Abstract
This work proposes a computationally efficient cell nuclei morphologic feature analysis technique to characterize the brain gliomas in tissue slide images. In this work, our contributions are two-fold: 1) obtain an optimized cell nuclei segmentation method based on the pros and cons of the existing techniques in literature, 2) extract representative features by k-mean clustering of nuclei morphologic features to include area, perimeter, eccentricity, and major axis length. This clustering based representative feature extraction avoids shortcomings of extensive tile [1] [2] and nuclear score [3] based methods for brain glioma grading in pathology images. Multilayer perceptron (MLP) is used to classify extracted features into two tumor types: glioblastoma multiforme (GBM) and low grade glioma (LGG). Quantitative scores such as precision, recall, and accuracy are obtained using 66 clinical patients’ images from The Cancer Genome Atlas (TCGA) [4] dataset. On an average ~94% accuracy from 10 fold cross-validation confirms the efficacy of the proposed method.
Keywords: Tumor grading, digital pathology images, morphologic feature, nuclei segmentation, GBM, LGG, TCGA, multilayer perceptron
Introduction
Tumor grading from microscopy tissue slide images is an invasive diagnosis process to assess tumor progression, proliferation, and invasion. Many automatic cyto-/histological image analysis methods have been reported in literature for classification of breast cancers, follicular lymphoma, bone marrow, and brain tumors. These cell nuclei segmentation methods can be primarily categorized into following types: seed detection-based, shape prior-based, blob detection-based and clustering-based method. The seed-based methods [5] need careful initialization of pixel-seed, while watershed methods [6] typically lead to over-segmentation. As alternatives to seed detection methods, several shape prior-based methods are proposed [7] [8], where polygonal approximation are used for ellipse fitting. These prior shape model based segmentations are biased to elliptical cell nuclei. To alleviate these problems, several blob detection-based methods are proposed in [9] [10]. However, these methods need cautious tuning of the filter parameters to minimize the over and under segmentation and hence show lack of generality. Using the Bayesian clustering Jung et al.[11] propose an unsupervised nuclei segmentation method which is again biased to its elliptical shape-prior. This work proposes a hysteresis thresholding and contrast enhancement of the hematoxylin stain for optimal seed detection and a bias free concave point based iterative process for clustered nuclei separation.
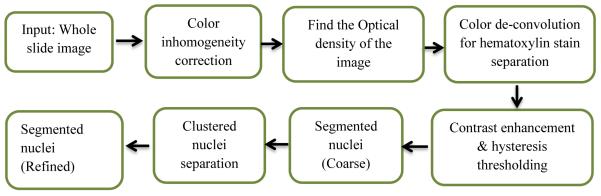
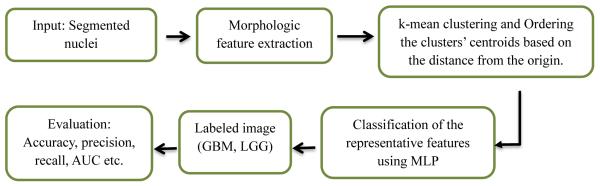
Among the recent automatic tumor classification works, Kong et al. [3] use a computationally extensive nuclear score (NS) based quantification of oligodendroglioma component (OC) population to classify GBM subtypes. Barker et al. [1] use local texture-tile based method, which is also computationally extensive and need more sophisticated image matching algorithm. Apart from the aforementioned feature based method, Xu et al. [2] use the deep convolution activation features to classify GBM and LGG. However, the method need extremely large number of training dataset for effective feature extraction. This work proposes a simple classification method using sophisticated features for tumor grading. The proposed segmentation method is shown in Figure 1(a), while the classification method in presented Figure 1(b), simply uses the k-mean clusters centroids of the morphologic features to avoid NS calculation, and search of a suitable candidate tile.
Figure 1.
(a): Flow diagram for nuclei segmentation
(b): Flow diagram for tumor classification
Methods and Materials
Our method consists of two main steps. In the first step, we segment the nuclei from the whole tissue slide images. In second step feature extraction and classification are performed. The overall flow diagram of the proposed method is shown in Figure 1.
Brief descriptions for each steps in the above flow diagram is given below.
The dataset used in this work includes two types of brain tumors: 38 images of GBM and 28 images from LGG. All the images are stained with hematoxylin and eosin. As the images are scanned with multi-resolution varies from 20X to 40X, we sample all images to 20X with bi-cubic interpolation.
Color inhomogeneity correction
Automatic contrast enhancement is applied to bring all the images with uniform color contrast.
Finding the optical density image
As the image intensities are of 8 bit depth, the maximum intensity, Imax is 256. The light absorbance of each pixel can be found by Beer-Lambert’s law [12],
| (1) |
where, I is the image intensity.
Color de-convolution
Since the optical density is proportionate to the stain’s concentration, we apply color de-convolution process on the optical density image. In this implementation the de-convolution matrix, M is defined
| (2) |
where, each row in matrix M indicates a specific stain and the columns represent the optical densities for the red, green and blue channels respectively. The color de-convolution is then performed with the following equation.
| (3) |
where, Y denotes the optical density vector, is the de-convoluted vector. The hematoxylin stain is the first channel of the de-convolved image.
Hysteresis thresholding
A contrast enhancement is done before the hysteresis thresholding. In this step, seeds are defined with upper threshold and connected component by the lower threshold. The threshold values are in between 0 to 1. Cell nuclei is the connected component at the seed regions.
Final nuclei segmentation
We remove the object pixels at the concave boundary to separate the clustered nuclei [13]. Finally the contour of the segmented nuclei is smoothed with linear interpolation of the boundary.
Morphologic feature extraction
Morphologic features like area, perimeter, eccentricity, circularity and major-axis length are extracted from the segmented nuclei.
k-mean clustering of the features
The above geometric features are clustered into 5 groups using k-mean clustering. Euclidean distance from the origin of the centroids are considered to determine the ascending order of the clusters. The centroids of the ordered clusters are used to characterize that individual image.
Classification using multi-layer perceptron
Using the WEKA toolbox [14], performance of different well-known classifiers’ for example SVM, Naïve Bayes, decision trees, MLP, linear regression etc. are observed. After intensive investigation we set MLP as the most effective classifier for this study.
Results and Discussion
In order to show the effectiveness of the proposed method, we perform 10 fold cross-validation. Out of 66 instances 62 (Table 1) are correctly classified with on an average 93.94 % accuracy. Details of the evaluation metrics are shown in Table 2. Although there are a lot of works on tumor classification from breast cancers, follicular lymphoma, bone marrow, sub-typing of brain glioblastoma, we notice quite a few works on brain tumor classification of GBM and LGG. Comparison of our results with the state-of-art works are shown in Table 3.
Table 1.
Confusion matrix; classification of 66 images.
| Original label | |||
|---|---|---|---|
| GBM | LGG | ||
| Pre- dicted |
GBM | 36 | 2 |
| LGG | 2 | 26 | |
Table 2.
Class wise and weighted average of the classifiers prediction.
| Class | TP rate | FP rate | Precision | Recall | AUC |
|---|---|---|---|---|---|
| GBM | 0.947 | 0.071 | 0.947 | 0.947 | 0.955 |
| LGG | 0.929 | 0.053 | 0.929 | 0.929 | 0.955 |
| Weighted Average |
0.939 | 0.063 | 0.939 | 0.939 | 0.955 |
Table 3.
State-of-art comparison of our results.
| Methods | Dataset / # of images | Features | Cross-validation accuracy |
|---|---|---|---|
| Barker et. al [1] | TCGA/total 45 (23 GBM, 22 LGG) |
Texture, color and shape of nuclei | 97.8 % |
| Xu et. al [2] | TCGA /total 45 (23 GBM, 22 LGG) |
Deep convolution activation features | 97.8 % |
| Our method | TCGA /total 66 (38 GBM, 28 LGG) |
Centroids from k-mean clustering of nuclei shape features |
94 % |
Accuracy of our results are comparable to other study reported in literature [1, 2]. Note we have used only nuclei’s geometric features to show the efficacy of the representative features which is very simple and computationally inexpensive. Moreover, our dataset is more unbalanced and may have impact on the outcomes. To investigate the robustness of these representative features, we vary the number of neurons in the hidden layer from 3 to 8 with fixed 0.3 learning rate and 0.2 momentum. We noticed that the accuracy remain constant in the range 5-8 (number of neurons) and a slight drop (92% accuracy) for the range 3-4 (number of neurons). This constant performance against the varying model parameters proves that the extracted feature are robust in classifying the tumor types. Overall on an average 94% accuracy suggests the efficacy of our algorithm.
Conclusion and Future Works
This work presents a computationally inexpensive method using morphological feature extraction of cell nuclei for brain tumor classification. Experimental results obtained from 66 patients images show that the proposed method is competitive when compared to recent works on tumor grading. In future, we plan to consider other useful features such as entropy, multi-fractal texture features [15], and color of nuclei in our pipeline to build a more effective classification scheme. We also plan to consider our prior structural MRI based method for tumor grading [16] and correlate that technique with the proposed digital pathology-based method in this study for robust tumor grading in large scale clinical cases.
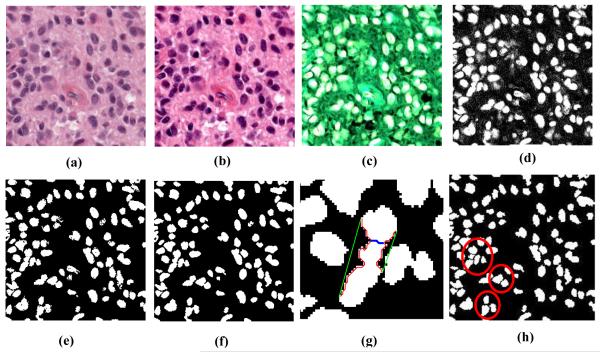
Figure 2.
Nuclei segmentation: (a) original image, (b) color normalized image, (c) Optical density image, (d) density of hematoxylin component, (e) hysteresis thresholding, (f) boundary smoothing of segmented nuclei (g) illustration of clustered nuclei separation; green (convex hull boundary), red (concave boundary), blue (cutting line found in a single iteration), (h) Final segmentation, clustered nuclei are separated (marked by red circle).
Acknowledgment
This work is partially supported through a grant from NCI/NIH (R15CA115464).
References
- [1].Barker J, Hoogi A, Depeursinge A, Rubin DL. Brain Tumor Digital Pathology Challenge. MICCAI; Boston, MA, USA: 2014. Automated classification of brain tumor type in digital pathology images using local texture patches; pp. 5–8. http://pais.bmi.stonybrookmedicine.edu/node/1. [DOI] [PubMed] [Google Scholar]
- [2].Xu Y, Jia Z, Zhang F, Ai Y, Lai M, I-Chao Chang E. Brain Tumor Digital Pathology Challenge. MICCAI; Boston, MA, USA: 2014. Deep convolutional activation features for large brain tumor histopathology image classification; pp. 25–29. http://pais.bmi.stonybrookmedicine.edu/node/1. [Google Scholar]
- [3].Kong J, Cooper LAD, Wang F, Gao J, Teodoro G, Scarpace L, Mikkelsen T, Schniederjan MJ, Morneo CS, Saltz JH, Brat DJ. Machine-based morphologic analysis of glioblastoma using whole-slide pathology images uncovers clinically relevant molecular correlates. PloS one. (ed. 81049) 2013;8(no.11) doi: 10.1371/journal.pone.0081049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].The Cancer Genome Atlas. http://cancergenome.nih.gov/
- [5].Malpica N, de Solorzano CO, Vaquero JJ, Santos A, Vallcorba I, Garcia-Sagredo JM, del Pozo F. Applying watershed algorithms to the segmentation of clustered nuclei. Cytometry. 1997 Aug 1;28(no. 4):289–297. doi: 10.1002/(sici)1097-0320(19970801)28:4<289::aid-cyto3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- [6].Veta M, van Diest PJ, Kornegoor R, Huisman A, Viergever MA, Pluim JPW. Automatic Nuclei Segmentation in H&E Stained Breast Cancer Histopathology Images. PLoS ONE. 2013;8(no. 7):e70221. doi: 10.1371/journal.pone.0070221. journal.pone.0070221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bai X, Sun C, Zhou F. Splitting touching cells based on concave points and ellipse fitting. Pattern Recogn. 2009;42:2434–2446. [Google Scholar]
- [8].Yan L, Jiang WX, Lee SR, Lee CY. Algorithm for splitting touching objects based on contour segments. Electron Lett. 2010;46:490–492. [Google Scholar]
- [9].Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng. 2010;57:841–852. doi: 10.1109/TBME.2009.2035102. [DOI] [PubMed] [Google Scholar]
- [10].Kong H, Akakin HC, Sarma SE. A generalized Laplacian of Gaussian filter for blob detection and its applications. IEEE Trans Cybern. 2013;43:1719–1733. doi: 10.1109/TSMCB.2012.2228639. [DOI] [PubMed] [Google Scholar]
- [11].Jung C, Kim C, Chae SW, Oh S. Unsupervised segmentation of overlapped nuclei using Bayesian classification. IEEE Trans Biomed Eng. 2010;57:2825–2832. doi: 10.1109/TBME.2010.2060486. [DOI] [PubMed] [Google Scholar]
- [12]. http://www.chemguide.co.uk/analysis/uvvisible/beerlambert.html.
- [13].Wienert S, Heim D, Saeger K, Stenzinger A, Beil M, Hufnagl P, Klauschen F. Detection and segmentation of cell nuclei in virtual microscopy images: a minimum-model approach. Scientific reports, 2(2012) doi: 10.1038/srep00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update; SIGKDD explorations. 2009;11(no. 1) [Google Scholar]
- [15].Islam A, Reza S, Iftekharuddin KM. Multi-fractal texture estimation for detection and segmentation of brain tumors. IEEE Transactions on Biomedical Engineering. 2013;60(no. 11):3204–15. doi: 10.1109/TBME.2013.2271383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reza SMS, Mays R, Iftekharuddin KM. Multi-fractal detrended texture feature for brain tumor classification. Proc. SPIE 9414, Medical Imaging, Computer-Aided Diagnosis, 941410; March, 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]