Abstract
Background
Multiple testing to understand global changes in gene expression based on genetic and epigenetic modifications is evolving. Chorionic villi, obtained for prenatal testing, is limited, but can be used to understand ongoing human pregnancies. However, optimal storage, processing and utilization of CVS for multiple platform testing has not been established.
Results
Leftover CVS samples were flash-frozen or preserved in RNAlater. Modifications to standard isolation kits were performed to isolate quality DNA and RNA from samples as small as 2–5 mg. RNAlater samples had significantly higher RNA yields and quality and were successfully used in microarray and RNA-sequencing (RNA-seq). RNA-seq libraries generated using 200 vs. 800 ng RNA showed similar biological coefficients of variation. RNAlater samples had lower DNA yields and quality, which improved by heating the elution buffer to 70°C. Purification of DNA was not necessary for bisulfite-conversion and genome-wide methylation profiling. CVS cells were propagated and continue to express genes found in freshly-isolated chorionic villi.
Conclusions
CVS samples preserved in RNAlater are superior. Our optimized techniques provide specimens for genetic, epigenetic and gene expression studies from a single small sample which can be used to develop diagnostics and treatments using a systems biology approach in the prenatal period.
Keywords: multiple platform testing, chorionic villi, CVS, RNAseq, DNA methylation, RNA, DNA, pregnancy, prenatal diagnostics
Introduction
Multiple platform testing, including RNA sequencing for gene expression profiles, CpG methylation arrays for epigenetic patterns, and genome-wide platforms to assess genetic contributions for single specimens can lead to a better understanding of the global genetic and epigenetic contributions leading to gene expression changes that contribute to disease. Limitations exist, as precious direct CVS specimens are small, frequently permitting only a single tiered approach to understand the complexities of biologic systems, with different specimens being utilized for each platform utilized. This limits the ability to control for specimen variability and understand how genetic and methylation changes are directly affecting gene expression.
Currently, chorionic villus sampling (CVS), used for prenatal diagnosis, determines genetic abnormalities in an ongoing pregnancy, early in gestation, at 10–13 weeks (1). Genetic testing is performed either by traditional g-banding karyotyping, fluorescence in situ hybridization (FISH) or microarray as well as PCR for single gene disorders (2–4). Despite advances, including the identification of microdeletions and copy number variants (5, 6), testing for disorders and adverse outcomes that manifest either through posttranslational modifications or gene expression changes have not been well studied, and may play crucial roles in pregnancy outcomes (7). For example, epigenetic regulation through DNA methylation, is a key factor by which environmental cues can regulate health and disease (8, 9). The most widely studied epigenetic phenomenon in the placenta is genomic imprinting (10). A number of imprinted genes are expressed during the preimplantation period, which may be vulnerable to disruption by environmental cues (11, 12), but the majority of studies to date have been performed later, using term placental and cord blood samples. Thus, we developed protocols and techniques that enabled us to properly store chorionic villi and isolate DNA and RNA of sufficient quantity and quality for use in multiple platform testing for epigenetic and gene expression studies. This represents a new method for identifying posttranslational mechanisms underlying adverse pregnancy outcomes early in the first trimester as a potential new means of prenatal diagnostic testing, through multiple platform testing. In addition, optimization techniques may also provide benefits to clinical laboratories by minimizing culture requirements for molecular based prenatal diagnostic testing, except when culture is required for cytogenetic karyotype testing.
Amniotic fluid is currently used for diagnostic testing for fetal well being such as assessment of fetal lung maturity (13–17). However, there are some disease states that are associated with placental function, such as IUGR and preeclampsia (18, 19) that affect fetal well being and CVS may be a way to understand fetal states early in gestation leading to the development of new diagnostic tools for noninvasive studies. One potential factor that may affect diagnostics using CVS is confined placental mosaicism (CPM) which is derived from errors specific to the outer layer of the blastocyst. However, we found that the overall the prevalence is only 0.9% (20) and CPM in general is also associated adverse fetal outcomes including growth retardation, poor fetal outcome, increased rate of miscarriage, and perinatal morbidity/mortality (21–26) which ultimately may be used for diagnostics of fetal well being.
In addition we explored the maintenance of cultured cells from CVS samples that can be used for future functional studies, similar to the HTR-8/SVneo cell line (27). We demonstrated that we can continue to passage CVS cells and they maintain expression of key genes expressed in trophoblast cells. Thus, these cells can potentially be used for functional studies which will be critical for studying the effects of altered gene expression on first trimester trophoblast function. Our approach could form the basis for personalized studies of the functional effects due to changes in genetics, epigenetics and gene expression patterns for the development of precision medicine in the prenatal stage.
Methods
Chorionic Villus Sample Procurement
Chorionic villus sampling (CVS) was performed at 11–13 weeks of gestation. Tissue not needed for genetic testing was collected under an IRB-approved protocol. Samples were flash-frozen in liquid nitrogen or placed in RNAlater RNA Stabilization Reagent (Qiagen, Valencia CA) and then stored at −80°C in our Prenatal Biorepository.
DNA and RNA Isolation from CVS samples
Genomic DNA and RNA were isolated from archived CVS samples that had been either flash-frozen or stored in RNAlater. For samples with 5–15 mg of villous tissue, RNA and DNA were isolated using AllPrep DNA/RNA Mini Kits (Qiagen). CVS samples were thawed on ice and 600 μl buffer RLT Plus with β-mercaptoethanol was added to each sample. The samples were homogenized by passing them at least 10 times through a 22-gauge needle fitted to an RNase-free syringe; at least 20 times through a 25-gauge needle; and then at least 10 times through a 27-gauge needle. The homogenates were then loaded onto AllPrep DNA spin columns and the remainder of the protocol was performed per the manufacturer’s instructions. RNAs were eluted using 30 μl RNase-free water, and the eluates were placed back on the columns and the elution process repeated a second time to increase RNA yields. For elution of DNAs, elutions were initially performed using 100 μl of room-temperature elution buffer (Buffer EB) for each elution. The eluates were placed back on the columns and the elution process repeated a second time to increase yields. In later experiments, the elution buffer was preheated to 70°C and 100 μl of preheated Buffer EB was used for each elution.
For samples with 2–5mg of villous tissue, the AllPrep DNA/RNA Micro Kit (Qiagen) was used. CVS samples were homogenized in 350 μl buffer RLT Plus with β-mercaptoethanol by passing them through 22-gauge, 25-gauge and 27-gauge needles as described above. The remainder of the protocol was followed per the manufacturer’s instructions. RNAs were eluted using 15 μl RNase-free water, and the eluates were passed through the columns a second time to increase RNA yields. DNA elutions were performed using 50 μl of Buffer EB which had been preheated to 70°C, and the eluates were put back on the columns and the elution process repeated to maximize DNA.
Sample Selection
Different samples were used when comparing RNAlater to flash-frozen specimens. Following superior RNA isolation with samples stored in RNAlater, subsequent experiments comparing heat elution versus no heat elution were performed using the RNAlater isolated samples. For purification studies, the same samples were compared pre and post purification.
Microarray and RNA sequencing analysis of gene expression
Microarray analysis was performed on RNAs isolated from CVS samples from 3 different pregnancy groups (N=3 per group) using Affymetrix Human Gene 1.0 ST Arrays. Results were analyzed using Partek Genomics Suite 6.5. Criteria for filtering differentially expressed genes (DEGs) was a 2-fold difference or greater change in gene expression plus group t-test significance at p<0.05.
RNA sequencing (RNA-Seq) libraries were constructed using Illumina TruSeq Stranded Total RNA LT with Ribo-zero kits (P/N RS-122-2201, Illumina, San Diego). For samples, libraries were generated using two different RNA amounts (200 ng and 800 ng respectively). The 12 libraries were then pooled at a 4 nM concentration and 2 × 75 bp paired-end reads were generated using Illumina NextSeq 500 using High Output 150-cycle flow cells (P/N FC-404-1002). The pooled libraries were run on three independent flow cells.
Methylation Profiling
Genomic DNA was isolated from CVS samples as described. Some DNAs underwent additional purification by ethanol precipitation. Genomic DNAs (500ng) were bisulfite-converted and submitted to genome-wide profiling using the Infinium HumanMethylation450K BeadChip (Illumina). Detection P values (<0.01) were calculated in the GenomeStudio Methylation module. Plate and batch effects were assessed visually by Principal Component Analysis (PCA), and data were normalized using the Chip Analysis Methylation Pipeline (ChAMP) package (implemented in R).
Culture of CVS cells and the HTR-8/SVneo cell line
CVS cells grown in culture for clinical genetic testing are kept for an additional 35 days. After this time period, these cells were obtained and cultured in Chang Medium In Situ (Irvine Scientific, T-104, Santa Ana, CA, USA). Cells were initially fed with fresh medium every 24 hours and were passaged at ~70% confluency three times and then harvested for RNA isolation.
The immortalized human first trimester trophoblast cell line HTR-8/SVneo, used as an in vivo model to study trophoblast invasion and migration (28–30), was provided by Dr. C. Graham (Queen’s University, Kingston, Ontario, Canada). HTR-8/SVneo cells were cultured in RPMI 1640 with Hepes buffer and L-Glutamine (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were plated on 6-well plates (Corning Life Sciences, Tewksbury, MA) at concentration of 2.5 × 105 cells/ml and grown until they reached ~80% confluency and then harvested for RNA isolation.
RNA isolation and reverse transcription PCR (RT-PCR) analysis for cultured cells
Total RNA was extracted from cultured CVS or HTR-8 cells using RNeasy minikits (Qiagen) according to the manufacturer’s instructions. Total RNA from CVS samples, cultured CVS cells or cultured HTR-8 cells was then used for cDNA synthesis. Briefly, 250 ng – 1 μg of RNA was reverse-transcribed using the iScript cDNA Synthesis kit (Bio-Rad Laboratories). The expression of FOS, END, HTRA1, INHBA, and TGFBI was then confirmed by PCR, using 18S RNA as a control. PCR was performed on an Eppendorf Mastercycler Gradient (Eppendorf International), using Taq DNA polymerase (Qiagen) and 1 μl of the RT reaction in a 25 μl PCR reaction. The PCR conditions were as follows: initial denaturation at 94°C for 3 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, extension at 72°C for 1 min, followed by final extension at 72°C for 10 min. The primers for human FOS, END, HTRA1, INHBA, TGFBI, and 18S RNA are listed in Table 1.
Table 1.
Primer sequence used for real-time PCR (RT-PCR) analyses.
| Gene Symbol | Sequence (5–3′) |
|---|---|
| FOS | F: TGTCTGTGGCTTCCCTTGATCTGA R: TGGATGATGCTGGGAACAGGAAGT |
| END | F: GGTGACAAGTTTGTCTTGCGCAGT R: ACAGGATATTGACCACCGCCTCAT |
| HTRA1 | F: ATTGACCCATAGGCAGAGGCATGA R: AAGAAGGATTCTCTTGGCCAGGGA |
| INHBA | F: ACATCGGCTGGAATGACTGGATCA R: AGCACGATTTGAGGTTGGCAAAGG |
| TGFBI | F: GTCCACAGCCATTGACCTTT R: ACCGCTCACTTCCAGAGAGA |
| 18S RNA | F: TCAACTTTCGATGGTAGTCGCCGT R: TCCTTGGATGTGGTAGCCGTTTCT |
Results
RNA isolation from CVS samples
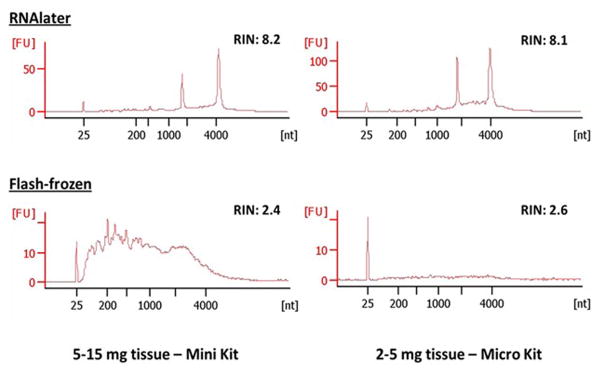
CVS samples with 2–5 mg tissue and 5–15 mg tissue that were flash-frozen or preserved in RNAlater (Qiagen) and stored at −80°C were tested for the quality of obtainable RNA and DNA. For samples with 5–15 mg tissue, RNA was isolated with AllPrep DNA/RNA Mini Kits (Qiagen) using our modified protocol (detailed in the Methods). Due to the consistency and small amount of tissue, homogenization of the tissue was necessary by passing them through increasing needle gauges, otherwise RNA yield was poor (data not shown). The yields and quality of RNAs isolated from 9 flash-frozen vs. 20 RNAlater samples were compared. An average RNA yield of 5.6 μg was obtained from flash-frozen CVS samples, compared to a significantly higher average RNA yield of 18 μg from RNAlater preserved CVS samples (Table 2). The average RNA Integrity Number (RIN) obtained from CVS samples preserved in RNAlater was 7.7 (range 6.6 to 9.8) (Table 2 and Figure 1). For flash-frozen CVS samples, several of the RNAs were too low in quality to produce a measureable RIN, but for those measured, the average RIN was significantly lower at 2.5 (Table 2 and Figure 1).
Table 2.
Quantities and qualities of RNAs isolated from CVS samples preserved in RNAlater vs. flash-frozen.
| RNA later | Frozen | P-value* | |
|---|---|---|---|
| 5–15 mg | (n =20) | (n = 9) | |
| RNA conc (ng/μl) | 621 ± 284 | 188 ± 231 | 0.0007 |
| RNA yield (ng) | 18,624 ± 8,517 | 5,650 ± 6,923 | 0.0007 |
| RIN | 7.7 ± 0.86 | 2.5 ± 1.1 | 0.0001 |
| 2–5 mg | (n = 7) | (n = 9) | |
| RNA conc (ng/μl) | 342 ± 278 | 28.4 ± 32.1 | 0.0008 |
| RNA yield (ng) | 5,715 ± 3,733 | 443 ± 469 | 0.0008 |
| RIN | 7.7 ± 0.30 | 3.0 ± 1.2 | 0.0027 |
Mann-Whitney test
Figure 1.
Qualities of RNAs isolated from CVS samples preserved in RNAlater vs. flash-frozen.
For samples with 2–5 mg tissue, 9 flash-frozen samples were compared to 7 RNAlater samples. RNA was isolated with an AllPrep DNA/RNA Micro Kit (Qiagen), using our modified protocol (detailed in the Methods). An average RNA yield of 443 ng was obtained from flash-frozen CVS samples, whereas a significantly higher average yield of 5.7 μg was obtained from RNAlater preserved CVS samples (Table 2). The average RIN obtained from RNAlater preserved CVS samples was 7.7 (range 7.3 to 8.1) (Table 2 and Figure 1). Again, some of the RNAs from flash-frozen CVS samples were too low in quality to produce a measureable RIN, but for those measured, the average RIN was significantly lower at 3.0 (Table 2 and Figure 1).
Utility of isolated RNA in gene expression studies
The utility of the higher-quality RNAs that were isolated from RNAlater samples was tested using gene expression microarrays and RNA sequencing. Microarray analysis was performed on RNAs isolated from CVS samples from 3 different pregnancy groups using Affymetrix Human Gene 1.0 ST Arrays, and the results were analyzed using Partek Genomics Suite 6.5. Using a cutoff of a 2-fold difference or greater change in gene expression plus group t-test significance at p<0.05, a number of DEGs were identified in the different pregnancy groups.
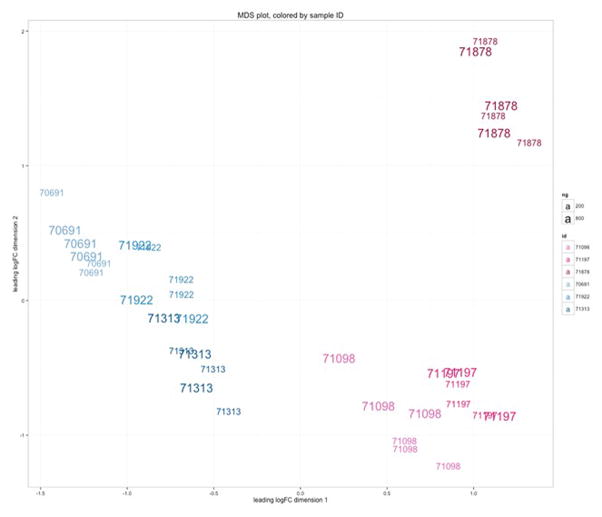
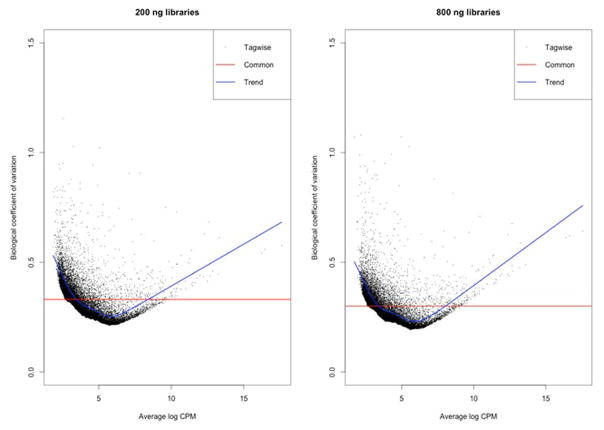
RNA sequencing (RNA-seq) was performed using six samples, and libraries were generated for each of these samples using two different RNA amounts (200 ng and 800 ng respectively). The total number of fragments sequenced and percent of fragments mapping to the hg19 transcriptome between RNA-seq libraries generated using 200 versus 800 ng of total RNA were highly similar (Supporting Data), despite variation in sequence yield and accuracy across the three flowcell sequencing runs. Multi-dimensional scaling plots confirmed this observation and revealed that the variation in expression due to library preparation was smaller than the effects of sample and sex (Figure 2; MDS plot). We then calculated the biological coefficient of variation (BCV; a measure of the biological variability of an individual gene) for all hg19 RefSeq genes across the 200 ng and 800 ng libraries. We observed a 2.5% increase in BCV in the 200 ng libraries (Figures 3–4). The difference was small relative to the mean BCV in both library types (30% for 800 ng, 33% for 200 ng, Figure 4a) and was consistent across both lowly and highly variable genes (Figure 4b).
Figure 2.
Multi-dimensional scaling (MDS) plot demonstrating that samples separate primarily by sex (males in blue on the left, females in pink on the right); then by sample identity; then by library prep (small vs. large numbers for 200 ng and 800 ng preps. respectively). The same 3 male and 3 female samples were used comparing 200 ng and 800 ng preps from 3 different flow cells.
Figure 3.
Plots of biological coefficient of variation (BCV) for 200 ng and 800 ng library preparations from the same samples.
Figure 4.
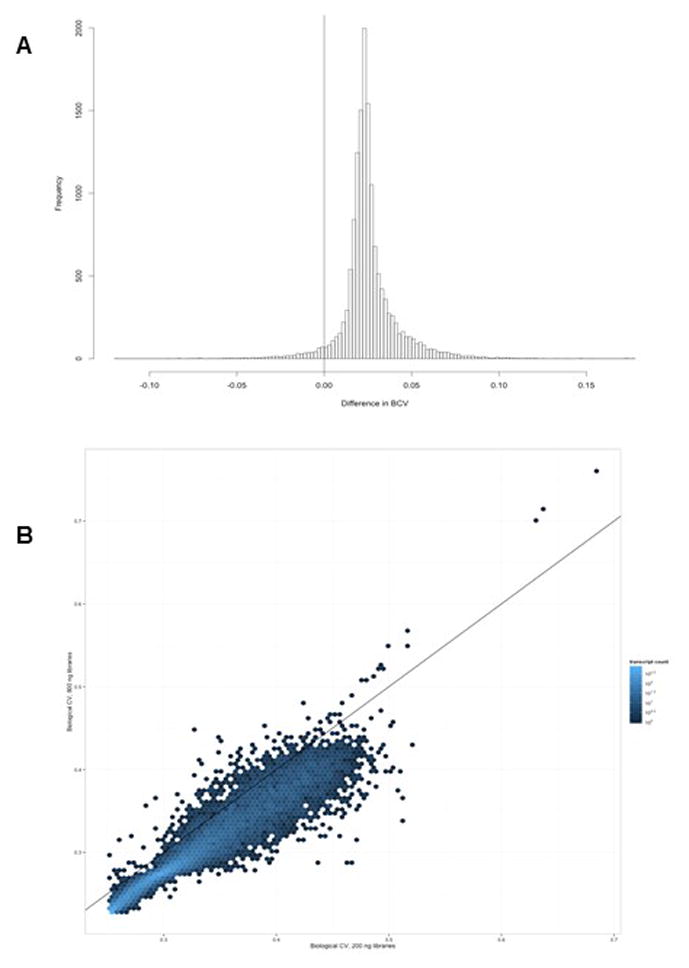
Variations in BCV in 200 ng vs. 800 ng library preparations from the same samples.
DNA isolation from CVS samples
For the CVS samples methods for DNA isolation were evaluated. For samples with 5–15 mg tissue, DNA was isolated with AllPrep DNA/RNA Mini Kits (Qiagen) using a modified protocol (detailed in the Methods). Initially for RNAlater preserved CVS samples, DNAs isolated using room temperature elution buffer resulted in average DNA yields of 1.74 μg (Table 3). The protocol was modified by preheating the elution buffer to 70°C. A significant increase in the average DNA yields to 4.44 μg was seen when comparing 8 samples without heat elution to 12 separate samples with heat elution (Table 3). For flash-frozen CVS, all elutions were performed using heated elution buffer, an average yields of 10.38 μg were obtained (data not shown). With respect to DNA quality, average A260/A280 and A260/A230 ratios of 1.86 and 0.06 respectively were obtained for samples preserved in RNAlater when room-temperature elution buffer was used, and these increased significantly to 1.94 and 0.11 respectively when heated elution buffer was used (Table 3). For flash-frozen CVS samples, all were isolated using heated elution buffer, average A260/A280 and A260/A230 ratios of 1.91 and 0.46 respectively were obtained (data not shown).
Table 3.
Quantities and qualities of DNAs isolated from CVS samples without and with heat elution.
| RNA later No heat elution | RNA later Heat elution | P-value* | |
|---|---|---|---|
| 5–15mg | (n = 8) | (n = 12) | |
| DNA conc (ng/μl) | 38.7 ± 30.1 | 50.5 ± 21.2 | 0.0372 |
| DNA yield (ng) | 1,740 ± 1,356 | 4,441 ± 2,010 | 0.0026 |
| A260/280 | 1.86 ± 0.11 | 1.94 ± 0.10 | 0.0274 |
| A260/230 | 0.06 ± 0.04 | 0.11 ± 0.06 | 0.0112 |
| 2–5mg | (n = 7) | ||
| DNA conc (ng/μl) | 56.7 ± 33,7 | ||
| DNA yield (ng) | 2,834 ± 1,684 | ||
| A260/280 | 1.92 ± 0.06 | ||
| A260/230 | 1.57 ± 1.23 |
Mann-Whitney test
For CVS samples with 2–5 mg tissue, DNA was isolated with an AllPrep DNA/RNA Micro Kit (Qiagen), using a modified protocol (detailed in the Methods) using preheated elution buffer. Average DNA yields were 2.83 μg for RNAlater preserved CVS samples (Table 3), and 5.07 μg for flash-frozen samples (data not shown). Average A260/A280 and A260/A230 ratios were 1.92 and 1.57 respectively for RNAlater preserved samples (Table 3), and 1.97 and 2.93 respectively for flash-frozen samples (data not shown).
Utility of isolated DNA in epigenetic studies
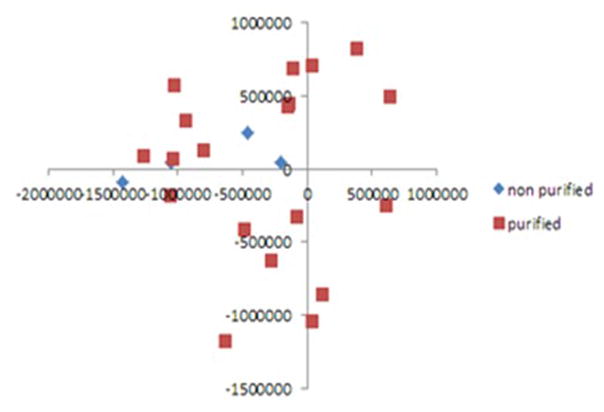
The utility of isolated DNA was tested in methylation profiling. As the A260/A230 ratios initially obtained were below those recommended for methylation profiling, 3 μg of selected DNAs were further purified by ethanol precipitation in 15 samples. For these samples, the average A260/A230 ratio improved from 0.31 to 2.46 for samples with 5–15 mg of starting tissue (Table 4). However, the average sample recovery was only 37% (Table 4), leaving less than the minimum 500 ng required for methylation profiling for some samples. For 15 samples with 2–5 mg of starting material, the average A260/A230 ratio did not improve from 2.36 to 2.30 but the average sample recovery was only 46%, Therefore, we compared the utility of purified and non-purified DNAs from flash-frozen and RNAlater samples. Purified and non-purified genomic DNAs from flash-frozen CVS samples and RNAlater preserved samples all underwent bisulfite conversion. All samples had >99.9% call rate, and two independent technical replicates were clustered tightly. Importantly, purified samples (n = 18) did not segregate from the same samples that were non-purified(n=3) on PCA analysis (Figure 5), and methylation patterns in purified and non-purified DNAs from the same samples were highly similar (Figure 6).
Table 4.
Effect of ethanol purification on DNA yields and quality.
| Before Purification | After Purification | P-value * | |
|---|---|---|---|
| 5–15mg | (n = 15) | (n = 15) | |
| DNA yield (ng) | 7760±5151 | 2207±731 | 0.0007 |
| A260/280 | 1.93±0.091 | 1.85±0.097 | 0.0258 |
| A260/230 | 0.31±0.20 | 2.46±0.68 | 0.0007 |
| 2–5mg | (n = 15) | (n = 15) | |
| DNA yield (ng) | 4086 ± 3038 | 1672 ± 946 | 0.0007 |
| A260/280 | 1.95 ± 0.86 | 1.86 ± 0.063 | 0.004 |
| A260/230 | 2.36 ± 2.12 | 2.30 ± 0.40 | 0.38 |
Wilcoxon sign rank test for paired non-parametric data
% recovery for 5–15mg = 37%
% recovery for 2–5mg = 46%
Figure 5.
Principal component analysis (PCA) of 18 purified and 3 non-purified DNA samples.
Figure 6.
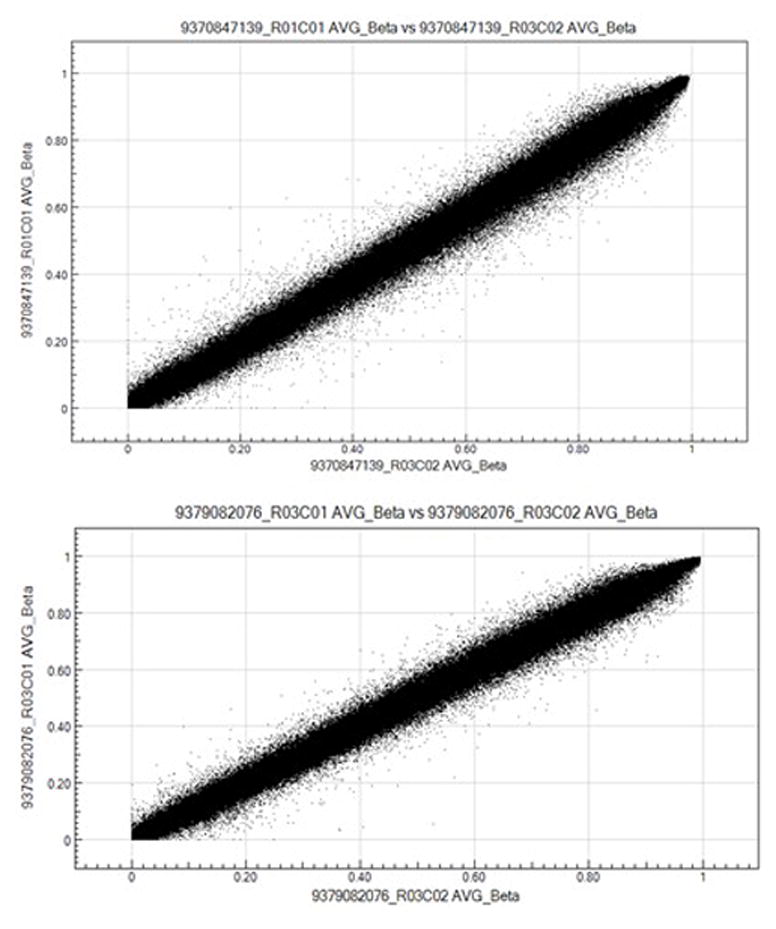
Scatter plots depicting the correlation coefficients for β methylation values from the same DNA samples with and without ethanol purification. Top panel: DNA from a female subject with and without ethanol purification, r2=0.9921. Bottom panel: DNA from a male subject with and without ethanol purification, r2=0.9933.
As purification of genomic DNA was not necessary for methylation profiling, we performed genome-wide methylation profiling on unpurified genomic DNAs from 3 different pregnancy groups using the Infinium HumanMethylation450K BeadChip. Detection P values (<0.01) were calculated in the GenomeStudio Methylation module. Plate and batch effects were assessed visually by Principal Component Analysis (PCA), and data were normalized using the Chip Analysis Methylation Pipeline (ChAMP) package (implemented in R). Significantly differentially methylated loci were then analyzed using Ingenuity Pathway Analysis (IPA) and identified a number of genes to be differentially methylated (31).
Utility of cultured CVS cells for future functional studies
Gene expression analysis
We also tested whether CVS cells can be grown in culture long term and maintain expression of key genes expressed in trophoblast cells. Under appropriate conditions (Methods), CVS cells can be maintained for at least an additional 4 passages after completion of clinical genetic testing. We compared expression patterns of key genes involved in trophoblast growth and development in these cultured CVS cells to the patterns in isolated CVS samples and the HTR8/SVneo cell line (28–30). Similar expression of human FOS, END, HTRA1, INHBA and TGFBI was detected in isolated CVS samples, HTR8/SVneo cells and our cultured CVS cells, but the same pattern of expression was not seen in an unrelated tissue type (liver) (Figure 7). The results shown are representative of three different experiments from 3 different subjects. This demonstrates that these cells maintain a similar signature to isolated CVS and the HTR8/SVneo cells and thus have the potential to be used for future functional studies.
Figure 7.
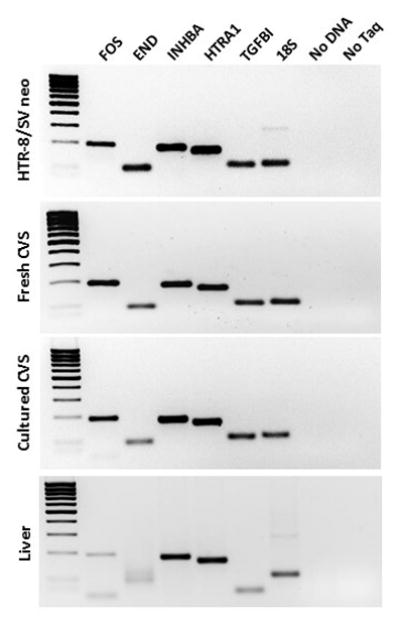
Gene expression patterns in the HTR-8/SVneo first trimester trophoblast cell line, a fresh CVS sample, and cultured CVS cells. Expression patterns of the same genes in liver are shown for comparison.
Discussion
We demonstrated that modifications to protocols can be used to isolate sufficient quantities of high-quality RNA and DNA for multiple array-based downstream applications including gene expression microarrays, RNA sequencing and methylation profiling, from small amounts of material, even as small as 2–5 mg, when properly preserved and stored. These refinements in techniques described for these precious small samples allows for prenatal diagnostic testing using a systems biology approach through multiple platform testing.
In addition, our findings demonstrate that utilization of leftover material following CVS can be used to study an ongoing human pregnancy and cultures derived from CVS samples can be grown for at least 4 additional passages and used for isolation of DNA and RNA, and the possibility for use in functional studies. These findings illustrate the importance of leftover CVS samples and cell cultures as a resource for research and additional prenatal diagnostic testing.
CVS samples are the earliest timepoint at which we can study first trimester placental trophoblasts in ongoing human pregnancies that can be followed to term. This is important as the biological foundation of pregnancy-associated pathologies is laid in the first trimester of pregnancy, as placentation occurs throughout the first trimester of pregnancy and aberrations leading to abnormal placentation are associated with adverse pregnancy outcomes (32–36). However, most studies of abnormal placentation have relied on term placental samples obtained at delivery to identify possible causative factors (32, 37), and it is unclear whether identified factors are involved in the early manifestations and actual development of disease, or are markers of disease progression. Our identification of techniques for the proper storage of CVS samples and isolation of RNA and DNA of sufficient quality and quantity for use in downstream gene expression and methylation profiling is significant as it allows for comparative analyses of samples from healthy pregnancies vs. those with specific adverse outcomes. It also provides for the identification of gene expression and methylation patterns during placentation in normal human pregnancies, rather than relying on animal models.
Techniques outlined may be utilized initially in the research setting to better understand placental function, particularly methylation changes and gene expression changes in the first trimester. We identified differences in methylation patterns in CVS samples from patients undergoing different types of fertility treatments and these patterns differ from those that are identified at term (31, 37, 38). Findings from research studies can ultimately be used to develop noninvasive diagnostic tools to understand placental function in normal and disease states, similar to already developed noninvasive diagnostics utilizing methylation alterations for identification of circulating fetal DNA for NIPT testing (39, 40). Furthermore, as fetal/placental RNA is also identified in the maternal circulation and associated with placental disease states, further identification of markers are necessary to identify disease early in gestation (18, 19).
Although high quality RNA can be obtained from multiple other tissues following flash freezing, leftover CVS samples yielded measurable quantities of RNA, however not of sufficient quality for use in downstream applications. In contrast, RNAlater preserved CVS samples yielded significantly higher-quality RNA that was successfully used for downstream microarray and RNA sequencing studies. In addition, average RNA yields from RNAlater preserved samples were significantly higher than flash-frozen samples. These differences in RNA yields and quality were observed regardless of whether the RNAs were isolated using the AllPrep DNA/RNA Mini or Micro Kit, supporting that preservation was the primary determinant of RNA yield and quality. Even samples with only 2 mg of villous tissue yielded over 1.5 μg of RNA, sufficient for microarray or RNA sequencing analysis, and should be preserved.
RNAs isolated from RNAlater preserved samples were successfully used in gene expression microarrays and RNAseq analyses, indicating that the quality of RNAs is sufficiently high for use in these applications. Given the small size of these samples, it is also encouraging that libraries built with as little as 200 ng of RNA could be successfully used for RNAseq. Although the mean BCV for the 200 ng libraries were slightly larger than that for the 800 ng libraries, the magnitude of the difference was around 0.025 BCV, which is less than 1/10th the average BCV seen in either library type, and was spread uniformly across the majority of the BCV spectrum and consistent across both lowly and highly variable genes. Overall, these data indicate that whole transcriptome RNAseq is feasible using 200 ng of total RNA from CVS samples, similar to quantities for formalin fixed paraffin-embedded (FFPE) tissue (41).
Although DNAs isolated from flash-frozen CVS samples, resulted in both higher average DNA yields and DNA quality (A260/A230 ratio) than RNAlater samples, our modifications, including preheating the elution buffer, significantly improved DNA yield and quality even from RNAlater samples, thus permitting us to use individual samples for multiple platform testing. Genomic DNAs from both flash-frozen and RNAlater samples successfully underwent bisulfite conversion and could be used for methylation profiling, suggesting slightly lower quality of DNAs from RNAlater samples did not significantly affect their utility. Given the importance of being able to isolate high quality RNA from CVS samples for downstream gene expression analyses, and the fact that preservation of RNAlater did not affect the utility of the isolated DNA, preservation in RNAlater is superior to flash-freezing. Thus, it continues to be important to consider proper initial storage of these precious samples.
An additional finding was that purification of DNA was not necessary. Purified and non-purified genomic DNAs all underwent bisulfite conversion and did not segregate on PCA analysis. This finding is extremely important as purification of the DNAs resulted in significant loss of DNA, with some less than the minimum 500 ng required to run the BeadChip after purification. In contrast, using non-purified samples allows us to perform BeadChip methylation analyses, potential repeat analyses and additional genomic testing. Given the relatively small number of samples available from pregnancies with specific adverse outcomes, this is tremendously important.
Another goal was to determine whether cultured trophoblasts from CVS samples can be maintained and have a similar signature to freshly isolated CVS cells and the HTR-8/SVneo cell line for functional studies in the future. We found that CVS cells kept for 35 days could be revived and maintained for an additional 4 passages. We also found key genes expressed in fresh CVS samples and the HTR8 first trimester trophoblast cell line showed the same pattern of expression in these cultured CVS cells. This finding suggests that it may be possible to use cultured CVS cells for functional studies of trophoblast migration, invasion and proliferation, and test how these may be affected by differences that occur early in gestation or assist in identification of pathways leading to adverse pregnancy outcomes. It might also be possible to differentiate these cells, similar to pluripotent stem cells or cytotrophoblast cells from term placenta (42, 43) to conduct functional studies in the future.
Conclusions
Our results demonstrate that CVS samples represent a tremendous resource for research and development of future diagnostics. When preserved in RNAlater, these samples can be used for the isolation of DNA and RNA of sufficient quality and quantity for multiple platform testing, including gene expression microarrays, RNA sequencing and methylation profiling, even with as little as 2 mg of villous tissue. These samples provide a window into the gene expression and epigenetic changes that occur in first trimester trophoblasts during the time when placentation occurs in human pregnancies that can be followed to term and beyond. Although CVS is clinically utilized for genetic testing, and there have been some studies demonstrating the utilization of CVS samples for either differential methylation or gene expression changes (44–46) none have been performed to utilize even small amounts of unused villi for multiple platform testing. Our refined techniques outlined give us the ability to conduct genetic, epigenetic and gene expression studies on a single sample. With the advent of personalized medicine, multiple platform testing, and the ability to culture trophoblasts from individual CVS samples for future functional studies, proper storage and processing with techniques outlined could be now used as a research tool and may be developed further for diagnostics using a systems biology approach and potentially for treatment of conditions that may manifest early in gestation.
Supplementary Material
What’s already known about this topic?
Chorionic villus sampling (CVS) tissue is used for diagnostic testing of genetic disorders.
Due to sample size, CVS specimens are limited to single platform testing.
What does this study add?
Our refined techniques outlined for proper storage and processing of direct CVS specimens gives us the ability to conduct genetic, epigenetic and gene expression studies on a single sample that can be used for the development of diagnostics and potentially treatments using a systems biology approach for conditions that manifest early in gestation.
Acknowledgments
Grant support. The research was supported the NICHD of the National Institutes of Health under award number R01HD074368, the American Society for Reproductive Medicine, the Helping Hand of Los Angeles, Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of Interest: none
Conference presentation: The Human Placenta Project (HPP) - Understanding Human Placental Structure and Function in Real Time, Incorporating omics and imaging into the HPP, 2nd Annual meeting. NIH, Bethesda, MD. April 2015.
Competing Interests: The authors declare no competing interests.
Authors’ Contributions:
MA carried out the tissue culture studies and drafted the manuscript; BL conducted the gene expression studies; GB carried out the tissue isolation techniques and devised the optimization of the techniques; NX and MG carried out the bisulfite conversion and methylation profiling; ER participated in the design of the study and performed the statistical analysis; AM conducted the RNA sequencing analysis; CF conducted the RNA sequencing; SR, JR, YC and SG participated in revising the manuscript critically for important intellectual content; MP conceived the study, and participated in its design and coordination and helped to draft and revised the manuscript. All authors read and approved the final manuscript.
The data set(s) supporting the results of this article is(are) included within the article.
References
- 1.Xanthopoulou AG, Anagnostopoulos AK, Thanasopoulou A, et al. The proteome of normal human chorionic villus sampling cells. In Vivo. 2011;25(6):945–61. [PubMed] [Google Scholar]
- 2.Dekker GA, Sibai BM. Etiology and pathogenesis of preeclampsia: current concepts. Am J Obstet Gynecol. 1998;179(5):1359–75. doi: 10.1016/s0002-9378(98)70160-7. [DOI] [PubMed] [Google Scholar]
- 3.Page NM, Kemp CF, Butlin DJ, et al. Placental peptides as markers of gestational disease. Reproduction. 2002;123(4):487–95. doi: 10.1530/rep.0.1230487. [DOI] [PubMed] [Google Scholar]
- 4.Keelan JA, Blumenstein M, Helliwell RJ, et al. Cytokines, prostaglandins and parturition--a review. Placenta. 2003;24(Suppl A):S33–46. doi: 10.1053/plac.2002.0948. [DOI] [PubMed] [Google Scholar]
- 5.Yatsenko SA, Davis S, Hendrix NW, et al. Application of chromosomal microarray in the evaluation of abnormal prenatal findings. Clin Genet. 2013;84(1):47–54. doi: 10.1111/cge.12027. [DOI] [PubMed] [Google Scholar]
- 6.Chu T, Yeniterzi S, Rajkovic A, et al. High resolution non-invasive detection of a fetal microdeletion using the GCREM algorithm. Prenat Diagn. 2014;34(5):469–77. doi: 10.1002/pd.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novakovic B, Saffery R. The ever growing complexity of placental epigenetics - role in adverse pregnancy outcomes and fetal programming. Placenta. 2012;33(12):959–70. doi: 10.1016/j.placenta.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg AP. Genome-scale approaches to the epigenetics of common human disease. Virchows Arch. 2010;456:13–21. doi: 10.1007/s00428-009-0847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinberg AP, Irizarry RA, Fradin D, et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci Transl Med. 2010;2(49):49ra67. doi: 10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monk D, Arnaud P, Apostolidou S, et al. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci U S A. 2006;103(17):6623–8. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monk M, Salpekar A. Expression of imprinted genes in human preimplantation development. Mol Cell Endocrinol. 2001;183(Suppl 1):S35–40. doi: 10.1016/s0303-7207(01)00575-5. [DOI] [PubMed] [Google Scholar]
- 12.Jirtle RL, Sander M, Barrett JC. Genomic imprinting and environmental disease susceptibility. Environ Health Perspect. 2000;108(3):271–8. doi: 10.1289/ehp.00108271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.American College of O, Gynecologists. ACOG Practice Bulletin No. 97: Fetal lung maturity. Obstetrics and gynecology. 2008;112(3):717–26. doi: 10.1097/AOG.0b013e318188d1c2. [DOI] [PubMed] [Google Scholar]
- 14.Neerhof MG, Haney EI, Silver RK, et al. Lamellar body counts compared with traditional phospholipid analysis as an assay for evaluating fetal lung maturity. Obstetrics and gynecology. 2001;97(2):305–9. doi: 10.1016/s0029-7844(00)01133-9. [DOI] [PubMed] [Google Scholar]
- 15.Winn-McMillan T, Karon BS. Comparison of the TDx-FLM II and lecithin to sphingomyelin ratio assays in predicting fetal lung maturity. American journal of obstetrics and gynecology. 2005;193(3 Pt 1):778–82. doi: 10.1016/j.ajog.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 16.Haymond S, Luzzi VI, Parvin CA, et al. A direct comparison between lamellar body counts and fluorescent polarization methods for predicting respiratory distress syndrome. American journal of clinical pathology. 2006;126(6):894–9. doi: 10.1309/8VXN5EM5L3831AT2. [DOI] [PubMed] [Google Scholar]
- 17.Karcher R, Sykes E, Batton D, et al. Gestational age-specific predicted risk of neonatal respiratory distress syndrome using lamellar body count and surfactant-to-albumin ratio in amniotic fluid. American journal of obstetrics and gynecology. 2005;193(5):1680–4. doi: 10.1016/j.ajog.2005.03.080. [DOI] [PubMed] [Google Scholar]
- 18.Mouillet JF, Ouyang Y, Coyne CB, et al. MicroRNAs in placental health and disease. American journal of obstetrics and gynecology. 2015;213(4 Suppl):S163–72. doi: 10.1016/j.ajog.2015.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox B, Leavey K, Nosi U, et al. Placental transcriptome in development and pathology: expression, function, and methods of analysis. American journal of obstetrics and gynecology. 2015;213(4 Suppl):S138–51. doi: 10.1016/j.ajog.2015.07.046. [DOI] [PubMed] [Google Scholar]
- 20.Huang A, Adusumalli J, Patel S, et al. Prevalence of chromosomal mosaicism in pregnancies from couples with infertility. Fertility and sterility. 2009;91(6):2355–60. doi: 10.1016/j.fertnstert.2008.03.044. [DOI] [PubMed] [Google Scholar]
- 21.Wilkins-Haug L, Roberts DJ, Morton CC. Confined placental mosaicism and intrauterine growth retardation: a case-control analysis of placentas at delivery. American journal of obstetrics and gynecology. 1995;172(1 Pt 1):44–50. doi: 10.1016/0002-9378(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 22.Dejmek J, Vojtassak J, Malova J. Cytogenetic analysis of 1508 spontaneous abortions originating from south Slovakia. European journal of obstetrics, gynecology, and reproductive biology. 1992;46(2–3):129–36. doi: 10.1016/0028-2243(92)90257-y. [DOI] [PubMed] [Google Scholar]
- 23.Johnson A, Wapner RJ, Davis GH, et al. Mosaicism in chorionic villus sampling: an association with poor perinatal outcome. Obstetrics and gynecology. 1990;75(4):573–7. [PubMed] [Google Scholar]
- 24.Wapner RJ, Simpson JL, Golbus MS, Ledbetter DH, Desnick RJ, et al. Chorionic mosaicism: association with fetal loss but not with adverse perinatal outcome. Prenatal diagnosis. 1992;12(5):347–55. doi: 10.1002/pd.1970120504. [DOI] [PubMed] [Google Scholar]
- 25.Kalousek DK, Barrett IJ, Gartner AB. Spontaneous abortion and confined chromosomal mosaicism. Human genetics. 1992;88(6):642–6. doi: 10.1007/BF02265289. [DOI] [PubMed] [Google Scholar]
- 26.Leschot NJ, Wolf H. Is placental mosaicism associated with poor perinatal outcome? Prenatal diagnosis. 1991;11(6):403–4. doi: 10.1002/pd.1970110611. [DOI] [PubMed] [Google Scholar]
- 27.Graham CH, Hawley TS, Hawley RG, et al. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res. 1993;206(2):204–11. doi: 10.1006/excr.1993.1139. [DOI] [PubMed] [Google Scholar]
- 28.Fitzpatrick TE, Graham CH. Stimulation of plasminogen activator inhibitor-1 expression in immortalized human trophoblast cells cultured under low levels of oxygen. Exp Cell Res. 1998;245(1):155–62. doi: 10.1006/excr.1998.4240. [DOI] [PubMed] [Google Scholar]
- 29.Huber AV, Saleh L, Bauer S, et al. TNFalpha-mediated induction of PAI-1 restricts invasion of HTR-8/SVneo trophoblast cells. Placenta. 2006;27(2–3):127–36. doi: 10.1016/j.placenta.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 30.Belkacemi L, Lash GE, Macdonald-Goodfellow SK, Caldwell JD, Graham CH. Inhibition of human trophoblast invasiveness by high glucose concentrations. J Clin Endocrinol Metab. 2005;90(8):4846–51. doi: 10.1210/jc.2004-2242. [DOI] [PubMed] [Google Scholar]
- 31.Xu NBG, Cui J, Wang ET, et al. Comparison of genome-wide and gene-specific DNA methylation profiling in first trimester chorionic villi from pregnancies conceived with infertility treatments. Reproductive Sciences. 2016 doi: 10.1177/1933719116675056. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brosens IA. Morphological changes in the utero-placental bed in pregnancy hypertension. Clin Obstet Gynaecol. 1977;4(3):573–93. [PubMed] [Google Scholar]
- 33.Khong TY, De Wolf F, Robertson WB, et al. Inadequate maternal vascular response to placentation in pregnancies complicated by pre-eclampsia and by small-for-gestational age infants. Br J Obstet Gynaecol. 1986;93(10):1049–59. doi: 10.1111/j.1471-0528.1986.tb07830.x. [DOI] [PubMed] [Google Scholar]
- 34.Kim YM, Chaiworapongsa T, Gomez R, et al. Failure of physiologic transformation of the spiral arteries in the placental bed in preterm premature rupture of membranes. Am J Obstet Gynecol. 2002;187(5):1137–42. doi: 10.1067/mob.2002.127720. [DOI] [PubMed] [Google Scholar]
- 35.Smith GC, Smith MF, McNay MB, et al. First-trimester growth and the risk of low birth weight. N Engl J Med. 1998;339(25):1817–22. doi: 10.1056/NEJM199812173392504. [DOI] [PubMed] [Google Scholar]
- 36.Robins JC, Heizer A, Hardiman A, et al. Oxygen tension directs the differentiation pathway of human cytotrophoblast cells. Placenta. 2007;28(11–12):1141–6. doi: 10.1016/j.placenta.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Katari S, Turan N, Bibikova M, et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Human molecular genetics. 2009;18(20):3769–78. doi: 10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novakovic B, Yuen RK, Gordon L, et al. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC genomics. 2011;12:529. doi: 10.1186/1471-2164-12-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan KC, Ding C, Gerovassili A, et al. Hypermethylated RASSF1A in maternal plasma: A universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clinical chemistry. 2006;52(12):2211–8. doi: 10.1373/clinchem.2006.074997. [DOI] [PubMed] [Google Scholar]
- 40.Ou X, Wang H, Qu D, et al. Epigenome-wide DNA methylation assay reveals placental epigenetic markers for noninvasive fetal single-nucleotide polymorphism genotyping in maternal plasma. Transfusion. 2014;54(10):2523–33. doi: 10.1111/trf.12659. [DOI] [PubMed] [Google Scholar]
- 41.Li P, Conley A, Zhang H, et al. Whole-Transcriptome profiling of formalin-fixed, paraffin-embedded renal cell carcinoma by RNA-seq. BMC genomics. 2014;15:1087. doi: 10.1186/1471-2164-15-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Adachi K, Sheridan MA, et al. Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proc Natl Acad Sci U S A. 2015;112(18):E2337–46. doi: 10.1073/pnas.1504778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Schust DJ. Isolation, purification and in vitro differentiation of cytotrophoblast cells from human term placenta. Reprod Biol Endocrinol. 2015;13(1):71. doi: 10.1186/s12958-015-0070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Founds SA, Conley YP, Lyons-Weiler JF, et al. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta. 2009;30(1):15–24. doi: 10.1016/j.placenta.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Founds SA. Bridging global gene expression candidates in first trimester placentas with susceptibility loci from linkage studies of preeclampsia. J Perinat Med. 2011;39(4):361–8. doi: 10.1515/jpm.2011.045. [DOI] [PubMed] [Google Scholar]
- 46.Eckmann-Scholz C, Bens S, Kolarova J, et al. DNA-methylation profiling of fetal tissues reveals marked epigenetic differences between chorionic and amniotic samples. PLoS One. 2012;7(6):e39014. doi: 10.1371/journal.pone.0039014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.