Abstract
Protozoan parasites of the genus Giardia are highly prevalent globally, and infect a wide range of vertebrate hosts including humans, with proliferation and pathology restricted to the small intestine. This narrow ecological specialization entailed extensive structural and functional adaptations during host-parasite co-evolution. An example is the streamlined mitosomal proteome with iron-sulphur protein maturation as the only biochemical pathway clearly associated with this organelle. Here, we applied techniques in microscopy and protein biochemistry to investigate the mitosomal membrane proteome in association to mitosome homeostasis. Live cell imaging revealed a highly immobilized array of 30–40 physically distinct mitosome organelles in trophozoites. We provide direct evidence for the single giardial dynamin-related protein as a contributor to mitosomal morphogenesis and homeostasis. To overcome inherent limitations that have hitherto severely hampered the characterization of these unique organelles we applied a novel interaction-based proteome discovery strategy using forward and reverse protein co-immunoprecipitation. This allowed generation of organelle proteome data strictly in a protein-protein interaction context. We built an initial Tom40-centered outer membrane interactome by co-immunoprecipitation experiments, identifying small GTPases, factors with dual mitosome and endoplasmic reticulum (ER) distribution, as well as novel matrix proteins. Through iterative expansion of this protein-protein interaction network, we were able to i) significantly extend this interaction-based mitosomal proteome to include other membrane-associated proteins with possible roles in mitosome morphogenesis and connection to other subcellular compartments, and ii) identify novel matrix proteins which may shed light on mitosome-associated metabolic functions other than Fe-S cluster biogenesis. Functional analysis also revealed conceptual conservation of protein translocation despite the massive divergence and reduction of protein import machinery in Giardia mitosomes.
Author Summary
Organelles with endosymbiotic origin are present in virtually all extant eukaryotes and have undergone considerable remodeling during > 1 billion years of evolution. Highly diverged organelles such as mitosomes or plastids in some parasitic protozoa are the product of extensive secondary reduction. They are sufficiently unique to generate interest as targets for pharmacological intervention, in addition to providing a rich ground for evolutionary cell biologists. The so-called mitochondria-related organelles (MROs) comprise mitosomes and hydrogenosomes, with the former having lost any role in energy metabolism along with the organelle genome. The mitosomes of the intestinal pathogen Giardia lamblia are the most highly reduced MROs known and have proven difficult to investigate because of their extreme divergence and their unique biophysical properties. Here, we implemented a novel strategy aimed at systematic analysis of the organelle proteome by iterative expansion of a protein-protein interaction network. We combined serial forward and reverse co-immunoprecipitations with mass spectrometry analysis, data mining, and validation by subcellular localization and/or functional analysis to generate an interactome network centered on a giardial Tom40 homolog. This iterative ab initio proteome reconstruction provided protein-protein interaction data in addition to identifying novel organelle proteins and functions. Building on this data we generated information on organelle replication, mitosome morphogenesis and organelle dynamics in living cells.
Introduction
Since the single endosymbiotic event leading to establishment of mitochondria approximately 2 billion years ago [1,2,3] these organelles have undergone massive changes and have evolved into highly specialized and essential subcellular compartments in all eukaryotes [4,5], with only one possible exception identified so far [6]. These changes comprise a dramatic size reduction, nuclear transfer of organelle genomes, and a renewal of the proteome, which is synthesized almost entirely as precursor proteins on cytosolic ribosomes [7,8,9,10,11,12,13,14] and imported from the cytoplasm [15]. Mitochondria have been remodeled and/or restructured to very different degrees in different species. Mitochondria-related organelles (MROs), i.e. hydrogenosomes and mitosomes [16,17,18,19,20] in some protists lacking canonical mitochondria represent extreme forms of reduction and/or divergence. The potential of highly diverged organelle-specific pathways as targets for intervention has sparked research into the evolution of MROs in single-celled organisms of all five eukaryotic supergroups [21,22]. Notably, the microaerophilic protozoan pathogens Entamoeba histolytica [20] and Giardia lamblia [23,24], as well as intracellular parasites such as Cryptosporidium parvum [25] and Encephalitozoon cuniculi [26] harbor mitosomes. Interestingly, recent investigation of MROs in Spironucleus salmonicida, a diplomonad and the closest relative of G. lamblia belonging to the Excavata super-group, revealed that these organelles are in fact hydrogenosomes [27]. Although it has been demonstrated that G. lamblia mitosomes do not produce hydrogen, this sheds a completely new light on the evolution of MROs in diplomonads.
Proliferating G. lamblia trophozoites contain 20–50 double membrane-bounded 100 nm spherical mitosomes [23,24] devoid of an organelle genome [28,29,30,31]. Although not proven experimentally, G. lamblia mitosomes are likely essential due to a subset of conserved mitochondrial proteins required for iron- sulphur (Fe-S) protein maturation [23,32,33,34,35]. Yeast genetic experiments suggested that Fe-S protein maturation, the only function currently ascribable to G. lamblia mitosomes, is in fact the minimal essential function of mitochondria [36]. Hence, these organelles have also attracted considerable interest as cell biological models to study extreme reductive evolution of MROs [23,37,38,39,40,41,42]. However, due to massive, albeit selective sequence divergence in G. lamblia, conventional data mining strategies for identification of mitosome proteins based on homology-based in silico searches fall short [26,28,32,43,44,45,46,47]. Moreover, classical, organelle enrichment-based proteome analyses approaches have had only limited success owing to the small size of the organelles and the omnipresence of contaminating endoplasmic reticulum (ER) and cytoskeleton elements in mitosome fractions [33,48,49].
Nevertheless, there is unambiguous experimental evidence for the functional conservation of the mitosomal protein import machinery [20,23,24,49]. The small subset of structurally conserved mitosome proteins such as G. lamblia IscU, ferredoxin, Cpn60, IscS and mtHsp70 are imported by transit peptide-dependent and -independent mechanisms [23]. However, the predicted components of the TOM/TIM import apparatus are diverged beyond recognition by state-of-the-art homology search tools. Indeed, the protein repertoire of the mitosomal outer membrane and its networks are scarcely characterized: only one subunit of the translocon in the outer mitochondrial (TOM) complex, a highly diverged Tom40 homologue (GlTom40), and [50] more recently a giardial Tim44 homologue [49], have been identified. Furthermore, there is no information on how mitosome homeostasis is achieved in terms of organelle size and number.
To address questions concerning protein networks at mitosomal membranes in association with mitosome homeostasis and to account for the extreme sequence divergence in G. lamblia, we implemented novel experimental approaches. We were successful to tag two outer membrane organelle proteins with GFP to show that these small organelles are immobilized, distinctive entities with no appreciable inter-organelle exchange or network character. Using a giardial TOM40 homolog as a starting bait we generated information on protein-protein interactions at the outer membrane as well as expanding the organelle proteome by identifying novel components. By using interactome targets validated by subcellular localization as baits for subsequent reverse co-IP rounds, we were able to extend this initial interactome beyond the outer membrane, including dually localized endoplasmic reticulum (ER) and mitosome proteins, as well as identifying previously described and novel imported organelle proteins. In addition to identification of two components with a role in mitosome morphogenesis and homeostasis the combined data revealed a core organelle membrane interactome composed of only 3 tightly-associated proteins. Furthermore, we tested constraints for import of nuclear-encoded mitosome proteins and could show conservation of this mechanism even in the highly diverged and reduced Giardia mitosome.
Materials and Methods
Giardia cell culture, induction of encystation, pulse-empty chase set-up and transfection
G. lamblia WB (line C6; ATCC catalog number 50803) trophozoites were grown and harvested using standard protocols [51]. Encystation was induced with the two-step method as described previously [40,52]. Transgenic parasites were generated according to established protocols by electroporation of linearized pPacV-Integ-based plasmid vectors prepared from E. coli as described in [42]. After selection for puromycin resistance, transgenic G. lamblia cell lines were cultured without puromycin.
Construction of expression vectors
All sequences of oligonucleotide primers for PCR used in this work are listed in S1 Table.
For cloning of C-terminally hemagglutinin (HA)-tagged proteins in Giardia, a vector PAC-CHA was designed on the basis of the previously described vector pPacV-Integ [42], where additional restriction sites were inserted [53].
A cyst wall protein 1 promoter (pCWP1)-driven G. lamblia ferredoxin (fd)-human dihydrofolate reductase (DHFR) chimeric gene was generated by fusing two genes by overlapping PCR: i) an intron-less fd mitosomal targeting signal (MTS) (MTSfdΔint) open reading frame (ORF) was generated using primer pair 33 (S1 Table) with G. lamblia cDNA as template, ii) a DHFR_HA minigene was generated using primer pair 34 (S1 Table) with a cloned human DHFR cDNA as template. The fused product was digested with SpeI and PacI and inserted in a PAC vector to yield construct pCWP1_MTSfdΔint-DHFR_HA.
A pCwp1_ MTSfdΔint-DHFR_Neomycin resistance construct (without HA tag) was generated for protein import block assays. Primer pair 35 (S1 Table) was used on pCwp1_ MTSfdΔint-DHFR_HA as a template. The amplified product was digested with NsiI and PacI and ligated into a vector containing a neomycin resistance cassette [51].
Co-immunoprecipitation with limited cross-linking
G. lamblia WBC6 and transgenic trophozoites expressing C-terminally HA tagged bait proteins were harvested and subjected to immunofluorescence assay to confirm correct subcellular distribution of bait proteins. Parasites were collected by centrifugation (900 x g, 10 minutes, 4°C), washed in 50 ml of cold phosphate buffer saline solution (PBS) and adjusted to 2 x107 cells.ml-1 in PBS (VWR Prolabo). The appropriate formaldehyde concentration for cross-linking (2.25%) was determined by a titration assay (S2 Fig). For the co-immunoprecipitation (co-IP) assays, 109 parasites were resuspended in 10 ml 2.25% formaldehyde (in PBS) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF; SIGMA, Cat. No. P7626) and incubated for 30 minutes at room temperature (RT). Cells were pelleted, washed once with 10 ml PBS, and quenched in 10 ml 100 mM glycine in PBS for 15 minutes at RT. The collected cells were then resuspended in 5 ml RIPA lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS, 10 mM EDTA) supplemented with 2 mM PMSF and 1 x Protease Inhibitor cocktail (PIC, Cat. No. 539131, Calbiochem USA) and sonicated twice using a Branson Sonifier with microtip (Branson Sonifier 250, Branson Ultrasonics Corporation) with the following settings: 60 pulses, 2 output control, 30% duty cycle and 60 pulses, 4 output control, 40% duty cycle. The sonicate was incubated on a rotating wheel for 1 h at 4°C, aliquoted into 1.5 ml tubes and centrifuged (14,000 x g, 10 minutes, 4°C). The soluble protein fraction was mixed with an equal volume detergent-free RIPA lysis buffer supplemented with 2% TritonX (TX)-100 (Fluka Chemicals) and 40 μl anti-HA agarose bead slurry (Pierce, product # 26181). After binding of tagged proteins to the beads at 4°C for 2 h on a rotating wheel, beads were pulse-centrifuged and washed 4 times with 3 ml Tris-Buffered Saline (TBS) supplemented with 0.1% TX-100 at 4°C. After a final wash with 3 ml PBS the loaded beads were resuspended in 350 μl PBS, transferred to a spin column (Pierce spin column screw cap, product # 69705, Thermo Scientific) and centrifuged for 10 s at 4°C. Elution was performed by resuspending beads in 30 μl of PBS. Dithiothreitol (DTT; 100mM; Thermo Scientific, Cat. # RO861) was added and samples were boiled for 5 min followed by centrifugation (14,000 x g, 10 minutes, RT).
Protein analysis and sample preparation for mass spectrometry-based protein identification
SDS-PAGE and immunoblotting analysis of input, flow-through, and eluate fractions was performed on 4%-12% polyacrylamide gels under reducing conditions, (molecular weight marker Cat. No. 26616, Thermo Scientific, Lithuania). Transfer to nitrocellulose membranes and antibody probing were done as described previously [54], using anti-HA (dilution 1:500; Roche) followed by anti-rat antibodies coupled to horseradish peroxidase (dilution 1:5000; Southern Biotech). Gels for mass spectrometry (MS) analysis were stained using Instant blue (Expedeon, Prod. # ISB1L) and de-stained with sterile water.
Mass Spectrometry, protein identification and data storage
Stained gel lanes were cut into 8 equal sections. Each section was further diced into smaller pieces and washed twice with 100 μl of 100 mM ammonium bicarbonate/ 50% acetonitrile for 15 min at 50°C. The sections were dehydrated with 50 μl of acetonitrile. The gel pieces were rehydrated with 20 μl trypsin solution (5 ng/μl in 10 mM Tris-HCl/ 2 mM CaCl2 at pH 8.2) and 40 μl buffer (10 mM Tris-HCl/ 2 mM CaCl2 at pH 8.2). Microwave-assisted digestion was performed for 30 minutes at 60°C with the microwave power set to 5 W (CEM Discover, CEM corp., USA). Supernatants were collected in fresh tubes and the gel pieces were extracted with 150 μl of 0.1% trifluoroacetic acid/ 50% acetonitrile. Supernatants were combined, dried, and the samples were dissolved in 20 μl 0.1% formic acid before being transferred to the autosampler vials for liquid chromatography-tandem MS (injection volume 7 to 9 μl). Samples were measured on a Q-exactive mass spectrometer (Thermo Scientific) equipped with a nanoAcquity UPLC (Waters Corporation). Peptides were trapped on a Symmetry C18, 5 μm, 180 μm x 20 mm column (Waters Corporation) and separated on a BEH300 C18, 1.7 μm, 75 μm x 150 mm column (Waters Corporation) using a gradient formed between solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). The gradient started at 1% solvent B and the concentration of solvent B was increased to 40% within 60 minutes. Following peptide data acquisition, database searches were performed using the MASCOT search program against the G. lamblia database (http://giardiadb.org/giardiadb/) with a concatenated decoy database supplemented with commonly observed contaminants and the Swissprot database to increase database size. The identified hits were then loaded onto the Scaffold Viewer version 4 (Proteome Software, Portland, US) and filtered based on high stringency parameters, i.e. 95% for peptide probability, a protein probability of 95%, and a minimum of 2 unique peptides per protein. Where indicated in the text, slightly relaxed filtering parameters were applied. Proteins identified in both bait-specific and control datasets were considered of interest if they were at least 5-fold enriched in the bait-specific datasets (in terms of spectral counts) based on high stringency parameters. Access to raw MS data is provided through the ProteomeXchange Consortium on the PRIDE platform [55].
In silico co-immunoprecipitation dataset analysis
Analysis of primary structure and domain architecture of putative mitosomal hypothetical proteins was performed using the following tools and databases: MITOPROT (https://ihg.gsf.de/ihg/mitoprot.html) and PSORTII (http://psort.hgc.jp/form2.html) for subcellular localization prediction, TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) for transmembrane helix prediction, SMART (http://smart.embl-heidelberg.de/) for prediction of patterns and functional domains, pBLAST for protein homology detection (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins), HHPred (http://toolkit.tuebingen.mpg.de/hhpred) for protein homology detection based on Hidden Markov Model (HMM-HMM) comparison, and the Giardia Genome Database (http://giardiadb.org/giardiadb/) to extract other/organism-specific information, e.g. expression levels of the protein, predicted molecular size and nucleotide/protein sequence. For functional domains predicted by SMART we used an e-value of 10e-5 as cutoff, and for protein homologies predicted by pBLAST we accepted alignment scores above 80. However, since G. lamblia homologs for eukaryotic proteins are highly diverged, we also considered functional domain predictions associated to a lower e-value. Alignment scores between 50 and 80 were accepted only when pBLAST predictions were consistent with HHPred output.
Immunofluorescence analysis (IFA) and microscopy
Preparation of chemically fixed cells for immunofluorescence and analysis of subcellular distribution of reporter proteins by wide-field and confocal microscopy were done as described previously [42,54]. Nuclear labelling was performed with 4',6-diamidino-2-phenylindole (DAPI). The HA epitope tag was detected with a monoclonal anti-HA antibody coupled to FITC (dilution 1:50; Roche) whereas GlIscU was detected with a self-made antibody (dilution 1:300) followed by an anti-mouse antibody coupled to Alexafluor 594 (dilution 1:300; Molecular Probes). To avoid any possibility for cross reaction, co-labelling experiments for IFA were performed by incubating first with the anti-GlIscU antibody, followed by the AF594-conjugated anti-mouse secondary antibody, and direct detection of the HA epitope tag with a FITC-conjugated rat anti-HA monoclonal antibody as a final step.
Live-cell microscopy and fluorescence recovery after photobleaching (FRAP)
Transgenic G. lamblia trophozoites expressing GFP-GlTom40 or Gl29147-GFP were harvested and prepared for imaging in PBS supplemented with 5 mM glucose (Cat. No. 49139, Fluka), 5 mM L-cysteine (Cat. No. C6852, Sigma) and 0.1 mM ascorbic acid (Cat. No. 95209, Fluka) at pH 7.1. FRAP and time-lapse series were performed as described previously [54,56].
Sample preparation for transmission electron microscopy
Transgenic trophozoites ectopically expressing wild type G. lamblia dynamin related protein (GlDRP) (ORF Gl50803_14373) or the constitutively active (GTP-locked) GlDRP-K43E variant under the control of the CWP1 promoter [56] were harvested 3 h post induction and analyzed by transmission electron microscopy (TEM) as described previously [56].
Sub-cellular fractionation analysis
For sub-cellular fraction experiments, 4.106 GlDRP-HA and GlDRP-K43E-HA- expressing transgenic cells were lysed by freeze-thawing and supernatant (soluble fraction) and pellet (membrane fraction) were prepared by centrifugation at 14’000 x g for 10 minutes at 4°C. The HA-tagged proteins were detected by SDS-PAGE and Western blot using a rat anti-HA mAb (clone 3F10, Roche) as described previously [54].
DHFR-MTX protein import block assay
The MTSfdΔint-DHFR fusion (see also above under “Constructs”) was expressed under the control of the inducible CWP1 promoter in a background transgenic line constitutively expressing HA- tagged 17030 (cell line Cwp1_MTSfdΔint-DHFR/Gl17030HA). DHFR expression was induced using the 2-step method [40] for 4 h and “chased” for 24 h by placing the cells again in standard growth medium in the presence or absence of 1 μM methotrexate (MTX). Total cell lysates were separated by SDS-PAGE and Western blot to detect processed and unprocessed forms of the Gl17030HA reporter. Subcellular distribution was analyzed by immunofluorescence assay (IFA) using wide field microscopy.
Results
G. lamblia mitosomes do not form dynamic networks and are associated to the single dynamin-related protein GlDRP
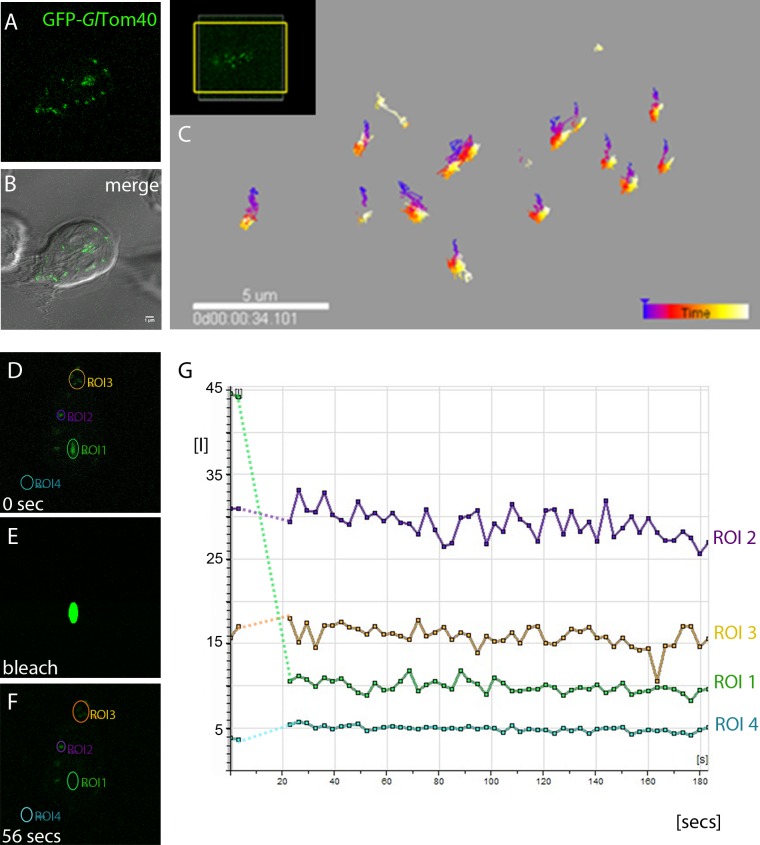
Mitochondria in higher eukaryotes are highly dynamic organelle networks that move in the cell via microtubules and microfilaments and undergo constant fission and fusion to meet the energy requirements of the cell [57,58]. IFA and TEM analyses suggest that G. lamblia mitosomes are very small spherical organelles with no evidence of network formation. In addition, the mitosome population in each cell can be divided into peripheral mitosomes (PM) distributed randomly in the cytoplasm and what has been dubbed the central mitosome complex (CMC) [23]. The latter consists of a grape-like cluster of individual organelles of the size and shape of peripheral mitosomes that is closely and permanently associated to the basal body complex between the two nuclei [23]. Interestingly, these organelles remain spatially distinct despite their close proximity. The motility of this central cluster is highly constrained and restricted to ordered segregation with the duplicated basal body complex during cell division [23]. Because green fluorescent protein (GFP) imported into the mitosome matrix is not fluorescent [23], GFP-tagging of mitosomes has not been possible until now. Martinkova et al. [59] have shown that mitosomes in trophozoites can be labeled for live cell microscopy using HaloTag markers [60]. However, no quantitative information on the spatial dynamics of peripheral mitosomes in the cytoplasm was presented in this report. We investigated organelle dynamics in living cells by performing time lapse microscopy of cells expressing GFP-tagged mitosome reporters for the outer membrane. Conditional expression of N-terminally GFP-tagged GlTom40 with 3 h of induction followed by “chasing” newly-synthesized GFP-Tom40 into mitosomes over 2–3 h in normal conditions was found suitable for labeling organelles in living cells (Fig 1A and 1B). Tracking of individual organelles over a period of >30 min showed no significant cytoplasmic movement or changes in number or morphology (Fig 1C), suggesting that organelles neither move randomly nor are they transported directionally in the cytoplasm along cytoskeleton structures. To test whether mitosome outer membrane proteins are exchanged between organelles we performed FRAP experiments on cells conditionally expressing GlTom40-GFP. Since GlTom40-GFP is membrane-anchored, FRAP addresses the question whether mitosomes are isolated organelles and whether they form membrane continuities which would allow exchange of outer membrane proteins. No recovery of fluorescence in bleached CMC or PM organelles was detected (Fig 1D–1G) suggesting that peripheral and CMC organelle membranes remain distinct.
Fig 1. G. lamblia mitosomes are immobilized and do not form dynamic networks.
(A-B) Detection of typical organelle distribution of GFP-tagged GlTom40 (green) in time-lapse microscopy. (B) Overlay showing the CMC between the two nuclei and PMs which show a canonical dispersed localization throughout the cell. (C) Tracking of organelles during a period of 30 min shows no significant movement of mitosomes in the cytosol. (D-G) FRAP experiments performed on cells conditionally expressing GFP-tagged GlTom40 suggest that outer membrane proteins are not able to move amongst/in between organelles. (E) Photobleaching of a single mitosome (region of interest 1 (ROI 1)) in a living cell is shown. (F-G) Fluorescence in a bleached organelle (green line in the graph) does not recover even after several minutes (>20 min). Purple and brown lines in the graph represent fluorescence in unbleached areas (ROIs 2 and 3). (G) Fluorescence micrographs from the image series at the start (0 sec) of the experiment, during bleaching, and at the beginning of the recovery phase (20 sec). Arbitrary units of fluorescence are indicated [I]. Broken lines connect pre- and post-bleaching values in the graph. Scale bar: 1 μM.
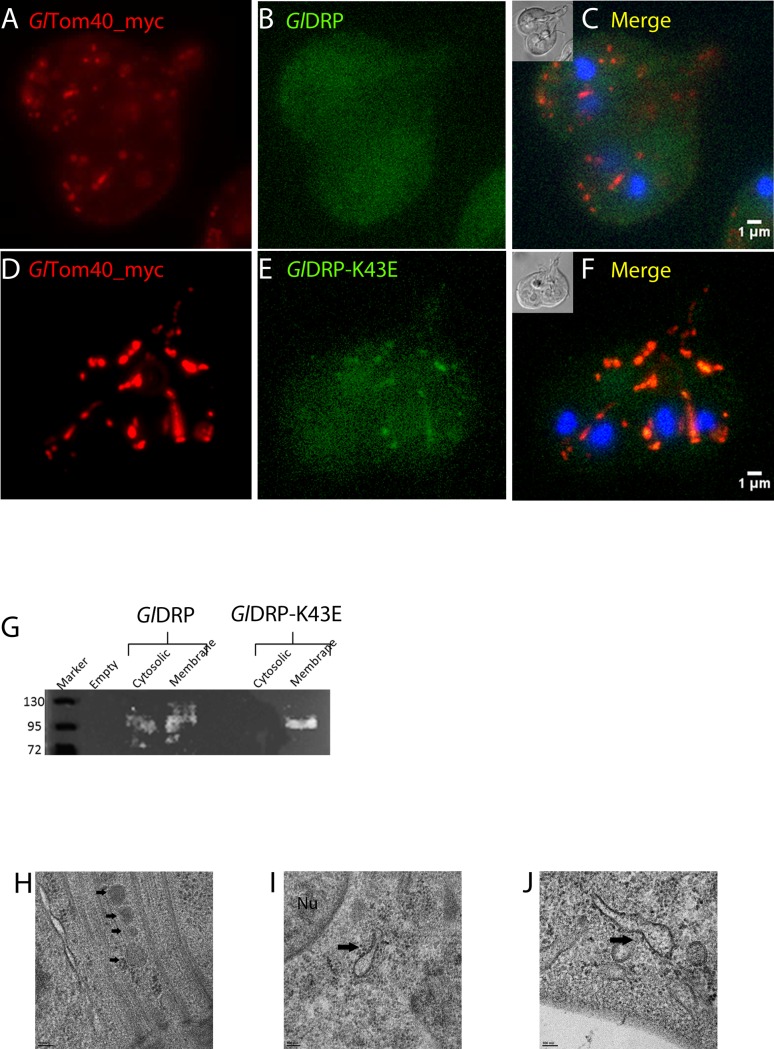
Despite intensive research in the field of MROs, little is known regarding factors required for their division. Dynamin-related proteins (DRPs) are implicated in mitochondrial and hydrogenosomal division in higher eukaryotes and in protozoa such as Trypanosoma brucei [61,62] and Trichomonas vaginalis [63]. G. lamblia harbors a single DRP (ORF Gl50803_14373) [56] with a previously documented role in trafficking of cyst wall material, and endocytic and exocytic organelle homeostasis [56]. To test for a hitherto unrecognized role of GlDRP in determining mitosome morphology and/number, we used a dual cassette expression vector [54] to express constitutive C-terminally myc-tagged GlTom40 as a reporter for mitosomes and inducible C-terminally HA-tagged wild-type (GlDRP-HA) or GTP-locked (GlDRP-K43E-HA) variants in trophozoites. In trophozoites expressing GlDRP-HA (Fig 2A–2C), IFA analyses demonstrated the typical random cytoplasmic distribution of PMs i.e. “dispersed” [23]. However, cells expressing the GTP-locked variant GlDRP-K43E-HA (Fig 2D–2F) presented a “clustered” mitosome phenotype, indicative of enlarged organelles. Consistent with this phenotype and in line with previous reports [56], the subcellular distribution of HA-tagged GlDRP remained mostly cytosolic (Fig 2B). Conversely, GlDRP-K43E-HA showed a punctate distribution (Fig 2E) and significant signal overlap with GlTom40-myc (Fig 2F), suggesting selective accumulation of GlDRP-K43E-HA on mitosome membranes. We tested whether this marked association of ectopically expressed GlDRP-K43E with organelle membranes compared to the wild type DRP variant in IFA could be corroborated in cell fractionation experiments. Separation by SDS-PAGE and immunoblot analysis revealed that GlDRP-HA was almost equally distributed between the “cytosolic” and “membrane” fraction, whereas the mutated variant GlDRP-K43E-HA was detected only in the “membrane” fraction (Fig 2G). These data were consistent with the microscopical analysis in Fig 2E and suggest increased association of GlDRP-K43E-HA with organelle membranes compared to wild-type GlDRP-HA. To characterize the nature of the GlDRP-K43E-HA-dependent phenotype in more detail, we performed transmission electron microscopy of induced transgenic cells. Cells expressing the GlDRP-K43E-HA variant frequently presented elongated and tubular mitosome structures (Fig 2I and 2J) compared to cells expressing wild type GlDRP-HA (Fig 2H).
Fig 2. Conditional expression of GlDRP-K43E elicits a mitosome morphogenesis phenotype.
(A-C) Subcellular localization of a C-myc tagged GlTom40 (red) by IFA in cells induced to express a wild type GlDRP (green) or GTP-locked variant GlDRP-K43E (D-F). Note the altered size and distribution of organelles labeled with Tom40-myc in GlDRP-K43E expressing lines. Nuclear DNA is stained with DAPI (blue). Insets: DIC images. Scale bar: 1 μM (G) -Cell fractionation experiments confirm fixed membrane localization of GlDRP-K43E. (H) TEM: normal morphology of mitosomes (black arrows) in the CMC in cells expressing wild type GlDRP whilst cells expressing GlDRP-K43E show enlarged dumbbell-shaped mitosomes (black arrows in I, J) indicative of defective organelle division. Nu: nucleus. Scale bars: 100 nm.
Taken together, these data show how mitosomes are immobilized in the cell and present no measurable outer membrane exchange in the conditions tested. Their morphogenesis is perturbed following conditional ectopic expression of a dominant-negative GTP-locked GlDRP variant, suggesting a previously unappreciated role for this GTPase in the maintenance of mitosome integrity and organelle morphogenesis in G. lamblia.
Co-IP with the G. lamblia Tom40 homolog identifies novel interacting proteins in the mitosome outer membrane
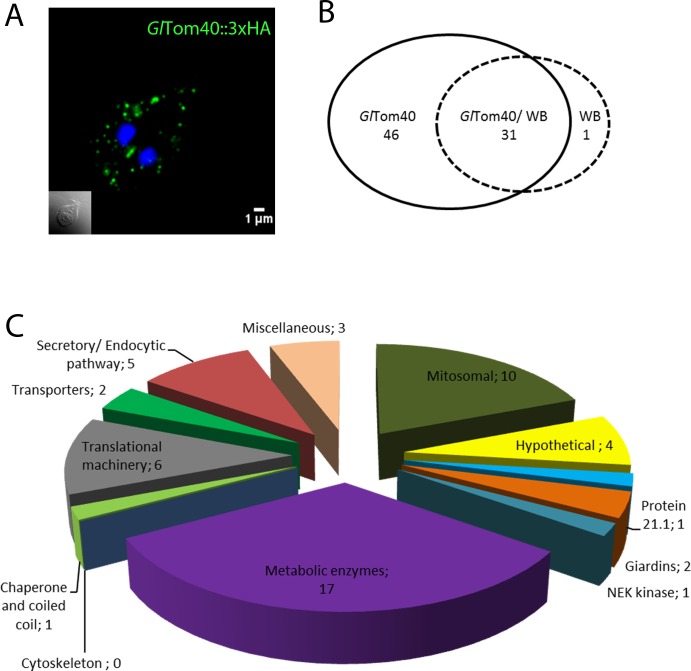
The aberrant mitosome morphology after conditional expression of GlDRP-K43E points towards mitosome-associated machinery at the organelle’s surface involved in organelle homeostasis. Despite efforts aimed at defining the protein content of mitosomes in Giardia [33,49,50], the composition of this organelle’s outer and inner membrane proteome remains sparsely characterized, with the exception of a highly diverged putative Tom40 homologue (GlTom40; ORF Gl50803_17161) and a structurally-conserved Tim44 [49,50]. To generate a robust mitosome outer membrane proteome we focused on GlTom40 as a point of origin and developed a tailored co-IP protocol with an HA-tagged variant as “bait”. A transgenic line GlTom40-HA constitutively expressing the epitope-tagged bait protein was generated; exclusive mitosome localization of the bait protein in transgenic cells was confirmed by IFA in co-labelling experiments with a newly-made anti-GlIscU antibody (Fig 3A and S1A Fig). To ensure solubilization of mitosomal membranes while avoiding disruption of Tom40-associated protein complexes, we used carefully titrated, formaldehyde-based cross-linking [64] to stabilize predicted protein-protein interactions in co-IP experiments during extraction with the option to reverse covalent bonds (S2 Fig; see also in Materials and Methods). Following MS analysis and data filtration using a control dataset obtained from non-transgenic cells (ctrl.co-IP) we identified a total of 52 proteins, 46 exclusive and 6 enriched in the GlTom40 co-IP dataset (Fig 3B). This protein set was parsed and subdivided into different metabolic and/or functional categories (Fig 3C). In the mitosomal protein category few detected four previously identified mitosome proteins namely: mitochondrial HSP70 (ORF Gl50803_14581), oxidoreductase 1 (GlOR1; ORF Gl50803_91252), proteins Gl50803_9296 and Gl50803_14939, recently named MOMP35 [33,49]. We extracted additional information from the GlTom40 co-IP data by relaxing stringency parameters to (95_1_95), obtaining a total of 150 proteins (FDR 3.4%). Of these, 109 hits were exclusive to the expanded GlTom40 co-IP dataset which contained 3 additional annotated mitosome proteins namely, chaperonin 60 (Cpn60; ORF Gl50803_103891), GlQb-SNARE 3 (putative Sec20, ORF Gl50803_5161) and GlIscU (NifU-like protein; ORF Gl50803_15196).
Fig 3. Co-IP with HA_tagged GlTom40 yields numerous candidate interacting proteins.
(A) Immunofluorescence microscopy: C-terminally HA-tagged GlTom40 (GlTom40-HA) is an exclusive marker for mitosomes (green). Nuclear DNA is stained with DAPI (blue). Inset: DIC image. (B) Venn diagram indicating 46 GlTom40 specific hits. (C) Parsing of 46 GlTom40-specific and 6 enriched proteins in metabolic categories based on available annotations in www.giardiaDB.org.
Imaging-based validation of the GlTom40 co-IP dataset
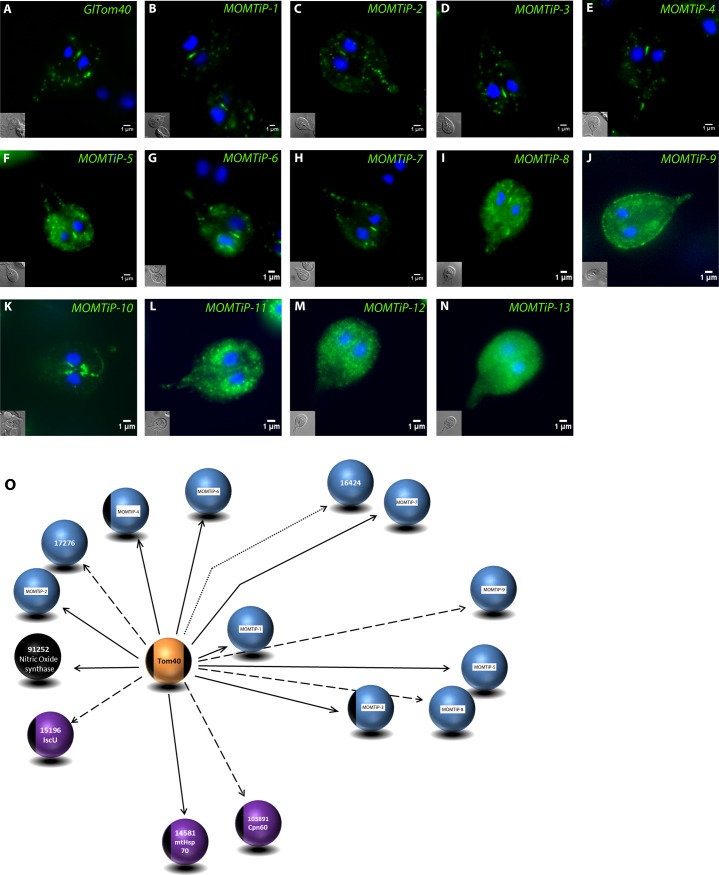
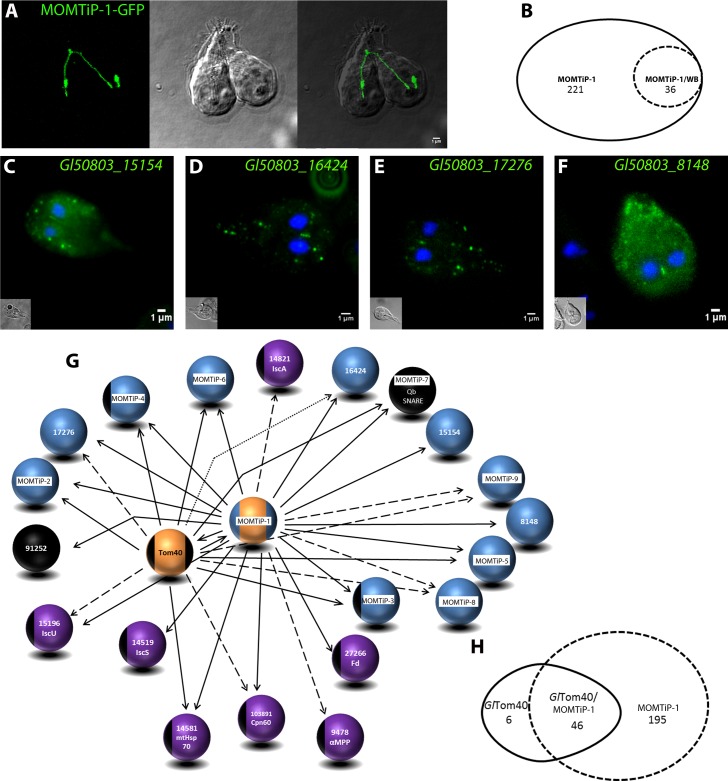
Limited chemical cross-linking in co-IP assays expands the range of discovery beyond primary interactions with the bait. We therefore performed an initial validation of the predicted GlTom40 interacting proteins in this dataset by subcellular localization of ectopically expressed, epitope-tagged candidates to mitosomes. We selected 13 of the 109 candidate Mitosomal Outer Membrane Tom40 interacting proteins (MOMTiP; Table 1) based on their spectral counts with high stringency parameters and/or protein domains identified with HHPred (S2 Table) and engineered endogenous promoter-driven, C-terminally HA-tagged variants for all. IFA analysis of corresponding transgenic lines showed mitosomal localization for 8 candidates (Fig 4A–4H), of which 4 proteins of unknown function (MOMTiP-5 to 8) presented dual localization (mitosome and ER) (Fig 4F–4I). The five remaining proteins of this set of 13 candidates (MOMTiP- 9–13; Fig 4J–4N) showed dispersed patterns of subcellular distribution and were not considered mitosome proteins. Fig 4O shows a consolidated depiction of a first GlTom40-centered mitosomal outer membrane interactome, which includes the 8 proteins localized to mitosomes described above, as well as 4 previously identified matrix proteins and 3 newly validated hypothetical proteins comprised in the list of GlTom40 interacting proteins. Taken together, the imaging data are in agreement with the protein-protein interaction data, and support limited chemical crosslinking as a suitable method to stabilize protein complexes during co-IP.
Table 1. Selected MOMTiP proteins for validation by light microscopy.
| ORF number | Assigned name | Peptide count in GlTom40-specific co-IP dataset (none detected in WB) |
|---|---|---|
| Gl50803_29147 | MOMTiP-1 | 30 |
| Gl50803_10971 | MOMTiP-2 | 63 |
| Gl50803_14939 | MOMTiP-3 (GiMOMP35) [49] | 21 |
| Gl50803_9296 | MOMTiP-4 | 11 |
| Gl50803_21943 | MOMTiP-5 | 13 |
| Gl50803_22587 | MOMTiP-6 | 8 |
| Gl50803_5785 | MOMTiP-7 (Qb-SNARE 4) [65] | 11 |
| Gl50803_9503 | MOMTiP-8 | 1 |
| Gl50803_7188 | MOMTiP-9 | 1 |
| Gl50803_114546 | MOMTiP-10 | 1 |
| Gl50803_113892 | MOMTiP11 | 43 |
| Gl50803_9719 | MOMTiP-12 | 47 |
| Gl50803_10822 | MOMTiP-13 | 3 |
Fig 4. Subcellular localization of co-precipitated GlTom40 interaction partners.
(A-N) Immunofluorescence microscopy: subcellular localization of C-terminally HA-tagged GlTom40 and 13 putative interaction partners (green) falls into 3 categories: Typical mitosome localization (A-E); dual localization to mitosomes and ER (F-I); no or ambiguous mitosome localization (J-N). Nuclear DNA is stained with DAPI (blue). Insets: DIC image. Scale bars: 1μm. (O) Partially validated GlTom40 interactome showing the bait protein (orange sphere), matrix proteins (purple), previously identified and mitosome-localized proteins (black), and mitosome-localized hypothetical proteins (blue). The stringency parameters used for detection (high, medium, and relaxed) are represented by bold, dashed, and dotted arrows, respectively.
Iterative reverse co-IP experiments expand the mitosomal protein interactome network beyond the outer membrane
IFA analysis of MOMTiP-1 to 13 indicated that the majority of these proteins are associated to mitosomes, thereby providing preliminary validation of the selected 13 candidates of the primary GlTom40-specific co-IP dataset. To further test the robustness of this primary interactome and expanding it beyond the mitosomal membrane, we performed a first reverse co-IP experiment using MOMTiP-1 (ORF Gl50803_29147) as bait. MOMTiP-1 was chosen because it presented the largest spectral count with high stringency parameters in the GlTom40 dataset and localized unequivocally to mitosomes (S1B Fig).
MOMTiP-1 is a Giardia-specific mitosome-localized protein of unknown function. In silico analysis using TMHMM robustly detected a 22 amino acid-long transmembrane helix in the N-terminal part of the protein followed by a large C-terminal domain predicted to be exposed to the cytosol on the mitosomal surface. To track this protein in vivo, we engineered MOMTiP-1 constructs for live cell imaging using GFP reporters. We have shown previously that GFP only fluoresces if exposed to the cytoplasm and never after import into mitosomes [23]. Therefore, the brightly fluorescing and mitosome-localized MOMTiP-1-GFP fusion supports the predicted topology for MOMTiP-1 as a type 1 transmembrane protein with respect to the outer mitosomal membrane. Surprisingly, many cells expressing MOMTiP-1-GFP showed a mitosome morphology dubbed “string” phenotype suggestive of extensive elongation of organelles to large tubules (Fig 5A; left). In many cases, virtually all PMs had been replaced by a single long organelle with a diameter that corresponded to that of an individual mitosome. Although the “string” mitosome phenotype was compatible with survival of the parasites, many trophozoites appeared to be delayed or even arrested in cytokinesis and had a typical heart-shaped appearance (Fig 5A; middle) previously observed in cells which are unable to complete cytokinesis [66]. Because the tubular organelles ran through the non-divided part connecting both daughter cells, we postulated that inability to divide mitosomes impairs completion of cytokinesis.
Fig 5. Expansion and validation of the GlTom40 interactome by reverse co-IP with MOMTiP-1.
(A) The “string” mitosome phenotype observed upon constitutive expression of GFP-tagged MOMTiP-1 (MOMTiP-1-GFP). Left panel: MOMTiP-1-GFP localized exclusively to mitosomes and in some cases virtually all of the peripheral organelles have been replaced by a single long tubular mitosome spanning both daughter cells length-wise. Middle panel: DIC image. Right panel: Overlay of the two channels. (B) Venn diagram showing MOMTiP-1-specific proteins identified after filtering the dataset. (C-F) Subcellular localization of selected C-terminally HA-tagged novel hypothetical proteins by IFA (green). Nuclear DNA is stained with DAPI (blue). Insets: DIC images. (G) Preliminary interactome of GlTom40 and MOMTiP-1 showing validated hits. Bait proteins (orange spheres), matrix proteins (purple), previously identified and localized proteins (black), and localized hypothetical proteins (blue). The stringency parameters used for detection (high, medium, and relaxed) are represented by bold, dashed, and dotted arrows, respectively. (H) Venn diagram showing the intersection of GlTom40 and MOMTiP-1datasets.
Co-IP with an HA-tagged variant of MOMTiP-1 yielded a large dataset of 221 exclusive hits (Fig 5B) which included GlTom40 detected at high stringency parameters, thereby confirming the strong interaction between GlTom40 and MOMTiP-1. The 221 MOMTiP-1 co-IP specific hits and an additional 20 enriched candidates were parsed according to different metabolic and/or functional categories (Fig 5B). In addition to GlTom40, the dataset contained several known mitosomal proteins, including matrix proteins HSP70 and GiOR1, cysteine desulfurase (IscS; Gl50803_14519), Cpn60, [2Fe-2S] ferredoxin (Gl50803_27266) and NifU-like protein, along with all 8 hypothetical proteins previously identified in the GlTom40 co-IP dataset and 4 additional non-annotated candidate mitosome proteins (Fig 5C–5F). Similarly to MOMTiP-1, one of these (Gl50803_17276) is also predicted to carry a TMD close to its N-terminus. Furthermore, this dataset contained two axoneme-associated GASP-180 proteins (Gl50803_137716 and Gl50803_16745) [67] detected with high stringency parameters, in line with association of the CMC to basal bodies.
Taken together, a first reverse co-IP analysis using the single-pass transmembrane MOMTiP-1 provided robust validation of the experimental approach used to identify mitosome membrane proteins, and has expanded the predicted mitosomal membrane and import machinery interactome to 22 proteins (Fig 5G).
Reverse co-IP using MOMTiP-1 as bait demonstrated that this protein and GlTom40 are strong interaction partners. We analyzed the intersection of their respective datasets to identify common candidate interaction partners and identified 27 proteins with high reliability (Fig 5H), 10 of which localized to mitosomes (Fig 4). Given MOMTiP-1’s predicted topology, strong interaction with GlTom40 and the interactome overlap, we postulated that MOMTiP-1 and GlTom40 exist in a core complex mostly likely involved in protein translocation across the outer mitosomal membrane. To characterize other components of this core interactome of the outer mitosomal membrane and to move beyond individual complexes to explore the boundaries of the growing protein interactome network (Fig 5G), we performed a series of additional reverse co-IP experiments using HA-tagged Qb-SNARE 4 (MOMTiP-7), GlIscS, protein Gl50803_9296 (MOMTiP-4) and protein Gl50803_14939 (MOMTiP-3) as baits. MOMTiP-7 (Qb-SNARE 4), MOMTiP-4 and MOMTiP-3 were chosen because they were identified either exclusively or in both the GlTom40- and MOMTiP-1 co-IP datasets, suggesting they may reside in the mitosomal outer membrane and could thus serve as tools for a lateral and outward expansion of this compartment’s interactome. On the other hand, GIIscS was chosen to extend the mitosomal proteome inwards towards the organellar matrix. Correct mitosomal localization for all 4 HA-tagged variants had been previously confirmed by IFA (Fig 4 and S1C–S1F Fig).
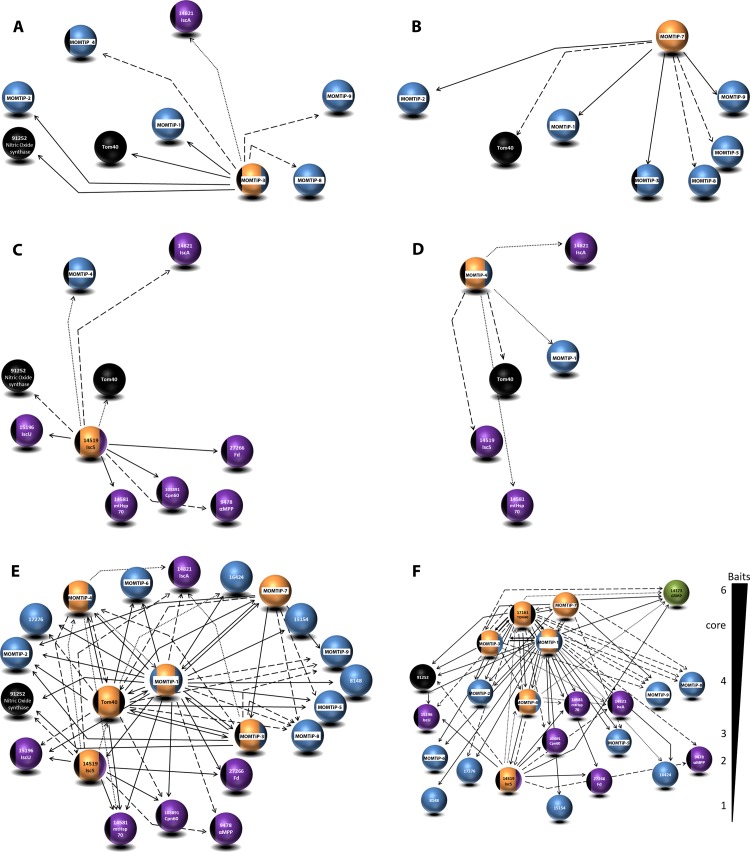
Expansion of the mitosomal membrane interactome network beyond and within the outer mitosomal membrane with MOMTiP-3, MOMTiP-4 and MOMTiP-7
MOMTiP-3, also known as MOMP35 [49], is predicted to contain 2 TMDs and was analyzed because of its potential role as a component of the import complex [49]. Co-IP using HA-tagged MOMTiP-3 as bait yielded 93 bait-specific candidate interactors. Both GlTom40 and G. lamblia oxidoreductase 1 (GiOR1) were detected in this dataset, in addition to several previously identified hypothetical mitosomal proteins (e.g. Gl50803_29147, Gl50803_10971 and Gl50803_7188) (Fig 6A), suggesting that MOMTiP-3 is a significant interacting partner of GlTom40 and MOMTiP-1.
Fig 6. Expansion of the core interactome beyond the outer mitosomal membrane with co-IPs of HA-tagged MOMTiP-7, MOMTiP-4, MOMTiP-3 and GlIscS.
Epitope (HA)-tagged (A) MOMTiP-3, (B) MOMTiP-7, (C) GlIscS and (D) MOMTiP-4 –derived interactomes. (E, F) Alternative depictions of the cumulative interactome of proteins localizing to mitosomes generated with 6 bait proteins. Note the tight association of GlTom40 with MOMTiP-1 and MOMTiP-3. (F) A socio-affinity depiction of protein-protein interactions derived from all co-IP experiments. Proteins are grouped according to how many baits they were significantly associated with. GlDRP (green sphere) is pulled down with all 6 bait proteins used in the co-IP assay at different stringency parameters. Bait proteins (orange spheres), matrix proteins (purple), previously identified and localized proteins (black), and localized hypothetical proteins (blue). The stringency parameters used for detection (high, medium, and relaxed) are represented by bold, dashed, and dotted arrows, respectively.
Consistent with its prediction as a SNARE protein, MOMTiP-7 presents dual localization, (mitosome and ER; Fig 4H and S1C Fig) and was identified as a strong interactor in both the GlTom40 and MOMTiP-1 co-IP datasets. We postulated that MOMTiP-7 may have a role in inter-organelle communication between mitosomes and the ER, possibly in protein/lipid transport [68,69]. We reasoned that identifying interaction partners could shed light on the nature of physical contacts between mitosomes and other membrane-bounded compartments. Analysis of co-IP using HA-tagged MOMTiP-7 as bait yielded 157 bait-specific proteins. Interestingly, the only 2 proteins in the dataset with high spectral counts were the bait itself and MOMTiP-3. Several non-annotated proteins, e.g. MOMTiP-2, MOMTiP-1, MOMTiP-9, GlTom40 as well as Type III DnaJ protein Gl50803_9751 were detected with relaxed stringency parameters.
MOMTiP-4 is a predicted soluble Giardia-specific protein of unknown function with a mitochondrial targeting signal. In line with this, HA-tagged MOMTiP-4 localized exclusively to mitosomes (Fig 4E and S1E Fig). MS data analysis performed with high stringency parameters of a MOMTiP-4 co-IP experiment (95_2_95; FDR 0%) yielded only 12 bait-specific hits, none of them known mitosome proteins. The bait protein itself was by far the most significant hit in the dataset. Analysis with more relaxed stringency parameters (90_1_90; FDR 6.2%) yielded 47 bait-specific identifications which included matrix proteins GlIscS, GlIscA, GlHsp70, and also GlTom40. The MOMTiP-4 co-IP dataset suggests that despite its clear-cut localization and considerable expression levels judged by the signal obtained in Fig 4E, this mitosomal protein has a limited interactome enriched mostly in matrix proteins. MOMTiP-1was identified only at very low stringency (20_1_20; FDR 51%) in this dataset, suggesting that MOMTiP-4 and MOMTiP-1 are not direct interactors but could be connected via bridging proteins. All identified MOMTiP-4-interacting proteins are depicted in Fig 6D.
Expansion of the the GlTom40 interactome towards the matrix with GlIscS
GlIscS is a mitosomal matrix protein and the central component of the Fe-S assembly machinery [70]. All mitosomal matrix proteins including GlIscS are translated in the cytoplasm and reach their final destination after unfolding and translocation across the mitosome double membrane. Thus, this trafficking route (cytoplasm–translocon–matrix) should be reflected in the protein-protein interactions of a co-IP dataset with GlIscS-HA as bait. Co-IP of HA-tagged GlIscS yielded 177 bait-specific protein hits. Among these, we identified all 5 known matrix proteins namely, NifU-like protein, HSP70, [2Fe-2S] ferredoxin, Cpn60, and GiOR1. GlTom40 as the sentinel protein for the outer membrane translocon was detected with relaxed stringency (50_1_50, FDR of 30%). Seventy out of 177 hits were enriched in the GlIscS-specific dataset, with ≥ 5 peptide counts. Eighteen of those (25%) belong to the Protein 21.1 family. The biological function of this protein family in G. lamblia [71] and the significance of its association to GlIscS is unknown.
In summary, we have generated an extensive mitosome-centered protein interaction network (Fig 6E) from 6 independent co-IP assays using epitope-tagged GlTom40 and 5 interaction partners (MOMTiP-1, MOMTiP-3, MOMTiP-7, GlIscS and MOMTiP-4) as baits based on i) spectral counts with high stringency parameters in the GlTom40 co-IP dataset and ii) confirmed localization of epitope-tagged variants to mitosomes by IFA. All 24 localized mitosome proteins (previously known and newly identified hypotheticals) were parsed according to molecular function and biological process (S3 Fig) using Blast2go (https://www.blast2go.com/). Metal ion, Fe-S, ATP, and protein binding were the major molecular functions associated with these proteins. Interestingly, other biological processes involving response to lipid and transmembrane transport were also identified with significant p-values. An additional 93 candidates annotated as hypothetical proteins (from all the 6 co-IP assays) were analyzed using Blast2go (S4 Fig). Binding and catalytic activities were the 2 major GO terms associated to this group. Interestingly, GlDRP was strongly overrepresented in 3 high-stringency co-IP datasets where mitosome membrane proteins were specifically used as bait. Moreover, with relaxed stringency parameters GlDRP was detected in all 6 co-IP datasets (Fig 6F), indicating that GlDRP is associated to mitosomes. These data are clearly in line with our previous observations concerning the perturbation of mitosome morphogenesis by mutant GlDRP (Fig 2D–2F). In Table 2, we have combined our data with data reported in [33,49] for a state-of-the-art overview of the main confirmed interactions within the mitosome-centered protein interactome.
Table 2. Detected and validated mitosomal proteins in this report and in [33,49,72].
| Assigned annotation on www.giardiaDB.org and/or ORF number | Detection of annotated and novel mitosomal proteins in this work, co-IPed with epitope-tagged | Detection of annotated and novel mitosomal proteins in Martincova et.al 2015 using BirA fusions to | Detection of annotated and novel mitosomal proteins in | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tom40 | MOMTiP-1 (GL50803_ 29147) | MOMTiP-3 (GL50803_ 14939) | MOMTiP-5 (GL50803_ 5785) | MOMTiP-4 (GL50803_9296) | GlIscS | Pam18 | Tim44 | Hsp70 | Tom40 | Gl14939 (GiMOMP35) | Jedelsky et. al, 2011 | Martincova et. al, 2105 | This work | |
| Tom40 (GL50803_17161) | ||||||||||||||
| GL50803_14939 (GiMOMP35) | ||||||||||||||
| Pam 18 (GL50803_300001) | ||||||||||||||
| Pam 16 (GL50803_19230) | ||||||||||||||
| Tim44 (GL50803_14845) | ||||||||||||||
| mtHSP70 (GL50803_14581) | ||||||||||||||
| Mge1 (GL50803_1376) | ||||||||||||||
| Isc S (GL50803_14519) | ||||||||||||||
| IscU (GL50803_15196) | ||||||||||||||
| IscA (GL50803_14821) | ||||||||||||||
| DnaJ (GL50803_17030) | ||||||||||||||
| DnaJ (GL50803_9751) | ||||||||||||||
| αMPP (GL50803_9478) | ||||||||||||||
| GiOR-1 (GL50803_91252) | ||||||||||||||
| Glutaredoxin5 (GL50803_2013) | ||||||||||||||
| Ferredoxin (GL50803_27266) | ||||||||||||||
| Cpn60 (GL50803_103891) | ||||||||||||||
| Cpn10 (GL50803_29500) | ||||||||||||||
| GrpE (GL50803_1376) | ||||||||||||||
| GL50803_29147 | ||||||||||||||
| GL50803_9296 | ||||||||||||||
| GL50803_10971 | ||||||||||||||
| GL50803_5785 | ||||||||||||||
| GL50803_7188 | ||||||||||||||
| GL50803_9503 | ||||||||||||||
| GL50803_21943 | ||||||||||||||
| GL50803_22587 | ||||||||||||||
| GL50803_17276 | ||||||||||||||
| GL50803_16424 | ||||||||||||||
| GL50803_8148 | ||||||||||||||
| GL50803_15154 | ||||||||||||||
The presence (in light green) of any given protein was based on stringency thresholds specific to each report. From darker to increasingly lighter shades of grey, proteins are putatively grouped in outer membrane, inner membrane, matrix and novel mitosomal components.
A pharmacologically-induced mitosome matrix-targeted DHFR complex inhibits processing of an imported endogenous reporter in mitosomes
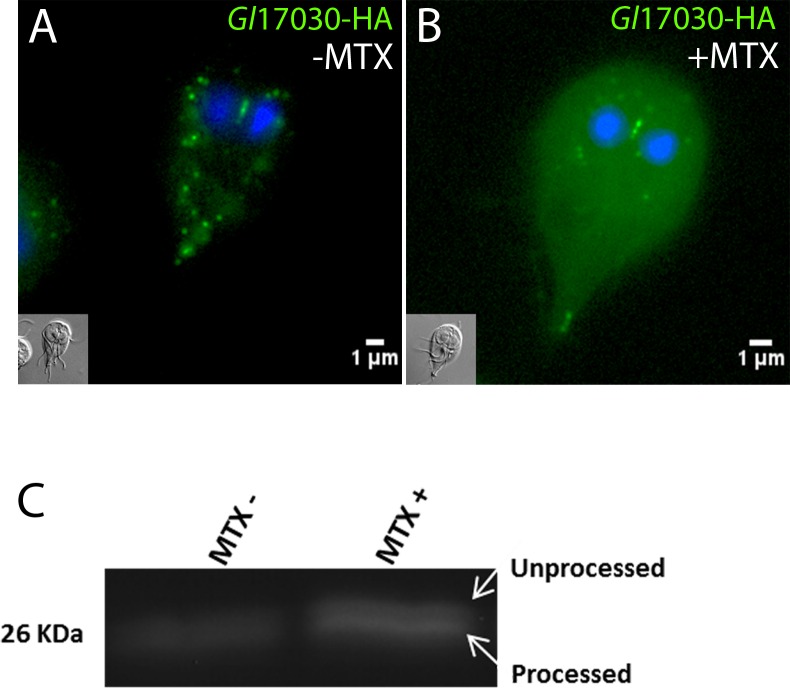
Evidence from extensive primary and reverse co-IP data combined with IFA analysis led us to postulate that GlTom40, MOMTiP-1 and MOMTiP-3 exist in an outer membrane core complex, likely involved in protein import. We probed the functional conservation of mitosomal import across the GlTom40 translocon with respect to the corresponding process in bona fide mitochondria by adapting the DHFR-folate analogue system [49,73] to G. lamblia. Pre-sequence directed DHFR is a classical substrate used in protein translocation studies due to its ability to fold irreversibly upon binding a folate analog, e.g. MTX. Complexed with MTX, DHFR becomes unsuitable as a substrate for import and blocks translocons, which results in a general blockage of organelle protein import [73]. Transfection of MTSfdΔint-DHFR into a Gl17030-HA background, i.e. a transgenic line expressing an HA-tagged MTS-directed mitosomal reporter, allowed testing of the general effects of MTX-induced import block. We reasoned that the presence of MTX in MTSfdΔint-DHFR expressing cells could lead to an import block due to jamming of the translocase. Localization of the reporter by IFA showed an increased cytosolic Gl17030-HA signal after addition of 1 μM MTX (Fig 7B) compared to parasites exposed to the solvent alone (Fig 7A). This suggested accumulation of the reporter in the cytosol in cells exposed to MTX as a result of a generalized import block. To test this we measured the ratio of the slightly larger Gl17030-HA reporter precursor protein and the imported and therefore processed form without the MTS by SDS-PAGE and Western blot using anti-HA antibodies. Consistent with the IFA data, unprocessed Gl17030-HA was strongly increased in the drug treated sample, whilst only the processed form was present in untreated controls (Fig 7C). Taken together the data support functional conservation of the highly diverged protein import machinery in G. lamblia mitosomes.
Fig 7. MTX treatment of cells expressing mitosome-targeted DHFR affects processing of a mitosomal matrix reporter.
Subcellular distribution of a matrix-targeted reporter (Gl17030-HA) without MTX (A) or after addition of 1μM MTX for 24 h (B) in transgenic cells expressing mitosome-targeted DHFR. Note the accumulation of HA-signal in the cytoplasm. Nuclear DNA is stained with DAPI (blue). Insets: DIC images. (C) Immunoblot analysis detects accumulation of unprocessed Gl17030-HA in the presence of MTX.
Discussion
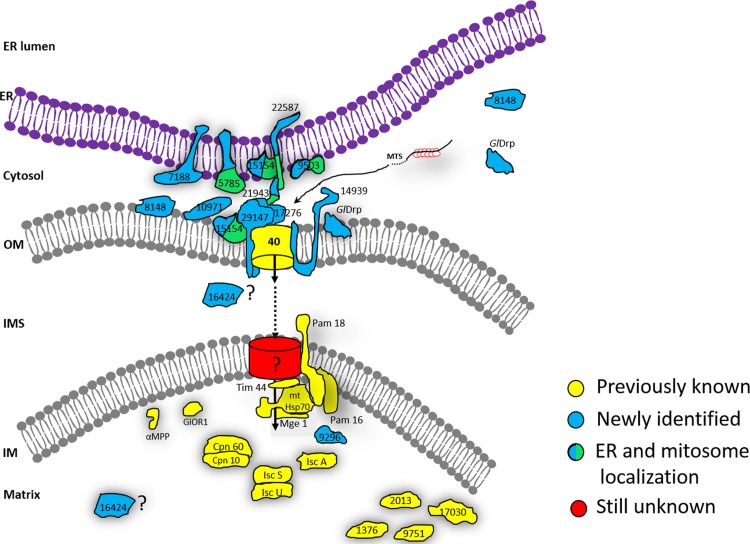
G. lamblia mitosomes remain the smallest known and least characterized MROs. Identification of protein components using shotgun proteomic analysis of enriched mitosome preparations has proven challenging primarily due to difficulties in isolating sufficient amounts of contaminant-free organelles [33,48]. Extensive sequence divergence prevents identification of organelle proteins via homology-based searches; a case in point is GlTOM40 whose sequence degeneration is so extensive that the identification of orthologues in Giardia, Entamoeba or Spironucleus remains tentative despite the constraints imposed by the beta barrel structure of these mitochondrial porins [44]. The function of candidate factors identified by other means and localized to organelle membranes usually cannot be deduced based on existing structural information from well-characterized mitochondrial homologs. A notable exception to this is a recently identified highly-diverged but structurally-conserved GlTim44 homologue [49]. Taken together, these challenges have frustrated attempts at systematizing intra- and inter- organelle mitosome-centered interactions, thereby limiting analysis to isolated complexes [49,50]. For example attempts at analyzing isolated GlTom40-containing protein complexes (presumably enriched mitosomal outer membrane translocons) using blue-native PAGE [50] detected no mitosomal proteins aside from GlTom40 and an unidentified 32kDa protein which could not be mapped to any known Giardia sequence. These data demonstrate how challenging it is to define novel GlTom40-interacting partners, probably due to the translocon complex being embedded in the outer membrane of the organelle. Here, we used epitope-tagged GlTOM40 [23,33] as first bait, and implemented an iterative co-immunoprecipitation approach to expand the mitosomal membrane interactome network beyond the few known components. With only 5 more bait proteins, this strategy allowed for building of a core membrane interactome and a complex interactome network extending inwards to the organelle matrix as well as outwards to components of the ER membrane, the axoneme cytoskeleton and the cytoplasm. The rationale is that with sufficient numbers of targeted reverse co-IP experiments using validated organelle proteins as baits, a comprehensive proteome interactome could be built, thereby achieving a systems-biological view of the giardial mitosome proteome. From a technical point of view, building an interactome network using a forward- and reverse co-IP approach allows isolation of “true” mutual interactions by validation in two completely independent co-IP experiments. Specifically, the 108 MS hits detected in the Tom40 co-IP dataset which include numerous non-specific interactions can be filtered with data from reverse co-IP assays to reveal actual protein-protein interactions (depicted in Fig 6E) that can be unambiguously distinguished from false-positive hits. Combined with imaging data and predicted topology this provides a robust platform to construct an integrated working model of all mitosome-associated protein interactome networks known to date (Fig 8). Blast2Go in silico enrichment analyses suggest that mitosomes may have a role beyond Fe-S protein maturation (S3 and S4 Figs) Only recently the major function of E. histolytica mitosomes was shown to be sulfate activation, and not Fe-S protein maturation as previously thought [44]. Although genes involved in this pathway are missing in other MRO-containing organisms such as G. lamblia, T. vaginalis, and C. parvum, the Entamoeba example points to a wider range of functions ascribable to mitosomes. This may even include general functions in stage-differentiation as recently shown in E. histolytica whose mitosomes are essential for the encystation process [74].
Fig 8. An integrated model for mitosome interactome networks.
Schematic representation of all proteins identified via protein-protein interaction data through serial coIP assays using 6 different mitosomal bait proteins are shown in a model. Previously identified/ known mitosomal proteins are depicted in yellow. Newly identified mitosome localized hypothetical proteins are shown in blue. Proteins with dual localization (mitosomes and ER) are shown in blue/green. As yet un-identified pore (translocase) in the inner membrane is shown in red. Positioning of these identified proteins on the model is based on in silico data (presence/ absence of (1) mitochondrial targeting sequence, (2) transmembrane domain), localization data and socio- affinity interaction of these proteins with their respective and other bait proteins.
GlTom40 and interaction partners MOMTiP-1 and MOMTiP-3: a minimized mitosome protein import apparatus?
Following its identification as a prominent GlTom40 interaction partner, the single pass membrane protein MOMTiP-1 was the first bait protein selected for reverse co-IP to expand the GlTom40 interactome. MOMTiP-1 as bait pulled down GlTom40 with the most abundant peptide counts. GFP-tagging and detection of MOMTiP-1::GFP on mitosomes suggests a membrane topology in line with characterized mitochondrial receptor proteins such as Tom20 [75]. Further support for MOMTiP-1’s membrane topology may derive from a definition of membrane orientation using alternative methods such as in situ proximity ligation or protease protection assays. The latter approach has proven useful in the determination of membrane topology for other mitosomal candidate proteins in Giardia [49]. The identification of MOMTiP-1 provides an exciting lead; however, a detailed functional characterization of this protein is required to provide independent evidence for the exact nature of this interaction and to test the hypothesis that MOMTiP-1 is a component of the GlTom40 complex with a receptor function. So far 20 proteins have been validated by localization to mitosomes, allowing for a significant expansion of the GlTom40/MOMTiP-1 interactome. This protein’s predicted topology combined with its exclusive mitosomal localization and the size and composition of its interactome, supports MOMTiP-1 as a GlTom40 accessory protein with a potential receptor function for protein import. To test this hypothesis, we engineered a truncated HA-tagged version of MOMTiP-1 consisting only of the predicted C-terminal domain (residues 31–133; C-MOMTiP-1). Ectopic expression of C-MOMTiP-1 showed a distinct cytosolic localization by IFA (S5A Fig). Native co-IP of C-MOMTiP-1 and analysis of the bait-specific dataset with medium stringency parameters (95_1_95) identified only 2 mitosomal proteins (Gl50803_16424 and MOMTiP-8) (S5B Fig). These data show that the soluble cytoplasmic MOMTiP-1 variant does not recapitulate the interaction properties of the full-length membrane-anchored protein, suggesting that capture of imported matrix proteins may require incorporation of the putative receptor domain into a fully-assembled TOM complex, complete with ancillary factors.
MOMTiP-3 was exclusively identified in the GlTom40 and the MOMTiP-1 co-IP datasets, suggesting that these 3 proteins may function in a tightly-knit complex, likely involved in protein import across the outer mitosomal membrane. TMHMM predicts two TMDs at MOMTiP-3’s N-terminus, followed by a large C-terminal domain. Powerful HMMER-based searches across several eukaryotic lineages, including the closely related diplomonad Spironucleus salmonicida [76], yielded no orthologues for MOMTiP-3, neither was there any predicted functional information available. Nevertheless, analysis of protease protection assays for this protein showed that MOMTiP-3 localizes at the outer mitosome membrane with its C- terminus in the cytosol [49]. These data, in combination with data on MOMTiP-1 predicted topology and interactomes developed herein, support a model for GlTom40, MOMTiP-1, and MOMTiP-3 for a minimized mitosomal import apparatus whose core import machinery is composed of only these 3 proteins. The dramatic perturbation of mitosomal homeostasis observed when either MOMTiP-1 (this work) or MOMTiP-3 [49] were constitutively overexpressed supports the hypothesis for their belonging to the same complex. Protein translocation across the outer mitosomal membrane through this highly reduced import apparatus would be conserved in its mechanism, given that MTX-induced complexing of mitosome-targeted DHFR caused accumulation of unprocessed i.e. untranslocated mitosome reporters (Fig 7C, [49]). Incidentally, these data also confirm that mitosome membrane translocation requires pre-proteins to remain in an unfolded state [49].
Mitosome-ER contact sites
Co-IP data combined with imaging of tagged variants identified 6 proteins with dual localization at mitosomes and ER (Fig 8). Contact between these organelles would serve at least two major functions, i.e. replication of mitosomes and transport associated to lipid biosynthesis. Thus far, we have identified five mitosome proteins with dual localization potentially involved in inter-organelle communication (Fig 4F). One of them is a transmembrane Qb-SNARE 4 (MOMTiP-7) [65] identified in GlTom40 and MOMTiP-1 co-IP datasets.
For their biogenesis, mitochondria and MROs rely on lipid transfer from the ER, the central site for phospholipid synthesis in the cell [77,78]. SNAREs are best known for mediating membrane fusion in vesicular transport [79] whereas in the context of mitochondria and the ER, they function as components of so called ER-mitochondria encounter structures (ERMES). In addition to being associated to mitochondrial protein import [80,81], ERMES fulfills an essential function in inter-organelle lipid transport [80]. Phosphatidylserine is shuttled from the ER to mitochondria through the ERMES complex where it is converted to phosphatidylethanolamine (PE) by a decarboxylation reaction that generates most if not all PE in mitochondria [80,82]. Unlike in the hydrogenosome-containing T. vaginalis [83], ERMES homologs have not been identified in G. lamblia, possibly due to extensive sequence divergence. Thus, whether this function is preserved in Giardia mitosomes is not known however, organelle biogenesis would necessarily depend on ER-derived lipids which are transported to mitosomes either by carrier proteins or via membrane contact sites. The latter requires tethering complexes to facilitate phospholipid exchange between the two organelles. Given that MOMTiP-7 is predicted to be a SNARE, we explored the idea that this protein is part of a larger complex mediating ER-mitosome interaction. Co-IP of MOMTiP-7 specifically detected, in addition to outer membrane proteins such as GlTom40, MOMTiP-1 and MOMTiP-3, 3 hypothetical proteins, two of which, MOMTiP-8 and MOMTiP-5 (both predicted soluble proteins), localized both to the ER and to mitosomes. In addition, a domain in MOMTiP-8 has similarity to a yeast “Maintenance of mitochondrial morphology” protein 1 (Mmm1) of the ERMES complex. Moreover, HHpred analysis revealed a link between MOMTiP-5 and a beta barrel lipid binding protein MLN64 (e-value 0.0006) in H. sapiens which facilitates cholesterol transport to mitochondria [84]. These preliminary data support the existence of an outer mitosomal membrane-associated complex in G. lamblia mitosomes possibly involved in generating ER—mitosome membrane contact sites (Fig 8).
Mitosome dynamics and a novel role for MOMTiP-1 and GlDRP in mitosome homeostasis
We had previously shown that replication and inheritance of the CMC is coordinated in a cell cycle-dependent manner, whereas PMs divided stochastically [23]. The lack of a system to track organelles in living trophozoites precluded addressing the question directly whether mitosomes were motile and constituted a dynamic network of organelles with measurable exchange. Development of two GFP-tagged reporters GFP-GlTom40 and MOMTiP-1-GFP (this study) allowed for time-lapse experiments to follow individual organelles in a cell. However, we found no evidence for motility of organelles, neither in the CMC nor in PMs, even after prolonged observation (1.5 h). This is consistent with the lack of motor proteins such as kinesins and dyneins in any of the mitosomal protein interactomes we generated. Moreover, FRAP experiments revealed no exchange of GFP-tagged membrane proteins between organelles during the period of observation (Fig 7F and 7G), which further corroborated the relative isolation of mitosomes within the cytosol. The lack of mitosomal motility and contact complicates investigation of their replication and morphogenesis. The two most plausible scenarios for this are currently the following: i) PMs are released from the CMC, which continuously produces new organelles by elongation and fission to maintain a constant number of organelles in a cell-cycle independent manner; ii) PMs and the CMC organelles replicate independently in a cell-cycle independent and -dependent manner, respectively [23]. Although time-lapse microscopy experiments did not provide evidence in support of either scenario, conditional expression of a dominant-negative, constitutively active GlDRP-K43E revealed a distinct morphogenesis phenotype (see also below) indicative of an organelle replication defect. As one of the key players in the regulation of mitochondrial fission, DRPs are mechano-enzymes conserved from yeast to vertebrates [85,86,87,88]. G. lamblia harbors a single dynamin homologue GlDRP shown to play a major role in this parasite’s endocytic pathway and stage conversion [56,89,90]. Transgenic parasites expressing the GlDRP-K43E variant exhibited larger and fewer mitosomes, compared to cells expressing the wild type GlDRP variant(Fig 6). This is in line with the dominant-negative effect on mitochondrial fission elicited by the corresponding mutation in DRPs in other organisms. To our knowledge, this is the first report on the involvement of GlDRP in mitosome homeostasis, supporting the (at least partial) functional conservation of mitochondrial and MRO fission [91,92,93,94]. The notion that G. lamblia mitosome fission is functionally conserved is further substantiated by the identification of MOMTiP-6 which presents dual localization to mitosomes and the ER. HMMER-based predictions relate MOMTiP-6 to human mitochondrial fission protein (Fis1, e-value 6.3E-05) which participates in the recruitment of dynamin-related protein 1 (Drp1) to the mitochondrial surface for organelle fission [95,96]. The distinctive “string” mitosome phenotype in cells expressing MOMTiP-1-GFP clearly demonstrated that mitosomes can assume an elongated, tubular morphology, which is a prerequisite for organelle division and replication. The implication is that G. lamblia mitosomes retain at least the machinery for fission in which the mechano-enzyme GlDRP and outer mitosomal membrane elements such as MOMTiP-1 and 3 [49] play central roles.
Conclusion
We used an iterative approach based on co-IP experiments to generate a GlTom40-centered interactome network. Ultimately this strategy should allow building a combined proteome, which delineates the full complement of organelle proteins, peripherally associated factors, as well as interfaces with the ER and the cytoskeleton. Although this strategy requires numerous rounds of sequential co-IP and validation, it is highly informative because it produces interaction data in addition to identifying novel organelle proteins. Combined with testing of epitope-tagged variants of candidate proteins for organelle localization as a straightforward validation criterion, serial co-IPs allow for unambiguous definition of the organelle-specific proteome, as well as interfaces with other cellular structures. This strategy also led to the discovery of MOMTiP-1, a strong GlTom40 interaction partner which plays a role in mitosomal morphogenesis. Together with GlDRP (this work) and MOMTiP-3 (MOMP35; [49]), these are the only proteins so far known to affect mitosomal homeostasis in G. lamblia.
Supporting Information
(XLSX)
(XLSX)
Immunofluorescence co-labelling and wide-field microscopy analysis of transgenic G. lamblia lines expressing (A) GlTom40, (B) MOMTiP-1, (C) MOMTiP-7, (D) GlIscS, (E) MOMTiP-4 and (F) MOMTiP-3, all used as HA-tagged baits in co-IP experiments (upper row, in green), in combination with the endogenous mitosomal marker GlIscU (middle row, in red). Nuclei were stained with DAPI (lower row, in blue). The central mitosome complex was clearly labelled by both fluorophores in all lines. Scale bar: 1μm.
(TIF)
With increasing concentrations of formaldehyde (0–4.5%), immuno-detection (Western blot) of the Tom40-HA reporter shows a shift from the monomeric form to higher molecular weight complexes,. Molecular size (kDa) marker (M) bands are indicated on the left axis. A concentration of 2.25% formaldehyde (arrowhead) was later adopted for all subsequent forward and reverse co-IPs.
(TIF)
(A) Data distribution pie chart for protein hits with either Blast2Go annotation (“B2G annotated”), or associated to gene ontology (GO) terms (“with GO mapping”) or to Blast annotation data (“with Blast hits”). (B) The top 20 GO terms distributed in the three root categories for “biological process” (BP), “molecular function” (MF) and “cellular component” (CC). (C) A direct count of GO terms associated to MF. (D) A direct count of GO terms associated to BP.
(TIF)
(A) Data distribution pie chart for protein hits with either Blast2Go annotation (“B2G annotated”), or associated to gene ontology (GO) terms (“with GO mapping”) or to Blast annotation data (“with Blast hits”). (B) The top 20 GO terms distributed in the three root categories for “biological process” (BP), “molecular function” (MF) and “cellular component” (CC). (C) A direct count of GO terms associated to MF. (D) A direct count of GO terms associated to BP.
(TIF)
(A) IFA and wide-field microscopy analysis of transgenic Giardia cells expressing C-MOMTiP-1 (in green), a truncated HA-tagged version of MOMTiP-1 consisting only of the predicted C-terminal domain (residues 31–133). C-MOMTiP-1 accumulates primarily in the cytosol. Nuclei are labelled with DAPI (in blue). Inset: DIC image. Scale bar: 1μm. (B) Venn diagram depicting the overlap of datasets derived from native co-IP of C-MOMTiP-1 and control WB cells.
(TIF)
Acknowledgments
We thank Therese Michel for technical assistance. Dr. Peter Hunziker and his team at the proteomics service facility of the Functional Genomics Center-Zürich are gratefully acknowledged for their support in tandem mass spectrometry performance and analysis. Prof. Robert Sinden at Imperial College, London (UK) is gratefully acknowledged for providing the human DHFR-encoding plasmid. Dr. Chandra Ramakrishnan is gratefully acknowledged for critical revision of the manuscript.
Data Availability
All co-IP datasets referenced in the manuscript were deposited to the PRIDE platform with the identifier PXD004583 where they can be accessed via ProteomeXchange.
Funding Statement
This work was partially supported by grant 31-140803/1, awarded to ABH by the Swiss National Science Fund (www.snf.ch). JPZ received grant 55080507 and CF received grant K-52201-06-01, all as fellowships from the Forschungskredit der Universität Zürich (www.researchers.uzh.ch). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gray MW, Burger G, Lang BF (1999) Mitochondrial evolution. Science 283: 1476–1481. [DOI] [PubMed] [Google Scholar]
- 2. Hedges SB, Blair JE, Venturi ML, Shoe JL (2004) A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol 4: 2 10.1186/1471-2148-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR (1985) Mitochondrial origins. Proc Natl Acad Sci U S A 82: 4443–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olsen GJ, Woese CR, Overbeek R (1994) The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol 176: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Viale AM, Arakaki AK (1994) The chaperone connection to the origins of the eukaryotic organelles. FEBS Lett 341: 146–151. [DOI] [PubMed] [Google Scholar]
- 6. Karnkowska A, Vacek V, Zubacova Z, Treitli SC, Petrzelkova R, et al. (2016) A Eukaryote without a Mitochondrial Organelle. Current biology: CB 26: 1274–1284. 10.1016/j.cub.2016.03.053 [DOI] [PubMed] [Google Scholar]
- 7. Bauer MF, Hofmann S, Neupert W, Brunner M (2000) Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol 10: 25–31. [DOI] [PubMed] [Google Scholar]
- 8. Jensen RE, Johnson AE (2001) Opening the door to mitochondrial protein import. Nat Struct Biol 8: 1008–1010. 10.1038/nsb1201-1008 [DOI] [PubMed] [Google Scholar]
- 9. Koehler CM, Merchant S, Schatz G (1999) How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem Sci 24: 428–432. [DOI] [PubMed] [Google Scholar]
- 10. Matouschek A, Pfanner N, Voos W (2000) Protein unfolding by mitochondria. The Hsp70 import motor. EMBO Rep 1: 404–410. 10.1093/embo-reports/kvd093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prokisch H, Scharfe C, Camp DG 2nd, Xiao W, David L, et al. (2004) Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol 2: e160 10.1371/journal.pbio.0020160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5: 1543–1554. 10.1021/pr050477f [DOI] [PubMed] [Google Scholar]
- 13. Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, et al. (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A 100: 13207–13212. 10.1073/pnas.2135385100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pfanner N, Truscott KN (2002) Powering mitochondrial protein import. Nat Struct Biol 9: 234–236. 10.1038/nsb0402-234 [DOI] [PubMed] [Google Scholar]
- 15. Gabaldon T, Huynen MA (2004) Shaping the mitochondrial proteome. Biochim Biophys Acta 1659: 212–220. 10.1016/j.bbabio.2004.07.011 [DOI] [PubMed] [Google Scholar]
- 16. Cerkasovová A, Lukasová G, Cerkasòv J, J K (1973) Biochemical characterization of large granule fraction of Tritrichomonas foetus (strain KV1). Journal of Protozoology 20. [Google Scholar]
- 17. Lindmark DG, Muller M (1973) Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J Biol Chem 248: 7724–7728. [PubMed] [Google Scholar]
- 18. Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, et al. (1999) Hsp60 is targeted to a cryptic mitochondrion-derived organelle ("crypton") in the microaerophilic protozoan parasite Entamoeba histolytica. Mol Cell Biol 19: 2198–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muller M (1993) The hydrogenosome. J Gen Microbiol 139: 2879–2889. 10.1099/00221287-139-12-2879 [DOI] [PubMed] [Google Scholar]
- 20. Tovar J, Fischer A, Clark CG (1999) The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol 32: 1013–1021. [DOI] [PubMed] [Google Scholar]
- 21. Shiflett AM, Johnson PJ (2010) Mitochondrion-related organelles in eukaryotic protists. Annu Rev Microbiol 64: 409–429. 10.1146/annurev.micro.62.081307.162826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, et al. (2012) The revised classification of eukaryotes. J Eukaryot Microbiol 59: 429–493. 10.1111/j.1550-7408.2012.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Regoes A, Zourmpanou D, Leon-Avila G, van der Giezen M, Tovar J, et al. (2005) Protein import, replication, and inheritance of a vestigial mitochondrion. J Biol Chem 280: 30557–30563. 10.1074/jbc.M500787200 [DOI] [PubMed] [Google Scholar]
- 24. Tovar J, Leon-Avila G, Sanchez LB, Sutak R, Tachezy J, et al. (2003) Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature 426: 172–176. 10.1038/nature01945 [DOI] [PubMed] [Google Scholar]
- 25. Riordan CE, Ault JG, Langreth SG, Keithly JS (2003) Cryptosporidium parvum Cpn60 targets a relict organelle. Curr Genet 44: 138–147. 10.1007/s00294-003-0432-1 [DOI] [PubMed] [Google Scholar]
- 26. Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, et al. (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414: 450–453. 10.1038/35106579 [DOI] [PubMed] [Google Scholar]
- 27. Jerlstrom-Hultqvist J, Einarsson E, Xu F, Hjort K, Ek B, et al. (2013) Hydrogenosomes in the diplomonad Spironucleus salmonicida. Nature communications 4: 2493 10.1038/ncomms3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, et al. (2004) Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304: 441–445. 10.1126/science.1094786 [DOI] [PubMed] [Google Scholar]
- 29. Leon-Avila G, Tovar J (2004) Mitosomes of Entamoeba histolytica are abundant mitochondrion-related remnant organelles that lack a detectable organellar genome. Microbiology 150: 1245–1250. 10.1099/mic.0.26923-0 [DOI] [PubMed] [Google Scholar]
- 30. Turner G, Muller M (1983) Failure to detect extranuclear DNA in Trichomonas vaginalis and Tritrichomonas foetus. J Parasitol 69: 234–236. [PubMed] [Google Scholar]
- 31. van der Giezen M, Sjollema KA, Artz RR, Alkema W, Prins RA (1997) Hydrogenosomes in the anaerobic fungus Neocallimastix frontalis have a double membrane but lack an associated organelle genome. FEBS Lett 408: 147–150. [DOI] [PubMed] [Google Scholar]
- 32. Dolezal P, Smid O, Rada P, Zubacova Z, Bursac D, et al. (2005) Giardia mitosomes and trichomonad hydrogenosomes share a common mode of protein targeting. Proc Natl Acad Sci U S A 102: 10924–10929. 10.1073/pnas.0500349102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jedelsky PL, Dolezal P, Rada P, Pyrih J, Smid O, et al. (2011) The minimal proteome in the reduced mitochondrion of the parasitic protist Giardia intestinalis. PLoS One 6: e17285 10.1371/journal.pone.0017285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lill R (2009) Function and biogenesis of iron-sulphur proteins. Nature 460: 831–838. 10.1038/nature08301 [DOI] [PubMed] [Google Scholar]
- 35. Craig EA, Voisine C, Schilke B (1999) Mitochondrial iron metabolism in the yeast Saccharomyces cerevisiae. Biol Chem 380: 1167–1173. 10.1515/BC.1999.148 [DOI] [PubMed] [Google Scholar]
- 36. Lill R, Kispal G (2000) Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends in biochemical sciences 25: 352–356. [DOI] [PubMed] [Google Scholar]
- 37. Ankarklev J, Jerlstrom-Hultqvist J, Ringqvist E, Troell K, Svard SG (2010) Behind the smile: cell biology and disease mechanisms of Giardia species. Nat Rev Microbiol 8: 413–422. 10.1038/nrmicro2317 [DOI] [PubMed] [Google Scholar]
- 38. Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, et al. (2007) Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317: 1921–1926. 10.1126/science.1143837 [DOI] [PubMed] [Google Scholar]
- 39. Davis-Hayman SR, Nash TE (2002) Genetic manipulation of Giardia lamblia. Molecular and biochemical parasitology 122: 1–7. [DOI] [PubMed] [Google Scholar]
- 40. Boucher SE, Gillin FD (1990) Excystation of in vitro-derived Giardia lamblia cysts. Infection and immunity 58: 3516–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abodeely M, DuBois KN, Hehl A, Stefanic S, Sajid M, et al. (2009) A contiguous compartment functions as endoplasmic reticulum and endosome/lysosome in Giardia lamblia. Eukaryot Cell 8: 1665–1676. 10.1128/EC.00123-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stefanic S, Morf L, Kulangara C, Regos A, Sonda S, et al. (2009) Neogenesis and maturation of transient Golgi-like cisternae in a simple eukaryote. J Cell Sci 122: 2846–2856. 10.1242/jcs.049411 [DOI] [PubMed] [Google Scholar]
- 43. Goldberg AV, Molik S, Tsaousis AD, Neumann K, Kuhnke G, et al. (2008) Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature 452: 624–628. 10.1038/nature06606 [DOI] [PubMed] [Google Scholar]
- 44. Mi-ichi F, Abu Yousuf M, Nakada-Tsukui K, Nozaki T (2009) Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci U S A 106: 21731–21736. 10.1073/pnas.0907106106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Putignani L, Tait A, Smith HV, Horner D, Tovar J, et al. (2004) Characterization of a mitochondrion-like organelle in Cryptosporidium parvum. Parasitology 129: 1–18. [DOI] [PubMed] [Google Scholar]
- 46. Sanderson SJ, Xia D, Prieto H, Yates J, Heiges M, et al. (2008) Determining the protein repertoire of Cryptosporidium parvum sporozoites. Proteomics 8: 1398–1414. 10.1002/pmic.200700804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsaousis AD, Kunji ER, Goldberg AV, Lucocq JM, Hirt RP, et al. (2008) A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature 453: 553–556. 10.1038/nature06903 [DOI] [PubMed] [Google Scholar]
- 48. Wampfler PB, Tosevski V, Nanni P, Spycher C, Hehl AB (2014) Proteomics of secretory and endocytic organelles in Giardia lamblia. PLoS One 9: e94089 10.1371/journal.pone.0094089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martincova E, Voleman L, Pyrih J, Zarsky V, Vondrackova P, et al. (2015) Probing the biology of Giardia intestinalis mitosomes using in vivo enzymatic tagging. Mol Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dagley MJ, Dolezal P, Likic VA, Smid O, Purcell AW, et al. (2009) The protein import channel in the outer mitosomal membrane of Giardia intestinalis. Mol Biol Evol 26: 1941–1947. 10.1093/molbev/msp117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hehl AB, Marti M, Kohler P (2000) Stage-specific expression and targeting of cyst wall protein-green fluorescent protein chimeras in Giardia. Mol Biol Cell 11: 1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morf L, Spycher C, Rehrauer H, Fournier CA, Morrison HG, et al. (2010) The transcriptional response to encystation stimuli in Giardia lamblia is restricted to a small set of genes. Eukaryotic cell 9: 1566–1576. 10.1128/EC.00100-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zumthor J, Cernikova L, Rout S, Kaech A, Faso C, et al. (2016) Static clathrin assemblies at the peripheral vacuole—plasma membrane interface of the parasitic protozoan Giardia lamblia. PLoS Pathog in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Konrad C, Spycher C, Hehl AB (2010) Selective condensation drives partitioning and sequential secretion of cyst wall proteins in differentiating Giardia lamblia. PLoS Pathog 6: e1000835 10.1371/journal.ppat.1000835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, et al. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: D447–456. 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gaechter V, Schraner E, Wild P, Hehl AB (2008) The single dynamin family protein in the primitive protozoan Giardia lamblia is essential for stage conversion and endocytic transport. Traffic 9: 57–71. 10.1111/j.1600-0854.2007.00657.x [DOI] [PubMed] [Google Scholar]
- 57. Chan DC (2006) Mitochondria: dynamic organelles in disease, aging, and development. Cell 125: 1241–1252. 10.1016/j.cell.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 58. Chen H, Chan DC (2009) Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Human molecular genetics 18: R169–176. 10.1093/hmg/ddp326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Martincova E, Voleman L, Najdrova V, De Napoli M, Eshar S, et al. (2012) Live imaging of mitosomes and hydrogenosomes by HaloTag technology. PloS one 7: e36314 10.1371/journal.pone.0036314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. England CG, Luo H, Cai W (2015) HaloTag technology: a versatile platform for biomedical applications. Bioconjugate chemistry 26: 975–986. 10.1021/acs.bioconjchem.5b00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Morgan GW, Goulding D, Field MC (2004) The single dynamin-like protein of Trypanosoma brucei regulates mitochondrial division and is not required for endocytosis. J Biol Chem 279: 10692–10701. 10.1074/jbc.M312178200 [DOI] [PubMed] [Google Scholar]
- 62. Chanez AL, Hehl AB, Engstler M, Schneider A (2006) Ablation of the single dynamin of T. brucei blocks mitochondrial fission and endocytosis and leads to a precise cytokinesis arrest. Journal of cell science 119: 2968–2974. 10.1242/jcs.03023 [DOI] [PubMed] [Google Scholar]
- 63. Wexler-Cohen Y, Stevens GC, Barnoy E, van der Bliek AM, Johnson PJ (2014) A dynamin-related protein contributes to Trichomonas vaginalis hydrogenosomal fission. Faseb J 28: 1113–1121. 10.1096/fj.13-235473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rajala N, Hensen F, Wessels HJ, Ives D, Gloerich J, et al. (2015) Whole cell formaldehyde cross-linking simplifies purification of mitochondrial nucleoids and associated proteins involved in mitochondrial gene expression. PLoS One 10: e0116726 10.1371/journal.pone.0116726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Elias EV, Quiroga R, Gottig N, Nakanishi H, Nash TE, et al. (2008) Characterization of SNAREs determines the absence of a typical Golgi apparatus in the ancient eukaryote Giardia lamblia. J Biol Chem 283: 35996–36010. 10.1074/jbc.M806545200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sonda S, Stefanic S, Hehl AB (2008) A sphingolipid inhibitor induces a cytokinesis arrest and blocks stage differentiation in Giardia lamblia. Antimicrobial agents and chemotherapy 52: 563–569. 10.1128/AAC.01105-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Elmendorf HG, Rohrer SC, Khoury RS, Bouttenot RE, Nash TE (2005) Examination of a novel head-stalk protein family in Giardia lamblia characterised by the pairing of ankyrin repeats and coiled-coil domains. International journal for parasitology 35: 1001–1011. 10.1016/j.ijpara.2005.03.009 [DOI] [PubMed] [Google Scholar]
- 68. Isenmann S, Khew-Goodall Y, Gamble J, Vadas M, Wattenberg BW (1998) A splice-isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondrial targeting signal. Mol Biol Cell 9: 1649–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Duman JG, Forte JG (2003) What is the role of SNARE proteins in membrane fusion? Am J Physiol Cell Physiol 285: C237–249. 10.1152/ajpcell.00091.2003 [DOI] [PubMed] [Google Scholar]
- 70. Bandyopadhyay S, Chandramouli K, Johnson MK (2008) Iron-sulfur cluster biosynthesis. Biochemical Society transactions 36: 1112–1119. 10.1042/BST0361112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Manning G, Reiner DS, Lauwaet T, Dacre M, Smith A, et al. (2011) The minimal kinome of Giardia lamblia illuminates early kinase evolution and unique parasite biology. Genome biology 12: R66 10.1186/gb-2011-12-7-r66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Martincova E, Voleman L, Pyrih J, Zarsky V, Vondrackova P, et al. (2015) Probing the Biology of Giardia intestinalis Mitosomes Using In Vivo Enzymatic Tagging. Mol Cell Biol 35: 2864–2874. 10.1128/MCB.00448-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Eilers M, Schatz G (1986) Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature 322: 228–232. 10.1038/322228a0 [DOI] [PubMed] [Google Scholar]
- 74. Mi-Ichi F, Miyamoto T, Takao S, Jeelani G, Hashimoto T, et al. (2015) Entamoeba mitosomes play an important role in encystation by association with cholesteryl sulfate synthesis. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dudek J, Rehling P, van der Laan M (2013) Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta 1833: 274–285. 10.1016/j.bbamcr.2012.05.028 [DOI] [PubMed] [Google Scholar]
- 76. Xu F, Jerlstrom-Hultqvist J, Einarsson E, Astvaldsson A, Svard SG, et al. (2014) The genome of Spironucleus salmonicida highlights a fish pathogen adapted to fluctuating environments. PLoS genetics 10: e1004053 10.1371/journal.pgen.1004053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. de Kroon AI, Dolis D, Mayer A, Lill R, de Kruijff B (1997) Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim Biophys Acta 1325: 108–116. [DOI] [PubMed] [Google Scholar]
- 78. Zinser E, Sperka-Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, et al. (1991) Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol 173: 2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kumar P, Guha S, Diederichsen U (2015) SNARE protein analog-mediated membrane fusion. Journal of peptide science: an official publication of the European Peptide Society 21: 621–629. [DOI] [PubMed] [Google Scholar]
- 80. Tamura Y, Sesaki H, Endo T (2014) Phospholipid transport via mitochondria. Traffic 15: 933–945. 10.1111/tra.12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yamano K, Tanaka-Yamano S, Endo T (2010) Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J Biol Chem 285: 41222–41231. 10.1074/jbc.M110.163238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tatsuta T, Scharwey M, Langer T (2014) Mitochondrial lipid trafficking. Trends Cell Biol 24: 44–52. 10.1016/j.tcb.2013.07.011 [DOI] [PubMed] [Google Scholar]
- 83. Wideman JG, Gawryluk RM, Gray MW, Dacks JB (2013) The ancient and widespread nature of the ER-mitochondria encounter structure. Mol Biol Evol 30: 2044–2049. 10.1093/molbev/mst120 [DOI] [PubMed] [Google Scholar]
- 84. Rigotti A, Cohen DE, Zanlungo S (2010) STARTing to understand MLN64 function in cholesterol transport. Journal of lipid research 51: 2015–2017. 10.1194/jlr.E008854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhao J, Lendahl U, Nister M (2013) Regulation of mitochondrial dynamics: convergences and divergences between yeast and vertebrates. Cell Mol Life Sci 70: 951–976. 10.1007/s00018-012-1066-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. van der Bliek AM, Shen Q, Kawajiri S (2013) Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Okamoto K, Shaw JM (2005) Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet 39: 503–536. 10.1146/annurev.genet.38.072902.093019 [DOI] [PubMed] [Google Scholar]
- 88. Elgass K, Pakay J, Ryan MT, Palmer CS (2013) Recent advances into the understanding of mitochondrial fission. Biochim Biophys Acta 1833: 150–161. 10.1016/j.bbamcr.2012.05.002 [DOI] [PubMed] [Google Scholar]
- 89. Marti M, Li Y, Schraner EM, Wild P, Kohler P, et al. (2003) The secretory apparatus of an ancient eukaryote: protein sorting to separate export pathways occurs before formation of transient Golgi-like compartments. Mol Biol Cell 14: 1433–1447. 10.1091/mbc.E02-08-0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. McArthur AG, Morrison HG, Nixon JE, Passamaneck NQ, Kim U, et al. (2000) The Giardia genome project database. FEMS Microbiol Lett 189: 271–273. [DOI] [PubMed] [Google Scholar]
- 91. Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM (1998) A human dynamin-related protein controls the distribution of mitochondria. The Journal of cell biology 143: 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Miyagishima SY, Nishida K, Mori T, Matsuzaki M, Higashiyama T, et al. (2003) A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15: 655–665. 10.1105/tpc.009373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nishida K, Takahara M, Miyagishima SY, Kuroiwa H, Matsuzaki M, et al. (2003) Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc Natl Acad Sci U S A 100: 2146–2151. 10.1073/pnas.0436886100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pan R, Hu J (2011) The conserved fission complex on peroxisomes and mitochondria. Plant Signal Behav 6: 870–872. 10.4161/psb.6.6.15241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Otera H, Mihara K (2011) Discovery of the membrane receptor for mitochondrial fission GTPase Drp1. Small GTPases 2: 167–172. 10.4161/sgtp.2.3.16486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lee H, Yoon Y (2014) Mitochondrial fission: regulation and ER connection. Molecules and cells 37: 89–94. 10.14348/molcells.2014.2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Immunofluorescence co-labelling and wide-field microscopy analysis of transgenic G. lamblia lines expressing (A) GlTom40, (B) MOMTiP-1, (C) MOMTiP-7, (D) GlIscS, (E) MOMTiP-4 and (F) MOMTiP-3, all used as HA-tagged baits in co-IP experiments (upper row, in green), in combination with the endogenous mitosomal marker GlIscU (middle row, in red). Nuclei were stained with DAPI (lower row, in blue). The central mitosome complex was clearly labelled by both fluorophores in all lines. Scale bar: 1μm.
(TIF)
With increasing concentrations of formaldehyde (0–4.5%), immuno-detection (Western blot) of the Tom40-HA reporter shows a shift from the monomeric form to higher molecular weight complexes,. Molecular size (kDa) marker (M) bands are indicated on the left axis. A concentration of 2.25% formaldehyde (arrowhead) was later adopted for all subsequent forward and reverse co-IPs.
(TIF)
(A) Data distribution pie chart for protein hits with either Blast2Go annotation (“B2G annotated”), or associated to gene ontology (GO) terms (“with GO mapping”) or to Blast annotation data (“with Blast hits”). (B) The top 20 GO terms distributed in the three root categories for “biological process” (BP), “molecular function” (MF) and “cellular component” (CC). (C) A direct count of GO terms associated to MF. (D) A direct count of GO terms associated to BP.
(TIF)
(A) Data distribution pie chart for protein hits with either Blast2Go annotation (“B2G annotated”), or associated to gene ontology (GO) terms (“with GO mapping”) or to Blast annotation data (“with Blast hits”). (B) The top 20 GO terms distributed in the three root categories for “biological process” (BP), “molecular function” (MF) and “cellular component” (CC). (C) A direct count of GO terms associated to MF. (D) A direct count of GO terms associated to BP.
(TIF)
(A) IFA and wide-field microscopy analysis of transgenic Giardia cells expressing C-MOMTiP-1 (in green), a truncated HA-tagged version of MOMTiP-1 consisting only of the predicted C-terminal domain (residues 31–133). C-MOMTiP-1 accumulates primarily in the cytosol. Nuclei are labelled with DAPI (in blue). Inset: DIC image. Scale bar: 1μm. (B) Venn diagram depicting the overlap of datasets derived from native co-IP of C-MOMTiP-1 and control WB cells.
(TIF)
Data Availability Statement
All co-IP datasets referenced in the manuscript were deposited to the PRIDE platform with the identifier PXD004583 where they can be accessed via ProteomeXchange.