, Mikulski Z, Dahlberg C, Miller AT, Crotty S. Activin A programs the differentiation of human TFH cells. Nat Immunol. 2016;17:976–984. doi: 10.1038/ni.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien J-P, de Val N, Cupo A, Potter CS, Klasse P-J, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CS, Avery DT, Chan A, Batten M, Bustamante J, Boisson-Dupuis S, Arkwright PD, Kreins AY, Averbuch D, Engelhard D, et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood. 2012;119:3997–4008. doi: 10.1182/blood-2011-11-392985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner-Nanan B, Deenick EK, Tangye SG. Early commitment of naive human CD4+; T cells to the T follicular helper (TFH) cell lineage is induced by IL-12. Immunology and Cell Biology. 2009;87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, et al. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J. Infect. Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- McElrath MJ, Corey L, Montefiori D, Wolff M, Schwartz D, Keefer M, Belshe R, Graham BS, Matthews T, Wright P, et al. A phase II study of two HIV type 1 envelope vaccines, comparing their immunogenicity in populations at risk for acquiring HIV type 1 infection. AIDS Vaccine Evaluation Group. AIDS Res. Hum. Retroviruses. 2000;16:907–919. doi: 10.1089/08892220050042846. [DOI] [PubMed] [Google Scholar]
- McGuire AT, Dreyer AM, Carbonetti S, Lippy A, Glenn J, Scheid JF, Mouquet H, Stamatatos L. HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science. 2014;346:1380–1383. doi: 10.1126/science.1259206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHeyzer-Williams LJ, Milpied PJ, Okitsu SL, McHeyzer-Williams MG. Class-switched memory B cells remodel BCRs within secondary germinal centers. Nat Immunol. 2015;16:296–305. doi: 10.1038/ni.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proceedings of the National Academy of Sciences. 2012;109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörner A, Douagi I, Forsell MNE, Sundling C, Dosenovic P, O'Dell S, Dey B, Kwong PD, Voss G, Thorstensson R, et al. Human immunodeficiency virus type 1 env trimer immunization of macaques and impact of priming with viral vector or stabilized core protein. J. Virol. 2009;83:540–551. doi: 10.1128/JVI.01102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape KA, Taylor JJ, Maul RW, Gearhart PJ, Jenkins MK. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331:1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, et al. CD4 T follicular helper cell dynamics during SIV infection. J. Clin. Invest. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SA. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad RR, Narasimhan R, Sankaran V, Veliath AJ. Fine-needle aspiration cytology in the diagnosis of superficial lymphadenopathy: an analysis of 2,418 cases. Diagn. Cytopathol. 1996;15:382–386. doi: 10.1002/(SICI)1097-0339(199612)15:5<382::AID-DC5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, Torrents de la Peña A, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223–aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau QJ. Immunogen design to focus the B-cell repertoire. Curr Opin HIV AIDS. 2014;9:217–223. doi: 10.1097/COH.0000000000000054. [DOI] [PubMed] [Google Scholar]
- Schmitt N, Bustamante J, Bourdery L, Bentebibel S-E, Boisson-Dupuis S, Hamlin F, Tran MV, Blankenship D, Pascual V, Savino DA, et al. IL-12 receptor β1 deficiency alters in vivo T follicular helper cell response in humans. Blood. 2013;121:3375–3385. doi: 10.1182/blood-2012-08-448902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert TA, Victora GD, Fooksman DR, Kamphorst AO, Mugnier MR, Gitlin AD, Dustin ML, Nussenzweig MC. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. Journal of Experimental Medicine. 2011;208:1243–1252. doi: 10.1084/jem.20102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, Nason MC, Montefiori D, Moldt B, Poignard P, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. Journal of Experimental Medicine. 2014;211:2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliepen K, Sanders RW. HIV-1 envelope glycoprotein immunogens to induce broadly neutralizing antibodies. Expert Rev Vaccines. 2016;15:349–365. doi: 10.1586/14760584.2016.1129905. [DOI] [PubMed] [Google Scholar]
- Tam HH, Melo MB, Kang M, Pelet JM, Ruda VM, Foley MH, Hu JK, Kumari S, Crampton J, Baldeon AD, et al. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proceedings of the National Academy of Sciences. 2016 doi: 10.1073/pnas.1606050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas JMJ, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill J-C, Reynaud C-A, Browne EP, et al. Visualizing antibody affinity maturation in germinal centers. Science. 2016;351:1048–1054. doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatovic D, Young P, Kochba E, Levin Y, Wong FS, Dayan CM. Fine-Needle Aspiration Biopsy of the Lymph Node: A Novel Tool for the Monitoring of Immune Responses after Skin Antigen Delivery. The Journal of Immunology. 2015;195:386–392. doi: 10.4049/jimmunol.1500364. [DOI] [PubMed] [Google Scholar]
- Verschoor EJ, Mooij P, Oostermeijer H, van der Kolk M, Haaft ten P, Verstrepen B, Sun Y, Morein B, Akerblom L, Fuller DH, et al. Comparison of immunity generated by nucleic acid-, MF59-, and ISCOM-formulated human immunodeficiency virus type 1 vaccines in Rhesus macaques: evidence for viral clearance. J. Virol. 1999;73:3292–3300. doi: 10.1128/jvi.73.4.3292-3300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West DJ. Clinical experience with hepatitis B vaccines. Am J Infect Control. 1989;17:172–180. doi: 10.1016/0196-6553(89)90213-7. [DOI] [PubMed] [Google Scholar]
- Xu Y, Fernandez C, Alcantara S, Bailey M, De Rose R, Kelleher AD, Zaunders J, Kent SJ. Serial study of lymph node cell subsets using fine needle aspiration in pigtail macaques. Journal of Immunological Methods. 2013;394:73–83. doi: 10.1016/j.jim.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Lynch RM, Gautam R, Matus-Nicodemos R, Schmidt SD, Boswell KL, Darko S, Wong P, Sheng Z, Petrovas C, et al. Quality and quantity of TFH cells are critical for broad antibody development in SHIVAD8 infection. Science Translational Medicine. 2015;7:298ra120. doi: 10.1126/scitranslmed.aab3964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.