Summary
As they proliferate, living cells undergo transitions between specific molecularly and developmentally distinct states. Despite the functional centrality of these transitions in multicellular organisms, it has remained challenging to determine which transitions occur and at what rates without perturbations and cell engineering. Here, we introduce Kin Correlation Analysis (KCA) and show that quantitative cell state transition dynamics can be inferred without direct, molecular-level observation from the clustering of cell states on pedigrees (lineage trees). Combining KCA with pedigrees obtained from time-lapse imaging and end-point single-molecule RNA-FISH measurements of gene expression, we determined the cell state transition network of mouse embryonic stem (ES) cells. This analysis revealed that mouse ES cells exhibit stochastic and reversible transitions along a linear chain of states ranging from 2C-like to epiblast-like. Our approach is broadly applicable and may be applied to systems with irreversible transitions and non-stationary dynamics, such as in cancer and development.
Introduction
In many multicellular contexts, cells switch among molecularly and phenotypically distinct states as they proliferate through repeated divisions (Figure 1A). Key biological functions often depend critically on the dynamics of these cell state transitions: on which transitions are forbidden or permitted, at what rates they occur, and whether they are stochastic or deterministic. For example, regulation of fat tissue depends on adipocyte differentiation and de-differentiation rates (Ahrends et al., 2014; Poloni et al., 2012); maintenance of intestinal crypts and the epidermis are governed by the relative rates of symmetric and asymmetric stem cell divisions (Simons and Clevers, 2011); development of the full repertoire of immune cell types is regulated by stochastic cell state transitions (Suda et al., 1984a; 1983; 1984b); and lineage commitment in embryonic development and later in trans- or de-differentiation depend critically on dynamic transitions (Dietrich and Hiiragi, 2007; Ohnishi et al., 2014; Slack and Tosh, 2001; Talchai et al., 2012; Tata et al., 2013; Yamanaka et al., 2010). Cell state transition dynamics are also important in disease, as their dysregulation can lead to type 2 diabetes (Talchai et al., 2012) and obesity (Ahrends et al., 2014; Ristow et al., 1998). Similarly, in cancer, the rates of transition between distinct cell states within a tumor impinges on the effectiveness of treatments (Gupta et al., 2011; Leder et al., 2014), and the likelihood of metastasis (Wagenblast et al., 2015).
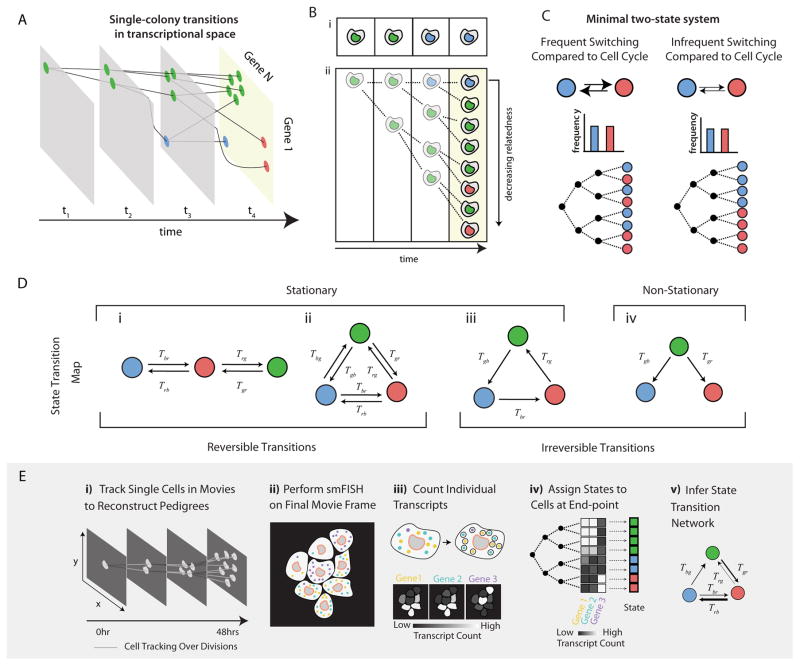
Figure 1. Cell state transition networks and the experimental platform for inferring transition rates.
(A) Trajectory of a proliferating colony of cells in gene expression space (schematic). At each time-point, a cell can independently and stochastically change its cell state (color) and corresponding gene expression profile. Following a division, both daughter cells inherit the state of the parent but then follow independent stochastic dynamic trajectories. (B) (i) Dynamics can be determined by directly observing state transitions in a single cell over time, neglecting cell proliferation. (ii) Proliferating colonies provide an indirect record of the history of cell state transitions. Here the cell of interest (top row) is in the blue state but is related to a sister and cousins that are in the green state, indicating a likely green to blue transition in its recent past. (C) Different dynamics give rise to different degrees of clustering on a pedigree (schematic). Frequent or infrequent switching between red and blue states leads to weak or strong clustering of cell states, respectively. The distribution of states is independent of the switching rates in this simple example (bar plots). (D) Cell state transition networks can be classified based on whether the population fraction of each state is constant (stationary) or changing over time (non-stationary). A subset of stationary networks also exhibit reversible dynamics. (E) Experimental approach: i) Live cells are tracked as they grow and divide using time-lapse microscopy. ii) After the movie, the cells are fixed and stained for smFISH. iii) Individual molecules of mRNA are detected and counted in each cell. iv) The pedigree reconstructed from (i) is combined with the smFISH measurements, and each cell is assigned an expression state. v) Using KCA, cell state transition dynamics are inferred across many of these state-associated pedigrees (see Box 1).
The notion of cell state can vary significantly depending on the particular biological system and the context of the study. Here, we consider cell states that satisfy certain criteria: first, a cell state must be heritable, such that after a cell division, the daughter cells by default remain in the same state as the parent cell unless a transition has occurred. This criterion excludes transient gene expression fluctuations. Second, different states should exhibit significant differences in the expression of multiple genes. Thus, although a single marker gene can be used to identify a particular cell state, the changes in the expression level of the marker gene must be correlated with that of other genes. Lastly, cell states should ideally possess distinguishing phenotypic properties such as morphological features (Thiery et al., 2009), chromatin structure (Kagey et al., 2010), developmental potential (Wu and Belmonte, 2015), or functional attributes (Duffy et al., 2012; Lu et al., 2015), although these may not always be readily apparent.
Mouse embryonic stem cells provide an important model system in which to study cell state transition dynamics. Multiple molecularly and phenotypically distinct ES cell states co-exist and stochastically interconvert in standard culture conditions (containing serum and leukemia inhibitory factor, LIF). In previous studies, these states were shown to be heritable and differ in gene expression, developmental potential, global epigenetic profiles, and other characteristics (Canham et al., 2010; Chambers et al., 2007; Falco et al., 2007; Hayashi et al., 2008; Macfarlan et al., 2012; Niwa et al., 2009; Singer et al., 2014; Singh et al., 2007; Yamaji et al., 2013; Yamanaka et al., 2010; Zalzman et al., 2010). Understanding the transitions among these cell states is important for applications seeking to control differentiation of these or other pluripotent cells for regenerative medicine applications. Recent improvements in single-cell profiling have enabled genome-wide analysis of ES cell states (Klein et al., 2015; Kumar et al., 2014). However, because these techniques do not track individual cells over time, they can’t provide information on transition dynamics. Other elegant studies have estimated transition dynamics of ES cells using either direct time-lapse microscopy, sorting of subpopulations, or by characterizing in static images the intra- and inter-colony gene expression heterogeneity (Canham et al., 2010; Chambers et al., 2007; Furusawa et al., 2004; Kalmar et al., 2009; Kumar et al., 2014; Rugg-Gunn et al., 2012; Singh et al., 2007; Suda et al., 1983; Toyooka et al., 2008; Zalzman et al., 2010).
Here, we describe a new approach to infer quantitative cell state transition dynamics in which cells transition stochastically and independently from one heritable gene expression state to another. This approach does not require sorting, perturbations or fluorescent reporters of gene expression. Rather than attempt to follow transitions directly in each individual cell over time, e.g. from the green to blue states in Figure 1Bi, we instead take advantage of cell division to infer dynamic information indirectly. Because sister cells start out in the same state, they generally provide independent realizations of transition dynamics starting from the same initial condition. Knowing the states of a cell’s sisters, cousins, and other relatives provides information about the likely history of that cell’s past transitions, as illustrated schematically in Figure 1Bii. Under some conditions, combining the lineage relationships, or pedigree, for a set of individual cells with their end-point states, can enable inference of cell state transition rates (Figure 1C). This approach is informative as long as cell states typically persist for durations longer than a cell cycle, but cannot access dynamics within a single cell cycle. This basic idea was recently described in Hormoz et al, 2015 for a special case, but is generalized and applied to embryonic stem cells here.
Using this approach to measure transition rates among multiple states can provide information about a cell’s overall state transition network, defined as the set of transitions between cell states that can occur in a given context. In principle, many different kinds of transition networks are possible, including all-to-all, chain-like, cyclical, or tree-like (Figure 1D). Some of these can produce stationary dynamics that maintain a constant distribution of states over time (Figure 1D, i–iii), and a further subset of these exhibit reversible transitions (Figure 1D, i–ii) (see Box 1 for definitions). Other networks, including binary fate trees, may include irreversible transitions (Figure 1D, iii–iv). Moreover, for any given network topology, the quantitative rates of each transition control the dynamic behavior of cells. These examples are idealized and natural systems can be more complex. For example, transition rates could depend on position in the tissue, e.g. through morphogen gradients, or on time. But for the embryonic stem cell system considered here, we show that this approach is sufficient to identify the cell state transition network and quantify its rates in a non-perturbative fashion.
Box 1. Kin-Correlation Analysis: Dynamics can be inferred from correlation functions of cellular states on pedigrees.
In this Box, we explain the KCA framework, which enables inference of cell state transition rates from the degree of clustering of cell states on pedigrees. We rely on the following definitions:
Lineage distance, u
The number of generations back to the common ancestor of two cells. u = 1 for sisters, u= 2 for first cousins, etc.
Transition matrix, T
A square N×N matrix, where N denotes the number of cell states, whose I,Jth element is the probability per cell cycle of a cell transitioning from state J to state I, T(I|J). Each column of this matrix sums to 1.
Reversible dynamics
The dynamics described by a transition matrix is reversible if for any pair of states A and B, the number of cells transitioning from A to B per unit time is equal to the number transitioning from B to A.
Two-cell correlation functions, C(u)
An N×N matrix, for each value of u, whose I,Jth element denotes the frequency of observing a pair of cells at lineage distance u in states I and J.
Three-cell correlation functions, C(u,v)
An N×N×N matrix that is a function of the two lineage distances u and v, which describe the degree of relatedness of three cells. u is the number of generations to the common ancestor of the two more closely related cells, while v specifies the number of generations to the common ancestor of all three cells. CIJK(u, v) denotes the frequency of observing the more distant relative in state I and the two more closely related cells in states J and K. Note that CIJK = CIKJ.
Here, our goal is to show how the transition matrix T can be inferred from the experimentally observable cell state correlation functions, C(u) or C(u, v). We first derive the equations for the case where all cell states are equally likely and the dynamics is reversible (or equivalently when T is symmetric), and then treat the more general case briefly here and in more detail in STAR Methods.
First, we compute the expected two-cell correlation function for a given transition matrix. Two cells at lineage distance u, shared a common ancestor in an unknown state M, u generations back. Subsequently, they each experienced u divisions and, potentially, zero or more state transitions independently of one another. Given the transition matrix for one generation, T, we can compute the resulting transition matrix for u generations by taking T to the uth power: Tu(I|M). It follows that the joint-probability of observing the two cells in states I an J is given by:
where the summation is over all possible states M of the ancestor. Here, by assumption, each state occurs with probability 1/N. The simplification in the last step follows because T is symmetric.
To infer the dynamics, we work backwards to recover matrix T by computing Tinferred(u) = C(u)1/(2u). For reversible dynamics, the transition matrix can be fully recovered by simply considering the two-cell correlation functions.
If the occurrence probability of the states is not a constant, we need to modify the equation for Tinferred(u). Assume that a given state I is observed in the population with frequency pI. The condition of reversibility requires that for any given pair of states, the forward and reverse fluxes must be equal: T(I|M)pM = T(M|I)pI. The two-cell correlation matrix is now given by,
To infer T, above equation can be rearranged to express the transition matrix in terms of the correlation matrix, by first defining a rescaled correlation matrix, . It then follows that, as in the simpler case, the transition matrix can be recovered by taking the appropriate root of the matrix C̃, Tinferred(u) = C̃(u)1/(2u).
Finally, for irreversible dynamics, T cannot be recovered from C directly because the assumption that T (I|M)pM= T (M|I)pI, no longer holds. Intuitively, the two-cell correlations are not sufficient, because they do not provide information about the directionality of state transitions. However, with a triplet of cells, the state of the ancestor of a cell pair is reflected in the state of its more distant relative. As a result, three-cell correlations do permit inference of directionality. The expected three-cell correlation functions can also be computed from the transition matrix T:
where S is summed over all possible states of the common ancestor of the two more closely related cells, and M is summed over all possible states of the common ancestor of all three cells. In the STAR Methods, we describe the full procedure for using three-cell correlations to infer T for irreversible dynamics.
Here, we describe an experimental platform which combines time-lapse movies to determine lineage relationships with single-molecule RNA-FISH (smFISH) of multiple genes to determine end-point gene expression states (Figure 1E). (For other elegant examples of combining time-lapse imaging with endpoint readout see (Filipczyk et al., 2015; Lee et al., 2014; Purvis et al., 2012)). Using this approach, we discovered that ES cells exhibit a distinct cell state transition network based on reversible stochastic transitions along a linear chain of states. We also generalize the previous theoretical framework to enable analysis of networks containing irreversible and non-stationary dynamics. Finally, because this is an inference approach, we provide a set of self-consistency checks to evaluate whether the assumptions of the underlying model are indeed valid. Thus, we believe the combined theoretical-experimental approach developed here should be applicable to other biological systems in which cells transition among multiple states.
RESULTS
Cell state transition networks can be inferred from clustering of states on pedigrees
To motivate the inference method, we first consider a simple minimal transition network (Figure 1C). In this example, cells stochastically transition between the two states at equal rates as they proliferate, such that the average population fraction of each state does not change over time. When transition rates per cell cycle are high, the states of sister cells rapidly become uncorrelated with one another, leading to no apparent clustering of states on the pedigree. By contrast, when transition rates per cell cycle are low and cells remain in the same state for multiple generations, closely related cells are more likely to be observed in the same state. As a result, different transition rates produce different degrees of clustering on the pedigree. Conversely, measurements of clustering between states of related cells can be used to infer transition rates. More specifically, clustering of cell states can be quantified by measuring how frequently related cells are observed to be in a given set of states, as a function of how long ago they shared a common ancestor. These correlations, computed between all pairs or, more generally, all triplets of cells over many pedigrees enable quantitative inference of the transition dynamics through an approach we term Kin Correlation Analysis (KCA), which is described briefly in Box 1 and in more detail in the STAR Methods. As derived in Box 1, dynamics of reversible transition networks can be inferred from the observed correlations between pairs of related cells (two-cell correlations), whereas inference of networks with irreversible transitions requires knowledge of the correlations between triplets of cells (three-cell correlations). To demonstrate that KCA can be used to infer the dynamics of the full range of transition networks depicted in Figure 1D, we simulated the transition dynamics of proliferating cells under different networks for physiologically relevant transition rates (STAR Methods), including reversible, irreversible, and non-stationary dynamics (Figure S1). Taken together, these results demonstrate that, at least under the idealized conditions of these models, KCA enables quantitative inference of diverse cell state transition networks. Limitations and self-consistency checks on the method are discussed more below.
To experimentally implement KCA, we developed a platform that combines two types of measurements: first, using time-lapse microscopy and custom software (STAR Methods), we track individual cells over multiple generations, as they grow from a single cell into a microcolony, and use this data to construct the pedigrees representing the lineage relationships among cells, with no gene expression measurements (Figure 1E,i). Second, at the end of the movie, we used single-molecule RNA-FISH (smFISH) (Femino et al., 1998; Raj et al., 2008; 2006) to measure the expression levels of multiple genes simultaneously, thereby determining each cell’s end-point state (Figure 1E,ii–v).
Kin Correlation Analysis (KCA) validation by comparing inferred two-state switching dynamics with direct time-lapse analysis
To experimentally apply KCA, we proceed in two stages. First, we validate the method by analyzing switching between two distinct states of Esrrb expression in mouse ES cells. Second, we broaden the analysis to determine the transition dynamics of a larger set of ES cell states.
Esrrb is a transcription factor central to maintaining the naïve pluripotent state, and it plays a critical role in the core pluripotency network (Festuccia et al., 2012; Martello et al., 2012; Singer et al., 2014; van den Berg et al., 2008). Esrrb up-regulation has also been shown to facilitate fibroblast reprogramming to the induced pluripotent state (Feng et al., 2009). Most importantly here, Esrrb expression in LIF+Serum culture conditions is bimodal, with cells switching between high and low expression states (Singer et al., 2014).
We constructed a knock-in fluorescent reporter for Esrrb expression (Figure 2A), and validated the reporter using smFISH (S2A–B). We acquired time-lapse movies (Fig 2B,i), using custom software to track individual cells over time and establish the pedigrees (lineage trees) of individual colonies (see STAR Methods). At the end of movie (~48 hours), we fixed the cells and acquired smFISH measurements of Esrrb expression (Figure 2B,ii–iiii). Finally, we combined the measurements, assigning smFISH Esrrb expression levels at the final time-point to the corresponding leaves of the tree (Fig 2D,i). Altogether, we analyzed 14 trees (299 cells) for this analysis.
Figure 2. Inference and direct validation of Esrrb dynamics.
(A) The Esrrb-H2B-mCitrine knock-in reporter (top), and PiggyBac integration construct for a palmitoylated-mTurquoise2 (bottom). (Bi) An example time-lapse movie showing H2B-mCitrine fluorescence in a proliferating colony of ES cells. Arrow indicates root cell in E. Scale bar, 10 μm (Bii) A composite image of the membrane-mTurquoise2 (white), DAPI (red), and Esrrb transcripts by smFISH (yellow dots). (Biii) Heat map showing Esrrb transcript counts for each cell in this colony. (C) The distribution of Esrrb transcript counts can be fit by a linear combination of two negative binomial distributions (solid lines), with indicated population fractions (percentages). (Di) Lineage tree (pedigree) from example movie shown in B. State assignments on leaves indicate the probability that the cells are in the E+ state (see STAR Methods). (Dii) The probability of observing a pair of cells both in the E+ state (red), both in the E− state (blue), and as a mixed E+E− pair (green), as a function of degree of relatedness of the two cells, u. Cell state transition rates were computed from the observed correlation functions for each value of u. (Diii–iv) The probability per cell cycle of transitioning from E− to E+ (blue) and from E+ to E− (red). Error bars were obtained by bootstrap (see STAR Methods). Inferred rates are (within statistical error) independent of u, consistent with stationary Markovian dynamics. (Ei–iii) The same pedigree as in D with branches displaying accumulation of mCitrine fluorescence in each cell cycle. Arrows indicate a significant, heritable change in the rate of fluorescence accumulation, corresponding to switches between Esrrb states. (Eiv) Essrb cell state transition rates measured from switching events in the time-lapse movies are consistent with inferred rates (cf. Div). (F) Histogram of the time of occurrence of state transitions (on-events, top panel; off-events, bottom panel) along the cell cycle in units of hours since the last cell division. (Gi) Empirically determined frequency of finding a pair of cells both Esrrb high (red points) or Esrrb low (blue points) as a function of their physical separation distance, d, in the colony (in units of average cell diameters). Error bars are s.d. determined by bootstrap (299 cells, 14 colonies). Dashed lines indicate expected cell state correlation as function of spatial separation distance. The observed spatial correlations are consistent with the correlations expected from shared lineage alone. (Gii) Spatial separation distance correlates weakly with lineage distance u. The distribution for each value of u is independently normalized to peak at 1.
Consistent with previous results, Esrrb exhibited a bimodal distribution of mRNA copy number by smFISH (Fig. 2C) (Kumar et al., 2014; Singer et al., 2014). To understand this distribution, we first note that a single state is expected to generate a distribution of mRNA copy numbers in individual cells, due to the stochastic, “bursty” nature of transcription and mRNA degradation, as shown previously (Elowitz et al., 2002; Friedman et al., 2006; Ozbudak et al., 2002; Peccoud and Ycart, 1995; Suter et al., 2011). For many genes and cell types, the distribution of mRNA copy number is well-fit by a negative binomial distribution (Friedman et al., 2006; Raj et al., 2006). This distribution is generated when there is a constant probability per unit time of initiating a transcriptional burst, and the number of mRNAs produced per burst follows an exponential distribution. For a gene with two expression states, we expect each state to generate a negative binomial distribution of mRNA with different burst rate and burst size parameters. These two distributions will, in general, overlap. Thus, the bimodal distribution can be explained as a linear combination of two negative binomial distributions, one for each expression state.
We fit the observed Esrrb distribution to a linear combination of two negative binomial distributions. Using this fit, we assigned each cell a probability of being in either the high or low Esrrb expression state given its observed transcript count (see STAR Methods). Thus we obtained a probabilistic endpoint state assignment for each cell on each pedigree (Figure 2D,i). Because of the overlap between the transcript count distributions of the two Esrrb states, many cells have approximately equal probability of being in either state. The KCA framework is compatible with these probabilistic state assignments. When computing the correlation matrix (Box 1), we account for probabilistic state assignments by summing over all possible pairs of states, for each pair of cells, weighting each state pair by its relative probability. When the state assignments are more ambiguous, a larger number of observations (pedigrees) is required to ensure accurate inference of transition rates (see STAR Methods for details).
To infer the rates at which cells switch between Esrrb states, we analyzed these trees with KCA. We first computed pair correlation matrices for lineage distances u, ranging from 1 to 4 (Fig. 2D,ii). As expected, the frequency of observing two cells in the same Esrrb state decreased with increasing lineage distance. Next, using KCA we computed the switching rates that would give rise to the observed correlation matrices. For stationary Markovian dynamics, these rates should not depend on the lineage distance from which the correlation matrix is computed (Box 1). The inferred rate of switching from the Esrrb low state to the Esrrb high state was 0.09±0.03 per cell cycle (errors are the standard deviation as estimated by bootstrap, see STAR Methods). The reverse rate was 0.08±0.02, per cell cycle. These rates remained constant across lineage distances of u = 1 to 4, consistent with stationary Markovian dynamics (Fig 2D,iii–iv). We note that the constant inferred rates imply an exponential waiting time between state transitions, a property of Markovian dynamics.
To independently validate these inferred Esrrb switching dynamics, we next analyzed data from the Esrrb knock-in fluorescent reporter. We extracted the total fluorescence from each cell over the duration of the movies. Because the H2B-mCitrine is stable, its abundance diminishes only through dilution during cellular division events. These dilution events correspond to approximately halving of the fluorescent readout from each cell across cell divisions, as evident in the saw-tooth pattern of the traces shown in Fig. 2Eii–iii. We therefore focused on the “promoter activity”, or the rate of accumulation of total fluorescence (slope of the fluorescence traces shown in Fig. 2Eii–iii), which should be proportional to the abundance of mRNA in the cell at any given time (Singer et al., 2014). Indeed, the Esrrb production rate in the final cell cycle of the movie was strongly correlated with Esrrb transcript counts measured at the end of the movie, but not with that of β-actin, a homogenously expressed housekeeping gene (Fig S2B), providing an internal validation of both readouts.
To classify Esrrb promoter activity as either high or low, we implemented a threshold on production rate at each time-point throughout the cells’ lineage history (Fig. 2E,i). Cell state transitions were defined as a change in the promoter activity across the threshold that persisted for at least one cell cycle (see STAR Methods). Examples of transitions can be observed in plots of total fluorescence trajectories, as shown in Fig 2E,ii–iii. Transitions from Esrrb low to high states, or high to low states, occurred with rates of 0.10±0.01 and 0.08±0.01 per cell cycle, respectively (Fig 2E,iv), consistent with the values inferred by KCA above (errors are the uncertainty in the observed frequencies due to finite number of observations). Finally, although this has no bearing on using KCA for inferring the transition rates, we also checked whether state transitions were more likely to occur at one particular point of the cell cycle. However, analysis revealed no strong cell cycle dependence in these data (Fig 2F). Together, these results suggest that the KCA method can correctly infer the reversible state switching dynamics of Esrrb, which appear to be consistent with a constant switching rate per unit time.
Transition rates cannot be explained by local intercellular signaling
One potential effect neglected in this cell-autonomous analysis is that neighboring cells could interact through a variety of signaling pathways, potentially impacting cell state changes in a non-cell-autonomous fashion. To test whether such effects play a significant role in the observed Esrrb transition rates, we computed the correlation of the Esrrb state for pairs of cells as a function of their spatial separation distance in the colony (Fig 2Gi). This is possible because in situ single-molecule RNA FISH measurements of cell state do not disrupt the spatial context of individual cells. We observed little cell-state correlations in space, mainly because the ES cells migrated frequently from one part of the colony to another (Fig 2Gii), as evident in our time-lapse movies (Movie S1). Nevertheless, because closely related cells are more likely to be located closer to each other in space and also share the same state as their common ancestor, we expect some degree of cell-state correlation in space from shared lineage history alone. To quantify this effect, we calculated the expected correlation of Esrrb state as a function of spatial separation distance from the inferred switching rates of Esrrb and the observed pedigrees (STAR Methods). This correlation fully explained the observed spatial correlations (Fig 2Gi), suggesting that, while local signaling interactions can and likely do occur, they are not required to explain the observed cell state transition dynamics in these conditions.
Characterization of ES cell states
Having established the inference framework and demonstrated its application experimentally, we set out to identify other ES cell states whose transitions could also be analyzed. In accordance with previous work (Falco et al., 2007; Ivanova et al., 2006; Lu et al., 2011; Macfarlan et al., 2012; Niwa et al., 2009; Singer et al., 2014; Singh et al., 2007; Toyooka et al., 2008; Weidgang et al., 2013; Zalzman et al., 2010), we selected Esrrb, Tbx3, and, Zscan4 as potential cell state markers (also see STAR Methods). To better characterize their expression distributions, we simultaneously measured the mRNA copy number of Esrrb, Tbx3, and, Zscan4, in single ES cells using 3-color smFISH. While RNA-seq enables classification of states by high dimensional transcriptional profiles, smFISH yields a higher resolution of quantitative, amplification-free measurements albeit with lower dimensionality, enabling estimation of in situ state assignments from fewer genes. The mRNA copy number distributions of Tbx3 and Zscan4 were long-tailed (Fig 3A) consistent with (Kumar et al., 2014; Zalzman et al., 2010). For these genes, we identified a threshold that optimally separated the mRNA distribution into high and low expression states, and ensured that subsequent results were robust to the choice of threshold (see STAR Methods and Figure S3). By contrast, Esrrb transcript counts exhibited a bimodal distribution, with overlapping modes (Figure 2C). While binary classification of three genes could in principle produce 23=8 possible states, three of these states were rare (<1% of the population) or did not occur, and were not considered further (Figure 3B).
Figure 3. Characterizing a set of mouse embryonic stem cell states.
(A) Distribution of the transcript counts of Esrrb, Tbx3, and Zscan4 in single cells as determined by smFISH. (B) Scatter plot of transcript counts by smFISH in 446 cells (individual dots). Color coding indicates assignment of each cell to one of five states. Blue-red gradations indicate probabilistic assignment of Esrrb expression states. (C) Example colonies showing groups of related cells in the same expression state for each of the three marker genes (for either the low or high state), consistent with cell states that persist over multiple generations. Yellow circles indicate transcripts detected by smFISH; red indicates DAPI stained nuclei; white is palmitoylated-mTurqoise2 demarcating cell membranes. (D) Sub-populations sorted on indicated marker genes (below columns) exhibit distinct RNA-seq profiles and broad differences in gene expression. FACS was performed based on distinguishable fluorescent reporter genes integrated at Esrrb and Tbx3 loci in the same cell (Fig. S2C), or, separately, based on a Zscan4 reporter integrated by PiggyBac transposition (right). Only genes showing statistically significant differential expression for the same cell line between sorted subpopulations are shown. (E) Zscan4+ cells exhibit a distinctive nuclear morphology compared to Zscan4− cells. DAPI stained nuclei (white, left); Zscan4 smFISH dots (yellow, right); membrane boundaries (red). (F) Nuclear morphology correlates with Zscan4 expression level (Pearson correlation coefficient = −0.15; p value= 0.002). The number of nuclear puncta detected in each cell plotted against the number of Zscan4 transcripts in the same cell. The color of each dot indicates the time since that cell’s last division, as determined by time-lapse microscopy.
Additional experiments supported the notion that these genes marked heritable cell states. First, many or all cells within individual colonies could be observed simultaneously expressing large numbers of transcripts for these genes, suggesting that their expression states are inherited across cell divisions. Conversely, whole colonies could also be observed expressing little to none of these transcripts (Fig. 3C). Second, gene expression analysis of sorted sub-populations exhibited broad differences in gene expression profiles. Using a double knock-in reporter for Esrrb and Tbx3 (Fig S2C), and a separate line with a PiggyBac promoter fragment reporter for Zscan4 (see STAR Methods), we sorted out Esrrb/Tbx3 negative (E−T−), Esrrb-positive/Tbx3-negative (E+T−), and Esrrb/Tbx3 positive (E+T+) cells, as well as Zscan4-positive (Z+) and –negative (Z−) cells, and performed RNA-seq on each sample. We observed hundreds of genes that were differentially expressed between these states (Fig. 3D), indicating that variations in marker gene expression do not simply reflect intrinsic noise, or fluctuations in the expression of individual genes, but rather indicate broad transcriptional changes. In particular, we observed decreasing expression levels of differentiation makers and signaling factors when going from E−T−, to E+T−, and finally to E+T+ cells, suggesting that Tbx3 could mark a more pluripotent state. Accordingly, Zscan4-positive cells displayed a unique nuclear morphology by DAPI-stain compared with Zscan4-negative cells: while Zscan4-negative cells appeared to have a larger number of distinct puncta, Zscan4-positive cells exhibited fewer but larger puncta (Fig 3E–F), potentially suggesting aggregation of presumed heterochromatin. This result further supports the notion that Zscan4-positive cells represent a distinct phenotypic state. Taken together, these results show that these three markers define heritable cell states with distinct gene expression profiles across multiple genes.
Cell state transitions are restricted
We set out to determine the transition dynamics of these states using KCA. We acquired ~48 hour movies, after which cells were fixed and stained for Esrrb, Tbx3, and Zscan4 mRNA in the same cells (Fig 4A–B) (see STAR Methods). Based on expression, we assigned each cell to one of the five states described above (Fig 3C). Altogether, we analyzed 41 pedigrees with a depth of 4.0±0.5 generations (mean ± s.d.) (see Fig 4D for examples, Figure S4 for all trees).
Figure 4. State-switching dynamics within a pluripotency network.
(A) (Left) Time-lapse movie used only for tracking cells to determine pedigrees. (Right) In the same cells, smFISH for Esrrb (cyan dots), Tbx3 (green dots), and Zscan4 (blue dots), as well as membrane-mTurquoise2 (white) and DAPI (red). (B) Segmented cells are color-coded by transcript count for each gene analyzed. (C) Pedigree reconstructed from cells tracked in A are plotted as a dendrogram, with state assignments and transcript counts for each of the three genes at the leaves. (D) Examples of other pedigrees and state assignments (see Fig. S4 for complete set). (Ei) Frequency of observation of each pair of states in sister cells (two-cell correlations). See Figure S5A for other lineage distances. (Eii) Using KCA, the transition rate matrix was computed from correlation matrices (see Box 1). (F) Inferred cell state transition network shows chain-like dynamics.
Inspection of these trees revealed cell state clustering, with closely related cells (e.g. sisters, first cousins) predominantly observed in the same state, implying that most states persist over multiple generations. In particular, Tbx3 and especially Zscan4 were expressed infrequently (population fractions of 31% and 8% respectively), but, once expressed, were typically observed to be ‘on’ in clades of 2 to 8 cells, consistent with extended (multi-generational) periods of expression. (For this reason, their long-tail mRNA distributions do not appear to represent brief stochastic bursts, as was previously hypothesized (Singer et al., 2014)). At the same time, most colonies contained cells in different states, demonstrating state-switching typically occurs multiple times in each colony during the movies. Notably, certain state combinations were more likely to be found together in the same pedigree. For example, the E−T−Z+ state was frequently found in the same pedigree with the E+T+Z− state but almost never with the E+T−Z− state (Figure S4). Together, these results indicate that the 48-hour timescale studied here can capture many transition events in ES cells, making the system amenable to analysis by KCA.
Mouse embryonic stem cells exhibit a chain-like state transition network
To extract the quantitative transition rates, we first computed the two-cell correlation matrices, which are plotted for sister cell pairs in Figure 4Ei and for more distantly related cell pairs in Figure S5A. From these correlations, we inferred the full set of transition rates between the five states using KCA (Fig 4Eii). These rates had two notable features: first, most states were stable over timescales of multiple cell cycles; all but one of the states showed an inferred half-life of ~6 generations. The exception was E−T+Z−, whose expected half-life is only ~1.7 generations. Second, many potential transitions occur at negligible rates (within the statistical error), suggesting they are either disallowed or extremely infrequent (Figure 4Eii). (Some of the negligible, but non-zero, rates could reflect ambiguities in state assignment when cell transitions from E−T+Z− to E−T−Z+ state are captured after deactivating Tbx3 but before activating Zscan4, or vice versa).
From this analysis, the full network of potential transitions effectively reduces to a linear chain, in which cells transition stochastically and reversibly only between adjacent states (Fig 4F). Cells traverse this chain by performing a random walk, hopping between adjacent states, but on average not moving in any particular direction. In the Discussion, we describe some implications of the chain-like cell state transition network in ES cells. For now, we note that the chain-like organization of states constrains the dynamic trajectories of ES cells. For example, it implies that transitions between 2C-like (Falco et al., 2007; Macfarlan et al., 2012) and epiblast-like states (Toyooka et al., 2008) pass through a specific set of long-lived intermediate states. Thus, to activate Tbx3 starting in the E−T−Z− state, cells must transition to the Esrrb high state first (E+T−Z−). Similarly, E−T−Z− cells must transition through the E+T+Z− state to reach the Zscan4 high state (E−T−Z+). The chain-like transition network also makes the prediction, which we validated directly and independently (see STAR Methods), that during the transition from E+T+Z− to E−T−Z+, Esrrb and Tbx3 should turn off almost simultaneously, closely followed by Zscan4 activation, over a time-scale comparable to the duration of a cell cycle. For validation of the inferred dynamics, see STAR Methods.
Self-consistency checks for applying KCA to other systems
Thus far, we have considered systems governed by cell-autonomous, time-independent, Markovian dynamics, in which sister cell transitions are independent of one another. However, many systems of biological interest may violate one or more of these conditions. A useful feature of KCA is the redundancy of different correlation measurements, which can be used to self-consistently check that these necessary conditions are satisfied in any particular system. Here, we consider several different potential violations and how they could be detected.
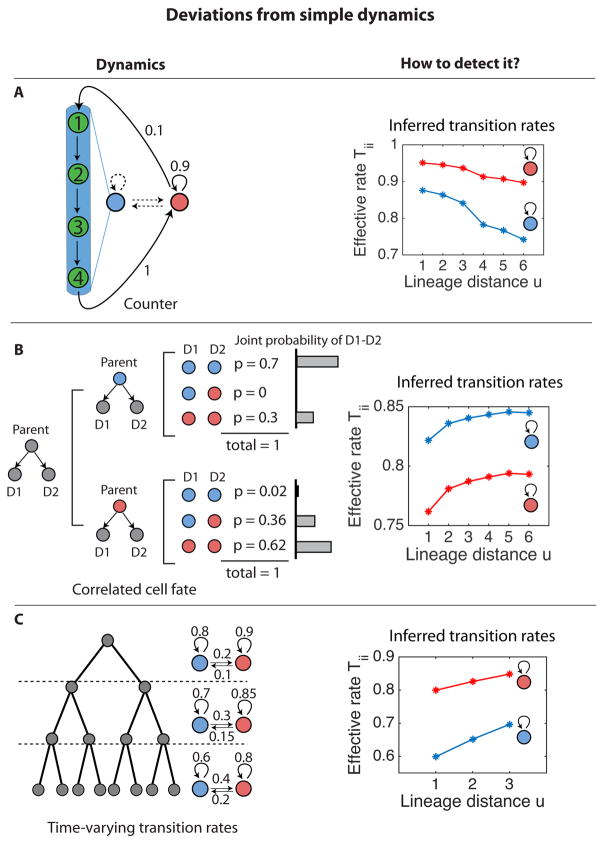
Some systems may exhibit non-Markovian dynamics, either because of “hidden” states, or because of “timed” transitions, in which cells spend a fixed amount of time or number of generations in a given state rather than exiting the state at a fixed stochastic rate (Norman et al., 2013). One way to detect such effects is to consider the effective transition rates, . Without hidden states, these rates are independent of the lineage distance, u, but with hidden states, they depend on u, as shown in Fig. 5A. Comparing effective transition rates determined at different lineage distances can thus be used to identify the existence of one or more hidden states or timers.
Figure 5. Detecting deviations from simple dynamics using self-consistency checks.
(A) ‘Hidden,’ states can produce apparent non-Markovian dynamics. In this example, the blue state is actually composed of multiple distinct states (labeled 1–4), which are not separately identifiable. The blue state is thus a counter that persists for exactly four generations. KCA applied to the apparent 2-state system generates inferred persistence rates which change systematically with lineage distance, u, especially near u = 4, causing the inferred transition rates (right) to depend on lineage distance. Transition rates are indicated on arrows. (B) Deviation from simple dynamics resulting from correlated transitions. In this example, distinct division patterns are indicated with corresponding probabilities, p. Values were chosen such that the joint probability of observing a pair of sister cells in a pair of states conditional on the state of their parent is not equal to the product of their marginal probabilities. In this case, inferred transition rates depend on lineage distance (right). (C) When transition rates vary with time (left), the inferred transition rates vary with lineage distance (right). In this example, this effect can be used to infer the time-varying transition rates (see STAR Methods).
Some systems, such as somatic stem cells in cycling tissues (Clayton et al., 2007; Snippert et al., 2010), exhibit correlated fate decisions in sister cells. Such correlations produce a deviation of the effective transition rates from a constant, particularly at lower values of u, where the correlations between the fates of sisters exhibit the largest effects (Fig. 5B). If it is already known that decisions are controlled through a specific class of models, as in (Klein and Simons, 2011; Lopez-Garcia et al., 2010), then this information may still be sufficient to infer the joint probability of sister fates conditional on the state of their parent (assuming other requirements of the method are met) (Hormoz et al., 2015). At the very least, this deviation can reveal that some assumption of the method is not satisfied.
Another potential issue is the possibility of time-dependent transition rates. This would lead to the inference of different effective transition rates from the two cell-correlation functions at different lineage distances, u. In principle, it is possible to infer time-dependent transition rates through measurement of two-cell correlations at all values of u. Finally, we note that three other potential deviations from simple Markovian dynamics were previously discussed above: irreversible transitions (Fig. S1C), non-stationary dynamics (Fig. S1D), and effects of local cell signaling (Fig. 2G).
To summarize, the KCA framework relies on measurement of the two-cell, three-cell, and potentially higher-order correlation functions at various lineage distances. Because the inference approach under-fits the measured correlation functions, the redundancy can be used to validate the assumptions of the model used for the inference through self-consistency checks. If this validation fails, the model can be extended to include additional factors, such as spatial signaling or correlated sister fates, and the process can be repeated.
Discussion
Although cell state transitions are central to biology, methods to measure their rates without cell line engineering, perturbations, or sorting have been lacking. The KCA approach implemented here with time-lapse movies and endpoint smFISH provides such a method.
Applying KCA to ES cells, we discovered a transition network consisting of a set of reversible transitions along a linear chain of metastable states (Figure 4F). These states are ordered in a sequence from 2C-like (totipotency) to the more differentiated epiblast-like state. Cells traverse this chain through stochastic reversible transitions, and pluripotency is therefore gained and lost by ES cells in a step-wise incremental way rather than continuously or all at once. The highly structured chain-like transition network dynamics discovered here contrasts with one prevailing view of ES cell heterogeneity as a noisy process consisting of random transitions among all states, as well as views in which it reflects independent noise in various genes. It also contrasts with the canonical binary unidirectional trees observed in many classic developmental systems. Moreover, because all transitions are reversible, this system can be accurately represented by a 1-dimensional energy landscape, in which each state can be characterized as a local minimum, and transitions can be thought of as stochastic hops to neighboring states along a reaction coordinate (Waddington, 1940); (Sokolik et al., 2015). The chain-like transition network could ensure that the ES cell culture is comprised primarily of cells whose recent ancestors were in the E−T−Z+ state, where telomere length may be extended (Zalzman et al., 2010), potentially enhancing the viability of the culture (see STAR Methods and Fig. S7).
The transition dynamics of ES cells appear consistent with a ‘memory-less’ Markov process, where transition rates depend only on the current state. Knowledge of these transition rates allows us to estimate the timescales required for colonies to reach an equilibrium distribution of cell states. Based on the measured rates, it should take about 25 generations for a single starting ES cell to yield an approximately equilibrium distribution of cell states. For smaller colonies, however, inter-colony variation is expected to dominate intra-colony variation. These considerations could help explain incomplete penetrance in directed differentiation protocols (Ieda et al., 2010; Suzuki et al., 2013; Vo and Daley, 2015), and reprogramming (Buganim et al., 2012; Hanna et al., 2009; Smith et al., 2010).
While powerful, KCA also has limitations. First, and most fundamentally, it applies only to transitions that occur at rates comparable to or slower than the cell cycle, since transitions that occur more rapidly leave no signature in the clustering of states on pedigrees. It is thus well-adapted to developmental and immunological processes but not well suited to analyze more rapid or transient physiological responses. Second, as with phylogenetic reconstruction, we can estimate the likelihood of a particular series of switching events on a given tree, but we cannot determine the exact histories of specific cells. Third, the technique requires previous identification of a set of distinct states, and corresponding marker genes. In the case of ES cells, inclusion of additional genes could reveal other states that might have been missed here (Klein et al., 2015; Kumar et al., 2014; Sasagawa et al., 2013). In this regard, emerging in situ single-cell transcriptomic techniques (Chen et al., 2015; Crosetto et al., 2015; Lubeck and Cai, 2012; Lubeck et al., 2014) are exciting, as they dramatically expand the number of genes that can be analyzed in the endpoint FISH-based measurement, enabling high-dimensional gene expression information with reduced a priori selection of genes. While we used thresholds on single genes for discrete state assignment here, such higher dimensional data could be used with the KCA framework to infer transition dynamics among a more continuous range of states, given a sufficient number of observations. Looking forward, the ability to quantify cell state transition networks should enable analysis of the effects of genetic perturbations on particular transition rates. Finally, we note that KCA can also work with alternative methods for obtaining lineage information (Behjati et al., 2014; Evrony et al., 2015; Jiang et al., 2013; Navin et al., 2011; Zong et al., 2012). Thus, we anticipate that the KCA framework will become more capable and broadly applicable in the future.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
The Lead Contact MBE is willing to distribute all materials (including constructs and engineered cell lines), datasets, software and analysis tools, and protocols used in the manuscript. Requests should be made directly to Michael B. Elowitz at melowitz@caltech.edu or by mail at California Institute of Technology. 1200 E. California Blvd., MC 114-96. Pasadena, CA 91125.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Line Construction and Tissue Culture
E14 cells (E14Tg2a.4) obtained from Mutant Mouse Regional Resource Centers were used as the base line for all cell line construction. Knock-In reporters were generated using CRISPR/Cas9 with guides targeting the C-terminus of the genes of interest (Supplementary Table 2), using donor vectors harboring ±300bp homology to the target locus flanking a T2A-H2B-XFP-P2A-PuroR. Single clones were first grown in 2i and isolated based on puromycin resistance and characterized for correct targeting using qPCR for genomic copy number, and then by a co-localization test of the endogenous targeted gene and XFP by smFISH (Figs. S2A and S2C). For the Zscan4 reporter, the 2570 base pairs upstream of the Zscan4c start codon were used as a promoter fragment reporter (as described in Zalzman, et al, 2010), to drive expression of H2B-mTurquoise2 on a PiggyBac integrated vector, which also contained a separate Blasticidin resistance cassette under an SV40 promtoer. Cells were maintained at 37°C and 5% CO2 in GMEM, 10% FBS, 2 mM L-glutamine, 100 units/ml penicillin, 100 ug/ml streptomycin, 1 mM sodium pyruvate, 1000 units/ml Leukemia Inhibitory Factor (LIF, Millipore), 1X Minimum Essential Medium Non- Essential Amino Acids (MEM NEAA, Invitrogen) and 50 uM β-Mercaptoethanol. Cell lines were also stably integrated with a PiggyBac-pGK-palmitoylated-mTurquoise2/HygroR to enable 3D segmentation of cell membranes.
METHOD DETAILS
Time Lapse Microscopy and single-molecule Fluorescence in situ Hybridization (smFISH) Imaging
For movies, cells were plated on Laminin-511 (BioLamina) in 24-well glass bottom plates (MatTek) six hours prior to the start of the movie at a density of 2000/well. Snapshots were taken at 12 minute intervals for ~48 hours, and tracked and segmented using home-grown Matlab scripts. Immediately following the end of the movie, cells were fixed in 4% Formaldehyde for five minutes at room temperature, and permeabilized in RNAse-free 70% ethanol and stored at −20°C overnight. The following day, cells were hybridized for smFISH overnight at 30°C, where genes of interest were simultaneously targeted with up to 48 20mer DNA oligos, with each gene’s probeset coupled to Alexa 555, 594, or 647 (Lifetech). Each 20mer oligo was used at ~3nM final concentration. The hybridization buffer was composed of 20% Formamide, 2X SSC, 0.1g/ml Dextran Sulfate, 1mg/ml E.coli tRNA, and 2mM Vanadyl ribonucleoside complex, in nuclease free water. After overnight incubation in hybridization buffer and probes, cells were washed once in 20% Formamide and 2X SSC at 30°C for 30min, twice in 2X SSC at room temperature, stained with DAPI, and finally imaged in 2X SSC.
smFISH imaging was performed on a Nikon Ti-E with Perfect Focus, Semrock FISH filtersets, Lumencor Sola illumination, 60x 1.4NA oil objective, and an Andor Zyla 4.2 sCMOS camera. Z-slices of DAPI, membrane-mTurquoise2 and smFISH were taken every 400nm through the sample. Segmentation of cellular boundaries was performed using the membrane targeted palmitoylated-mTurquoise2 with a 3D watershed algorithm. Dots were detected by thresholding on the distribution of local maxima of Laplacian-of-Gaussian kernel responses performed on each z-slice, with local-maxima defined around a 26-connected-pixel 3D region. Automatic image registration was performed in Matlab between the fluorescent protein in the final frame of the movie and DAPI stained image collected during smFISH imaging.
RNA-seq
On two separate days for biological replicates, ~500,000 were sorted of each subpopulation. Only the top 2% of reporters cells were collected in the positive gate for Zscan4c, while the lowest 50% were collected for the negative gate. For the Esrrb/Tbx3 double reporter (described above), 4% of the population made up the sorted double-negative population, 14% made up the sorted double positive population, and 63% made up the sorted Esrrb only population. The remaining unsorted cells made up buffer regions between subpopulations. Consistent with smFISH results, no Esrrb−/Tbx3+ population was observed. Differences in population fractions from smFISH-estimated population fractions is due to the half-life of the long-lived H2B-fused fluorescent proteins which only dilute by cellular division. Immediately following the sort, RNA was extracted using the Qiagen RNEasy Mini kit. 100 base single-end reads were generated on a HiSeq 2500. Galaxy was used to process RNAseq reads, using the Cufflinks package with default options. Briefly, reads were mapped using default Tophat parameters against the mouse mm10 genome. Cufflinks was used to estimate transcript abundance, and Cuffdiff was used to identify differentially expressed genes within E−T−, E+T−, and E+T+ sets, and then separately between Z+ and Z− sets.
QUANTIFICATION AND STATISTICAL ANALYSIS
Kin Correlation Analysis (KCA) applied to non-uniform cell state distributions
In Box 1 of the main text, we derived a formula that related the two-cell correlation matrices to the transition matrix, under the assumption that the dynamics was reversible (or equivalently that it satisfied the condition of detailed balance) and briefly generalized the result to the case where all the states are not equally likely. Here, we will derive in full detail a formula for inferring the transition matrix from the observed two-cell correlation matrices assuming only that the dynamics is reversible. Then, in section 3 below, we will further relax the assumption of reversibility, and derive a more general expression using three-point correlation functions to infer transition dynamics.
As in Box 1 of the main text, consider a transition matrix T(I|M) that represents the probability of observing a daughter cell in state I given that the parent cell was in state M. We assume that a given state I is observed in the population with frequency pI.
With detailed-balance, for any given pair of states, the forward and reverse fluxes must be equal:
The simpler condition used in Box 1 that T is a symmetric matrix, T(I|J) = T(J|I) is a special case of this expression, valid when pI = pJ. A similar condition must hold going from a parent cell in state M to a descendent in state I after two generations:
where s is summed over all possible state of the intermediate cell between the parent cell and its grand-daughter. The summation is equivalent to matrix multiplication and can be rewritten as,
More generally, for a cell in state M and its descendent u generations later in state I, the following condition must be satisfied:
The joint probability of observing two cells at lineage distance u in states I and J is given by,
Reversibility of the dynamics implies that Tu(J|M)pM = Tu(M|J)pJ. Making this substitution, we have,
| (1) |
To infer, we observe the correlation matrix C(u), and solve for the transition matrix T. Equation (1) can be rearranged to express the transition matrix in term of the correlation matrix, by first defining a rescaled correlation matrix,
| (2) |
It then follows that the transition matrix can be recovered by taking the appropriate root of the matrix C̃,
| (3) |
Kin Correlation Analysis (KCA) applied to time-varying transition rates
Previously, we assumed that transition rates remain constant over time. However, in a developmental context they could change systematically with time or generation number. In this subsection, we extend the above results to such cases. We still assume that the dynamics are stationary and reversible. As shown below, it is possible to fully recover time-varying dynamics by using the two-cell correlation functions at all lineage distances.
Consider a time-varying transition matrix, T(u), where u denotes the number of generations back from the final time-point. u generations back, the probability of observing a daughter cell in state I conditional on the state M of its parent is given by the I, Mth element of T(u). For an example of such dynamics, see Figure 5C in the main text.
The two-cell correlation matrix for a pair of cells at lineage distance u takes the form,
where S is an effective transition matrix given by Su = T(1)T(2)···T(u), and pJ denotes the endpoint frequency of cells in state J. From a measurement of the two-cell correlation matrix C(u), Su can be inferred using Eqs. 2 and 3, namely, define . It follows,
To recover the time-varying transition rates T(u), we start at u = 1 and work our way backwards to larger values of u. The transition rates are given by,
Lastly, we note that the above framework can be in principle extended to the case of continuous dynamics where the transition matrix is a continuous function of absolute time back to the common ancestor, i.e. where transitions have a probability per unit time (rather than per generation) of occurring, and where this probability itself changes with absolute time. To do so, the effective transition matrix, Su, is computed by taking the product integral of the continuous transition rate matrix, T́, from the current time, t = 0, back to the time of the common ancestor, t = tc, namely, . This formulation accounts for biologically relevant cases in which the durations of cell cycles vary from cell to cell and/or over time.
Kin Correlation Analysis (KCA) with probabilistic state assignment
We show that the distribution of the Esrrb transcript counts in the low (E−) and high (E+) states overlapped significantly (Fig. 2C), such that it was not possible to assign a definite Esrrb state (either E− or E+) to a cell given a readout of its Esrrb transcript count. Here, we explain how we assigned probabilistic Esrrb states to each cell, and how the KCA framework is applied to probabilistic states.
First, we fit a sum of two negative binomial distributions to the distribution of Esrrb transcript counts in single cells (black lines in Fig. 2C). Let’s denote the distribution of transcript counts of the E− state as D−(x) and that of the E+ state as D+(x), where x is an integer denoting the transcript count in a given cell. More specifically, D−(x) is the probability that a cell in the E− state will have x Esrrb transcripts. The fit also has a free parameter that reflects the population fraction of each state. We will denote the population fraction of the E− state as f− and the population fraction of the E+ state as f+. It follows that f− + f+ = 1.
The probability that a cell with x Esrrb transcripts is in the E+ state is given by,
The probability that the cell is in the E− state is simply p− = 1 − p+. For large transcript counts, e.g. x = 200, f−D−(x) ≈ 0, which implies, p+ ≈ 1. Alternatively, for some intermediate values of transcript counts, e.g. x = 75, f−D−(x) = f+D+(x), which implies p+ ≈ 0.5, or that the cell is equally likely to be in the E− or the E+ state.
Correlation matrices can be computed using probabilistic states in a similar manner as with definite states. However, whereas with definite state assignments, each pair of cells in states I and J contributes 1 to the I,Jth element of the correlation matrix and 0 to all the other elements, with probabilistic state assignments, each pair of cells contributes PIPJ to element CIJ of the correlation matrix.
Lastly, the switching rates can be inferred from the correlation matrices computed using probabilistic states in a similar way as outlined in the previous section. However, in cases where the overlap between the two distributions is not symmetric, i.e. when it is more likely to misclassify a cell in the E− state as E+ than vice-versa, we need to first adjust the correlation matrices for incorrect assignment of states.
On average, the probability that a cell in state J is assigned to state I is given by,
where the integration runs over all possible values of transcript counts, x, and the summation K is over all states. Q is effectively a transition matrix satisfying the same properties as T; for example, columns of Q sum to 1. However, unlike T, Q does not capture actual cell state transitions, but rather effective state transitions caused by measurement errors (for example, ambiguous mapping from transcript counts to cell state). Thus, we can imagine the dynamics as follows: the state of a cell u generations after its ancestor is given by the appropriate power of the transition matrix, namely, Tu. The measurement error, at the endpoint, results in one additional mixing of states as given by matrix Q. Put together, the probability of observing a given state conditional on the state of the ancestor is given by the matrix QTu.
To infer the actual transition matrix T, we must first remove the contribution of Q. The actual population fraction of the states, p̄, can be calculated from the measured population fractions, p, as follows,
where Q̂ is the inverse of the matrix Q. Similarly, the actual correlation matrix can be calculated from the measured correlation matrix as follows
where denotes the inverse of the transpose of matrix Q. The corrected populations fractions, p̄, and correlation matrix, C̄, can be used directly in Equations 2 and 3 above instead of p and C to infer the actual transition matrix.
Computing the three-cell correlation functions for a general transition matrix with irreversible dynamics
Here, we derive the general expression for the three-point correlation functions in terms of the transition matrix. As in the previous section, consider a transition matrix T(I|M) that represents the probability of observing a daughter cell in state I given that the parent cell was in state M. Unlike the previous section, we do not require that T(I|M) satisfies the condition of detailed balance, enabling analysis of cell state transition networks containing irreversible transitions.
We would like to calculate the joint probability of observing three cells in states I, J, and K. The degree of relatedness of three cells is characterized by two lineage distance: u, the number of generations back to the common ancestor of the two more closely related pair of cells (observed to be in states J and K), and v, the number of generations back to the common ancestor of all three cells (see Box 1 for a schematic).
The three-cell correlation function takes the form,
| (4) |
where the summation over S is over all possible states of the common ancestors of the two cells at lineage distance u. The summation over M is over all the possible states of the common ancestor of the three cells. pM is the expected probability of observing the common ancestor of all three cells in state M.
For non-stationary dynamics, the probability of observing the common ancestor in a given state pM changes from generation to generation. However, pM is still related to the transition matrix in a self-consistent way. Namely, the probability that a cell u generations back will be in state M is given by
| (5) |
where Tu0−u(M|N) denotes element M, N of the transition matrix taken to the power of u0 − u. u0 is the number of generations back to the root of the tree, which is in state N with probability pN(u0). Equation (5) captures how the population fraction of each state changes over time as a function of the initial distribution of the states and the transition matrix.
Equations (2) and (3) are general and do not require reversible (detailed balance) or stationary dynamics. Although an analytical solution for T in terms of CIJK(u, v) is not possible, we can solve the inference problem by considering the elements of the transition matrix as fitting parameters. We then calculate the expected three-cell correlation functions (Eq. 4) and fit them to the observed three-cell correlation functions (see Methods in the main text for the numerical implementation).
Simulating KCA for various types of dynamics
Using KCA, we were able to accurately infer the underlying cell state transition network and transition rates in simulations by observing 30 cell pedigrees of 5 generations (Figure S1). For reversible dynamics like those shown in Figure S1A,B, the transition network was inferred from the two-cell correlation functions. For networks with irreversible dynamics, like those shown in Figures S1C,D, we used the three-cell correlations for the inference. For example, in Figures S1B and S1C, which differ only in the reversibility of their transitions, the two-cell correlation functions are identical, but the three-cell correlation functions are different and can be used to infer the directionality of the transitions. Furthermore, we analyzed a previously published model of a 3-state system containing irreversible transitions among cancer cell states (Gupta et al., 2011), and verified that KCA with three-cell correlations could indeed infer the previously determined rates (Figure S1E). Finally, we asked whether the KCA framework could be applied to non-stationary, branching cell fate determination networks, similar to those frequently observed in development and immunology. We simulated a 3-level branched cell fate tree with specific transition rates, applied KCA, and recovered the correct rates within statistical error (Figure S1D). This indicates that accurate inference is possible for branching fate trees with feasible amounts of experimental data.
Here, we describe the details of the simulations used to generate the results shown in Figure S1. Simulated pedigrees of 5 generations each were generated using Matlab. For S1A–C, the state of the root was selected randomly from the stationary distribution of cell states. In Figure S1D, the root was always set to the green state, resulting in a non-stationary distribution of cell states over the generations. At each generation, every node gave rise to two daughter nodes, whose states were selected randomly and independently from the probability distribution set by the state of the parent and the transition matrix. The two-cell and three-cell correlations were directly computed from the simulated pedigrees by measuring the frequency of occurrence of pairs and triplets of cell states at a given lineage distance over all pedigrees. We simulated 30 pedigrees for the plots in Figure S1A to C. For Figure S1D, we simulated 100 pedigrees. KCA using two-cell correlation functions was conducted on simulated data as outlined above using the framework in Box 1 and Supporting Information without any fitting. To infer the rates for irreversible dynamics, we used a set of fitting parameters, corresponding to the independent entries of a general asymmetric transition matrix. We then fit the three-cell correlation functions predicted from this transition matrix (see STAR Methods) to the observed three-cell correlation functions. A non-linear least square fitting algorithm was used (implemented in Matlab) to minimize the residual.
Direct measurement of Esrrb switching dynamics
We tracked and segmented each cell in time-lapse movies of colony growth using automated software and manual corrections, similar to previously described (Singer et al., 2014). By integrating the background corrected pixel intensity in the nucleus of each cell, we obtained the accumulated level of H2B-mCitrine fluorescence at every point along each cell cycle. The rate at which fluorescence accumulated was used to estimate the promoter activity of Esrrb. To identify changes in promoter activity that corresponded to state switching, we fit either a single line, or two piece-wise linear segments to the fluorescence read-out of each cell using a least-squares method, implemented in Matlab. The first and last hour of each cell cycle was discarded to ensure reliable fluorescence read-out despite cell division. We used two criteria to identify state switching events: 1) the change in the slope across a division or between the segments of the two-line fit had to exceed a significance threshold. 2) A significant change in the slope (increase or decrease) had to persist into the subsequent cell cycle after division. The candidate switching events were identified automatically using a script implemented in Matlab and then verified manually.
Computing the predicted spatial correlation functions for Esrrb
We calculated the expected correlation of Esrrb state as a function of spatial separation distance from the inferred switching rates of Esrrb and the observed pedigrees as follows,
where q(r|u) is the empirically determined probability of observing two cells at lineage distance u at spatial separation distance r; q(r|u) is plotted in Fig. 2Gii. p(u) is the probability that two randomly chosen cells will be at lineage distance u. This was empirically computed using the set of observed pedigrees. The expected and directly observed spatial correlation are plotted in Fig 2Gi.
Selecting marker genes for the ES pluripotency states
Previous studies have revealed that ES colonies exhibit a heterogeneous set of states potentially related to early embryonic cell types. For example, recent evidence identified a subpopulation of cells that express Zscan4, potentially corresponding to the totipotent 2 cell (2C)-state (Falco et al., 2007; Macfarlan et al., 2012). This state is also associated with telomere-elongation, essential for long-term culture in vitro (Zalzman et al., 2010) (although Zscan4 has also been shown to be activated by DNA damage responses and PI3K signaling (Storm et al., 2014)). Furthermore, representing slightly later stages of development, both inner cell mass (ICM)-like and epiblast-like stages can be identified and distinguished in culture by the high or low expression, respectively, of a cluster of correlated genes that includes Rex1, Nanog, and Esrrb. The totipotent state, marked by Zscan4 expression, shows low Rex1/Nanog/Esrrb expression (Singer et al., 2014), potentially defining a sub-population among Rex1/Nanog/Esrrb-low cells. Finally, we identified a complementary sub-population within the Rex1/Nanog/Esrrb-high population, marked by expression of Tbx3. Tbx3 has been shown to destabilize pluripotency when lost or over-expressed. It also appears critical for mesendoderm specification, and its expression may change the global levels of DNA methylation in mouse ES cells (Dan et al., 2013; Ivanova et al., 2006; Lu et al., 2011; Niwa et al., 2009; Weidgang et al., 2013). However, it remains unclear how Tbx3 expression emerges dynamically from these states.
As described in the main text, to verify that changes in the expression levels of the marker genes corresponded to collective changes in expression levels of multiple genes, we performed RNA-seq on subpopulations of cells sorted using fluorescent reporters for the three marker genes described above. We sorted out Esrrb/Tbx3 negative (E−T−), Esrrb-positive/Tbx3-negative (E+T−), and Esrrb/Tbx3 positive (E+T+) cells, as well as Zscan4-positive (Z+) and –negative (Z−) cells, and observed hundreds of genes that were differentially expressed between these states (Fig. 3D), indicating that variations in marker gene expression do not simply reflect intrinsic noise, or fluctuations in the expression of individual genes, but rather indicate broad transcriptional changes. More specifically, compared with E+T− cells, E−T− cells expressed lower levels of pluripotency regulators (Fig 3Di), and higher levels of differentiation markers and signaling proteins (Fig 3Dii). In contrast, E+T+ cells showed reduced expression levels of signaling proteins and differentiation markers and increased levels of pluripotency genes compared to E+T−, suggesting that Tbx3 could mark a more pluripotent state. Moreover, we observed increased expression levels of 2C-associated genes like Tmem92, Tcstv3, Tdpoz3/4, and Zfp352 in the Zscan4-positive cells compared with the Zscan4-negative cells (Fig 3Diii). This result is consistent with Zscan4 marking the previously reported 2C-like state (Macfarlan et al., 2012).
Assigning cells to the pluripotent states
The probabilistic assignment of the Esrrb state is presented in Figure 2C and the STAR Methods. T+ state was defined as Tbx3 transcript counts larger than 15. A threshold was obtained by comparing transcript counts to the direct observation of the promoter activity of Tbx3 gene in individual ES cells that had a knock-in fluorescent reporter for both endogenous loci of the Tbx3 gene (see Fig S2Ci, S2D). Zscan4 expression levels were observed to be largely binary (Figure 3A). We used a threshold of 50 transcripts to assign cells to the Z+ state. Cells that were in the Z− and T+ states but were also in the E− state with a confidence level of at least 80% were assigned to the E−T+Z− state. We discarded any cells that were in the Z+ state but were also in the T+ state and/or the E+ state with a confidence level of 80% or higher (composing <1% of observed cells). The inference results were not sensitive to changes in the thresholds used in defining the pluripotency states (see Figure S3).
Inferring the transition rates
In Figure 4F, we directly inferred the transition rates from the two-cell correlation matrices and the population fractions of each state – without fitting – using the formalism in Box 1, and its generalization to non-uniform population fractions outlined in the STAR Methods. Each transition rate in Figure 4F is the rate that had the smallest statistical error of the three rates inferred for the same transition from the two-cell correlation matrices at distances u=1 to u=3. The statistical error of the inferred transition rates was computed by bootstrapping over individual colonies: we randomly selected with replacement the same number of colonies as in the original data set, with the probability of selecting a given colony proportional to the number of cells that it contained. KCA was performed on the resampled data 1000 times to estimate the variability in the inferred rates.
Self-consistency checks on the inferred transition dynamics
In this section, we validate the inferred ES cell state dynamics presented in the Results section of the main text and Figure 4.
First, the inference assumes that ES cell state dynamics are accurately represented as a stationary Markovian process with reversible transitions. If true, transition rates inferred from correlations between cells at different lineage distances (e.g. sisters vs. cousins) should produce the same result. In fact, this was observed within statistical error (Figure S5B).
Second, it is often of great biological interest to know whether a given transition is reversible or irreversible, for example in the context of “de-differentiation” (see main text, Introduction). To test whether the system exhibits irreversible transitions we next computed the three-cell correlations (Main Text, Box 1). Recall that reversible dynamics can be correctly inferred using only the two-cell correlation functions. For such dynamics, the three-cell correlations should not contain any additional information about the dynamics beyond that found in the two-cell correlations. Therefore, observing the two-cell correlations should be sufficient to predict the three-cell correlations. Indeed, the three-cell correlation functions measured on the observed trees were consistent with the predicted values, validating the reversibility of the transitions (Figure S6).
Third, we tested the specific qualitative prediction that Zscan4-positive cells are generated from E+T+Z− cells that inactivate Esrrb and Tbx3 in close succession right before activating Zscan4. To test this prediction, we acquired time-lapse movies of Esrrb and Tbx3 using the double reporter described above, and then used smFISH to measure the endpoint expression levels of Zscan4 in the same cells. In all cells where Zscan4 was high by smFISH at the movies’ end (11 trees had at least one Z+ cell), Esrrb and Tbx3 reporters were observed to turn off within a single cell cycle of one another, as predicted. Examples are shown in Figure S5D.
Fourth, we verified that, where they overlapped, our results agreed with previous work on ES cell dynamics. For example, the rates of transition between the high and low metastable states of Nanog or Zscan4 matched those observed from measurements of engineered reporter lines using re-equilibration following sorting or direct movie-based analysis (Chambers et al., 2007; Macfarlan et al., 2012; Miyanari and Torres-Padilla, 2012; Zalzman et al., 2010). Taken together, the self-consistency of the method, the direct experimental validation, and the agreement with previous work strongly support the notion that the KCA approach developed here can correctly infer state transition dynamics.
The two-state model of Esrrb Figure 2 is consistent with the inferred chain-like model in Figure 4
In the main text, we inferred the transition rates between Esrrb low (E−) and high (E+) states and showed that Esrrb switching dynamics is well approximated by a two-state Markovian process (Fig. 2). We also analyzed transitions across a more general network including 5 states, which is also well-approximated as a Markovian stochastic process (Fig. 4F). How can the same cellular dynamics be compatible with both models?
The existence of internal sub-states is not generally consistent with Markovian dynamics among the two Esrrb states. A two-state Markovian model implies that during any interval of time, there is a constant probability of transitioning from one state to another (for example, E− to E+). However, in the presence of sub-states, the probability of exiting the E− or E+ state will depend on which sub-state the cell is in. The five-state transition network need not be compatible with effective two-state Markovian dynamics for Esrrb. However, we show here that for the specific transition rates in this case, the two-state reduction of the five-state model is still well-approximated by a two-state Markovian model within the statistical limitations of our finite data sets. More specifically, we simulated the five-state model (Fig. 4F) for 14 trees of 4 generations (same number of trees and generations as the data in Fig. 2D,E). We then assigned states solely based on the Esrrb state, effectively ignoring the Tbx3 and Zscan4 expression levels. That is, E−T−Z− and E−T−Z+, and E−T+Z− cells were assigned to the E− state, while E+T−Z− and E+T+Z− cells were assigned to the E+ state. Note that this is equivalent to the measurements in Fig. 2, where only Esrrb expression level is used to designate cell state, with the expression levels of the other genes ignored.
We then used the KCA framework to infer the transition matrix of Esrrb dynamics. The simulations were repeated 10,000 times to estimate the statistical uncertainly of the inference. Within statistical error, the transition rates inferred from pairs of cells at all lineage distances from u=1 to u=4 were equivalent, consistent with a two-state Markovian model. In fact, we needed to measure more than 5,000 end-state cells before a statistically significant deviation from the two-state Markovian model could be observed. In that case, the five-state model fits the data significantly better than the two-state model. The key point is that although the five-state model is a more accurate description of Esrrb dynamics, by correctly accounting for the internal sub-states within the E− and E+ states, the two-state model remains approximately valid within the statistical limitations of our finite data set.
Potential benefits of chain-like state transition networks
The inferred cell state transition network in ES cells is a linear chain of 5 states. What implications does this type of transition network have for the maintenance of ES cells in culture, and for other systems that might utilize similar transition networks?
Previous work has suggested that to maintain a healthy population of ES cells in culture, each cell must transition through the E−T−Z+ state at some minimum frequency (Zalzman et al., 2010). This requirement was shown to be related to processes of telomere extension via telomere sister chromatid exchange (Zalzman et al., 2010) and global epigenetic resetting (Akiyama et al., 2015), which occur specifically in the Zscan4-positive state and helps maintain genomic stability and normal karyotype, and in turn the cell’s potential to proliferate. Based on this, we asked how the different stationary networks shown in Fig. 1D compare in the frequency with which cells visit the E−T−Z+ state.
To elucidate the differences between network architectures, we consider a simplified five state network where the last state (green color in Fig. S7) plays a role equivalent to that of the E−T−Z+ state. That is, the viability of each cell depends on the number of generations that have elapsed since it last occupied this state. The other four states (red color) have no bearing on viability. For each of the three networks, we set all inter-state transition rates to identical values, ensuring equal population fractions for each state (Fig. S7). Moreover, we selected the transition rates to ensure that the flux of the cells into the green state was the same for all three networks.
Every time a simulated cell visited the green state, we tabulated the number of generations elapsed since its most recent ancestor left that state. This represents the distribution of “waiting times” between consecutive visits to the green state. Since we selected the rates to ensure equal population fractions and equal fluxes into the green state, the mean waiting time for the three networks is identical.
Strikingly, however, the distributions of waiting times are very different for the different networks. In particular, the linear chain network results in a long-tailed distribution, where most cells return to the green state after a brief number of generations, but a relatively few cells spend a much larger amount time between consecutive visits, and therefore exhibit waiting times much longer than the average (Fig. S7A). The short waiting time for most cells is balanced by the exceptionally long waiting times of a minority. This is also reflected in the difference between the mean and median waiting time. By contrast, in the cycle motif (Fig. S7B), almost all cells spend approximately the same amount of time between consecutive visits to the green state (close to the mean waiting time). Finally, the all-to-all network (Fig. S7C) has a long-tailed waiting time distribution similar to that of the chain-like motif. However, the difference between its median and mean is not as pronounced.
What are the implications of these differences in waiting time distributions? In the chain, the relatively large discrepancy between the median and the mean implies that most cells would have relatively recently visited the Zscan4-positive state, increasing their viability. This is achieved at the expense of a relatively small fraction cells that experience significantly longer waiting times, and therefore presumably would show reduced viability. In this way, the chain-like network could be advantageous in the cell culture context. Similar effects could also make this type of network advantageous in other contexts where a system may need to optimize for the largest fraction of cells entering into a critical state over time.
DATA AND SOFTWARE AVAILABILITY
All the analysis software, including those used for movie tracking, FISH dot detection/counting, and KCA analysis is available upon request (see CONTACT FOR REAGENT AND RESOURCE SHARING). The data visualization package is also hosted on the Elowitz lab website: http://www.elowitz.caltech.edu/.
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE86417.
ADDITIONAL RESOURCES
The complete end-point FISH data and the associated lineage relationships is available on the Elowitz lab website (http://www.elowitz.caltech.edu/) for interactive viewing using a novel visualization tool, CellLines, developed by the Elowitz Lab in collaboration with the Caltech Data Visualization Program.
Supplementary Material
Acknowledgments
We thank Jordi Garcia-Ojalvo and David Sprinzak for helpful comments on the manuscript. We thank Fred Tan for construction of the Zscan4 reporter, and members of the Elowitz Lab for fruitful discussions. This work was supported by the National Institutes of Health grants R01HD075605A, R01GM086793A, and P50GM068763; the Weston Havens Foundation; Human Frontiers Science Program, and in part by the National Science Foundation under Grant No. NSF PHY11-25915. BIS also acknowledges support from NIH Grant R01-GM086793. This work is funded by the Gordon and Betty Moore Foundation through Grant GBMF2809 to the Caltech Programmable Molecular Technology Initiative.
Footnotes
Author Contributions
S.H., Z.S.S., and M.B.E. designed experiments. S.H. and Z.S.S. performed experiments and analyzed data. J.M.L. and Z.S.S. constructed cell lines with help from S.H.. Y.E.A. developed the movie tracking system. B.I.S. and M.B.E. supervised research. S.H., Z.S.S., and M.B.E. wrote the manuscript with substantial input from all authors.
Bibliography
- Ahrends R, Ota A, Kovary KM, Kudo T, Park BO, Teruel MN. Controlling low rates of cell differentiation through noise and ultrahigh feedback. Science. 2014;344:1384–1389. doi: 10.1126/science.1252079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Huch M, van Boxtel R, Karthaus W, Wedge DC, Tamuri AU, Martincorena I, Petljak M, Alexandrov LB, Gundem G, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canham MA, Sharov AA, Ko MSH, Brickman JM. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8:e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupé DP, Klein AM, Winton DJ, Simons BD, Jones PH. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015;16:57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Duffy KR, Wellard CJ, Markham JF, Zhou JHS, Holmberg R, Hawkins ED, Hasbold J, Dowling MR, Hodgkin PD. Activation-induced B cell fates are selected by intracellular stochastic competition. Science. 2012;335:338–341. doi: 10.1126/science.1213230. [DOI] [PubMed] [Google Scholar]
- Elowitz M, Levine A, Siggia E, Swain P. Stochastic Gene Expression in a Single Cell. Science. 2002;297:1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Evrony GD, Lee E, Mehta BK, Benjamini Y, Johnson RM, Cai X, Yang L, Haseley P, Lehmann HS, Park PJ, et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron. 2015;85:49–59. doi: 10.1016/j.neuron.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MSH. Zscan4: A novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Developmental Biology. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JCD, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb Is a Direct Nanog Target Gene that Can Substitute for Nanog Function in Pluripotent Cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipczyk A, Marr C, Hastreiter S, Feigelman J, Schwarzfischer M, Hoppe PS, Loeffler D, Kokkaliaris KD, Endele M, Schauberger B, et al. Network plasticity of pluripotency transcription factors in embryonic stem cells. Nat Cell Biol. 2015;17:1235–1246. doi: 10.1038/ncb3237. [DOI] [PubMed] [Google Scholar]
- Friedman N, Cai L, Xie XS. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys Rev Lett. 2006;97:168302. doi: 10.1103/PhysRevLett.97.168302. [DOI] [PubMed] [Google Scholar]
- Furusawa T, Ohkoshi K, Honda C, Takahashi S, Tokunaga T. Embryonic stem cells expressing both platelet endothelial cell adhesion molecule-1 and stage-specific embryonic antigen-1 differentiate predominantly into epiblast cells in a chimeric embryo. Biol Reprod. 2004;70:1452–1457. doi: 10.1095/biolreprod.103.024190. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic State Transitions Give Rise to Phenotypic Equilibrium in Populations of Cancer Cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, de Lopes SMCS, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormoz S, Desprat N, Shraiman BI. Inferring epigenetic dynamics from kin correlations. Proc Natl Acad Sci USA. 2015 doi: 10.1073/pnas.1504407112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nat Cell Biol. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He XS, Dekker CL, Zheng NY, Huang M, Sullivan M, et al. Lineage Structure of the Human Antibody Repertoire in Response to Influenza Vaccination. Sci Transl Med. 2013;5:171ra19–171ra19. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. - PubMed - NCBI. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar T, Lim C, Hayward P, Muñoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Arias AM. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Simons BD. Universal patterns of stem cell fate in cycling adult tissues. Development. 2011;138:3103–3111. doi: 10.1242/dev.060103. [DOI] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RM, Cahan P, Shalek AK, Satija R, DaleyKeyser AJ, Li H, Zhang J, Pardee K, Gennert D, Trombetta JJ, et al. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature. 2014;516:56–61. doi: 10.1038/nature13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder K, Pitter K, Laplant Q, Hambardzumyan D, Ross BD, Chan TA, Holland EC, Michor F. Mathematical modeling of PDGF-driven glioblastoma reveals optimized radiation dosing schedules. Cell. 2014;156:603–616. doi: 10.1016/j.cell.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee REC, Walker SR, Savery K, Frank DA, Gaudet S. Fold Change of Nuclear NF-κB Determines TNF-Induced Transcription in Single Cells. Mol Cell. 2014;53:867–879. doi: 10.1016/j.molcel.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- Lu R, Yang A, Jin Y. Dual functions of T-box 3 (Tbx3) in the control of self-renewal and extraembryonic endoderm differentiation in mouse embryonic stem cells. Journal of Biological Chemistry. 2011;286:8425–8436. doi: 10.1074/jbc.M110.202150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Xue Q, Eisele MR, Sulistijo ES, Brower K, Han L, Amir EAD, Peer D, Miller-Jensen K, Fan R. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc Natl Acad Sci USA. 2015;112:E607–E615. doi: 10.1073/pnas.1416756112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat Meth. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Meth. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Göttgens B, Niwa H, Smith A. Esrrb Is a Pivotal Target of the Gsk3/Tcf3 Axis Regulating Embryonic Stem Cell Self-Renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Norman TM, Lord ND, Paulsson J, Losick R. Memory and modularity in cell-fate decision making. Nature. 2013;503:481–486. doi: 10.1038/nature12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Huber W, Tsumura A, Kang M, Xenopoulos P, Kurimoto K, Oleś AK, Araúzo-Bravo MJ, Saitou M, Hadjantonakis AK, et al. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat Cell Biol. 2014;16:27–37. doi: 10.1038/ncb2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Peccoud J, Ycart B. Markovian modeling of gene-product synthesis. Theoretical Population Biology. 1995;48:222–234. [Google Scholar]
- Poloni A, Maurizi G, Leoni P, Serrani F, Mancini S, Frontini A, Zingaretti MC, Siquini W, Sarzani R, Cinti S. Human dedifferentiated adipocytes show similar properties to bone marrow-derived mesenchymal stem cells. Stem Cells. 2012;30:965–974. doi: 10.1002/stem.1067. [DOI] [PubMed] [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Meth. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA Synthesis in Mammalian Cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Müller-Wieland D, Pfeiffer A, Krone W, Kahn CR. Obesity associated with a mutation in a genetic regulator of adipocyte differentiation. N Engl J Med. 1998;339:953–959. doi: 10.1056/NEJM199810013391403. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Cox BJ, Lanner F, Sharma P, Ignatchenko V, McDonald ACH, Garner J, Gramolini AO, Rossant J, Kislinger T. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev Cell. 2012;22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, Ueda HR. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14:R31. doi: 10.1186/gb-2013-14-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons BD, Clevers H. Strategies for Homeostatic Stem Cell Self-Renewal in Adult Tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Singer ZS, Yong J, Tischler J, Hackett JA, Altinok A, Surani MA, Cai L, Elowitz MB. Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol Cell. 2014;55:319–331. doi: 10.1016/j.molcel.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Slack JM, Tosh D. Transdifferentiation and metaplasia--switching cell types. Current Opinion in Genetics & Development. 2001;11:581–586. doi: 10.1016/s0959-437x(00)00236-7. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010 doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Sokolik C, Liu Y, Bauer D, McPherson J, Broeker M, Heimberg G, Qi LS, Sivak DA, Thomson M. Transcription factor competition allows embryonic stem cells to distinguish authentic signals from noise. Cell Systems. 2015;1:117–129. doi: 10.1016/j.cels.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda J, Suda T, Ogawa M. Analysis of differentiation of mouse hemopoietic stem cells in culture by sequential replating of paired progenitors. Blood. 1984a;64:393–399. [PubMed] [Google Scholar]
- Suda T, Suda J, Ogawa M. Single-cell origin of mouse hemopoietic colonies expressing multiple lineages in variable combinations. Proc Natl Acad Sci USA. 1983;80:6689–6693. doi: 10.1073/pnas.80.21.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Suda J, Ogawa M. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc Natl Acad Sci USA. 1984b;81:2520–2524. doi: 10.1073/pnas.81.8.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Yamazaki S, Yamaguchi T, Okabe M, Masaki H, Takaki S, Otsu M, Nakauchi H. Generation of engraftable hematopoietic stem cells from induced pluripotent stem cells by way of teratoma formation. Mol Ther. 2013;21:1424–1431. doi: 10.1038/mt.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- van den Berg DLC, Zhang W, Yates A, Engelen E, Takacs K, Bezstarosti K, Demmers J, Chambers I, Poot RA. Estrogen-Related Receptor Beta Interacts with Oct4 To Positively Regulate Nanog Gene Expression. Mol Cell Biol. 2008;28:5986–5995. doi: 10.1128/MCB.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo LT, Daley GQ. De novo generation of HSCs from somatic and pluripotent stem cell sources. Blood. 2015;125:2641–2648. doi: 10.1182/blood-2014-10-570234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Organisers & genes. The University Press; 1940. [Google Scholar]
- Wagenblast E, Soto M, Gutiérrez-Ángel S, Hartl CA, Gable AL, Maceli AR, Erard N, Williams AM, Kim SY, Dickopf S, et al. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature. 2015 doi: 10.1038/nature14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidgang CE, Russell R, Tata PR, Kühl SJ, Illing A, Müller M, Lin Q, Brunner C, Boeckers TM, Bauer K, et al. TBX3 Directs Cell-Fate Decision toward Mesendoderm. Stem Cell Reports. 2013;1:248–265. doi: 10.1016/j.stemcr.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Belmonte JCI. Dynamic Pluripotent Stem Cell States and Their Applications. Stem Cell. 2015;17:509–525. doi: 10.1016/j.stem.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.