SUMMARY
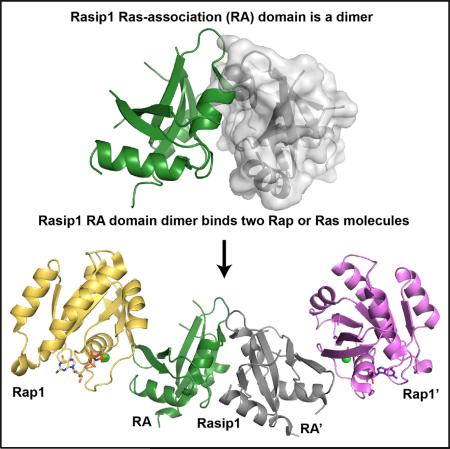
Ras-interacting protein 1 (Rasip1) is an endothelial-specific Rap1 and Ras effector, important for vascular development and angiogenesis. Here, we report the crystal structure of the Rasip1 RA domain (RRA) alone, revealing the basis of dimerization, and in complex with Rap1 at 2.8 Å resolution. In contrast to most RA domains, RRA formed a dimer that can bind two Rap1 (KD = 0.9 μM) or Ras (KD = 2.2 μM) molecules. We solved the Rap1-RRA complex and found that Rasip1 binds Rap1 in the Switch I region, and Rap1 binding induces few conformation changes to Rasip1 stabilizing a β strand and an unstructured loop. Our data explain how Rasip1 can act as a Rap1 and Ras effector and show that Rasip1 defines a subgroup of dimeric RA domains that could mediate cooperative binding to membrane-associated Ras superfamily members.
Graphical abstract
INTRODUCTION
Endothelial cells (EC) line the entire circulatory system, and regulation of endothelial barrier function is critical for homeostasis because increased endothelial permeability causes edema or contributes to inflammation. Furthermore, breakdown of the endothelial barrier contributes to diseases such as multiple sclerosis, cancer, and heart disease. Ras-interacting protein 1 (Rasip1), also known as Rain, is an endothelial-specific protein essential for endothelial barrier function, vascular integrity, and development (Wilson et al., 2013; Xu et al., 2009, 2011). It possesses a Ras-association (RA) domain and was identified as an effector of Rap and Ras (Mitin et al., 2004). A pool of Rasip1 localizes to EC cell-cell junctions in a Rap1-regulated manner, and the cytoplasmic tail of heart of glass (HEG1) transmembrane receptor mediates this Rap1-dependent recruitment of Rasip1 (de Kreuk et al., 2016; Wilson et al., 2013). Rasip1 cooperates with its paralog, Radil, to inhibit RhoA and its effector, Rho Kinase (ROCK) (Post et al., 2013). Radil is another Rap1 effector required for Rap1-induced cell spreading that directly interacts with the RhoGAP ArhGAP29. Rap1 activity induces independent translocation of Rasip1 and a Radil-ArhGAP29 complex to the plasma membrane, and subsequently a multimeric complex is formed with Rap-Rasip1-Radil-ArhGAP29 (Post et al., 2015).
Here, we used protein crystallography to determine the structural basis of the Rasip1 RA (RRA)-Rap1 interaction. We find that RRA forms a dimer in solution with many unstructured loops. The crystal structure of the RRA alone reveals the basis of dimerization where an atypical three-residue antiparallel β sheet not found in other RA/UBQ domains is formed at the dimer interface. Surprisingly, the crystal structure of the RRA-Rap1 complex shows a dimer bound to two Rap1 molecules, one at each end, forming an elongated complex. Comparison of the two structures shows that Rap1 binding induces a small conformational change in the Rasip1 dimer by stabilizing a β strand and one of the unstructured loops, but that most of the loops remain exposed for molecular interactions with other proteins. Furthermore, analysis of the Rasip1-Rap1 binding interface does not dictate a set of key residues providing Rap or Ras specificity. Our biochemical studies show that RRA can indeed bind both Rap1 and Ras (KD = 0.9 and 2.2 μM, respectively). We also find that Radil RA domain behaves similarly to Rasip1; it is a dimer in solution and it can bind both Rap and Ras (KD = 1.2 and 3.9 μM, respectively). These data show that both Rasip1 and Radil have an atypical ubiquitin-like fold and form a novel subgroup of dimeric RA domains that can engage either Rap or Ras, thereby mediating signaling from both GTPases.
RESULTS
Rasip1 Ras-Association Domain Has an Extended C Terminus and Forms a Stable Dimer in Solution
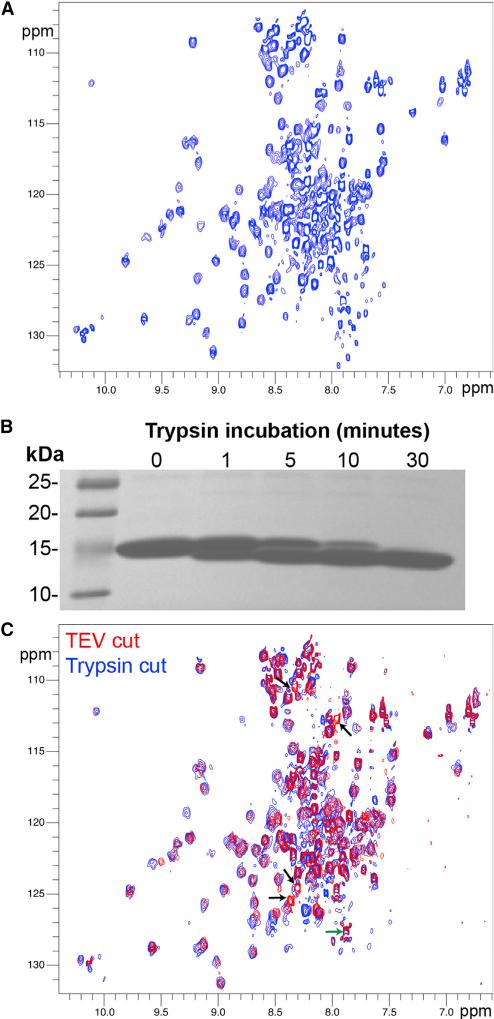
Small GTP-binding proteins bind RA motifs contained within ubiquitin-like (UBQ) domains, and Rasip1 was predicted to contain such a fold. When we tried to express soluble protein fragments based on the human Rasip1 RA (RRA) domain boundaries, soluble protein was only obtained in high yield after extending the domain C terminally to the original RA domain boundary definition (UniProt: Q5U651, residues 144–259). The isolated extended RRA (134–285) forms a dimer in solution as judged by size exclusion chromatography (SEC) (de Kreuk et al., 2016). Our initial attempts to crystallize RRA failed. To assess the reason for this failure, we purified the 15N-labeled protein for nuclear magnetic resonance (NMR) analysis. The SoFast-HMQC (sfHMQC) showed two peak populations; the majority were broad with good chemical shift dispersion, and approximately 30 were very sharp with poor dispersion. The broad peaks sharpened at higher temperatures and a satisfactory sfHMQC could be obtained at 35°C (Figure 1A), suggesting that the protein dimer and overall fold were very stable. NMR linewidths pointed to an elongated dimer in solution: the increase in linewidth of the RA dimeric module was greater than would be expected for a rigid globular domain of a similar size. Judging by the number of sharp peaks, we predicted that approximately 30 residues were in unstructured regions. We then performed limited proteolysis on the Rasip1 RA domain and observed that, after treatment with trypsin for 30 min, a stable fragment accumulated (Figure 1B). We purified the dimeric RRA fragment after trypsin cleavage and observed that at least four sharp resonances disappeared from the NMR spectrum and possibly a few more due to clustering of overlapped peaks around the middle of the spectrum (arrowheads in Figure 1C). Based on the small change in mass by SDS-PAGE, we predicted that the protein was cleaved after Arg140 at the N terminus; (1) the C terminus was not affected (green arrow in Figure 1C), and (2) the Trp side-chain resonances were preserved after cleavage, indicating that Trp281 at the C terminus was intact. Thus, the Rasip1 RA domain has an extended C terminus, is a stable dimer in solution, contains many unstructured loops, and a short unstructured N-terminal fragment can be released by trypsin cleavage.
Figure 1. Rasip1 RA Domain Is a Stable Dimer in Solution with Many Unfolded Loops.
(A) 1H,15N-sfHMQC spectrum (308K, 600 MHz) of 217 μM Rasip1 RA domain. Two populations of peaks were observed; the majority broad with good dispersion and many sharp with poor dispersion.
(B) Incubation of the Rasip1 RA domain with trypsin (250:1 w/w) at different time intervals. A stable fragment was accumulating after 30 min (~2 kDa smaller).
(C) 1H,15N-sfHMQC spectra (298K, 600 MHz) of 300 μM Rasip1 RA domain after cleavage with TEV (red) or trypsin (blue). The green arrow highlights the C terminus resonance and the black arrows the resonances that disappeared after trypsin treatment.
Crystal Structure of the Atypical Rasip1 RA Domain Dimer
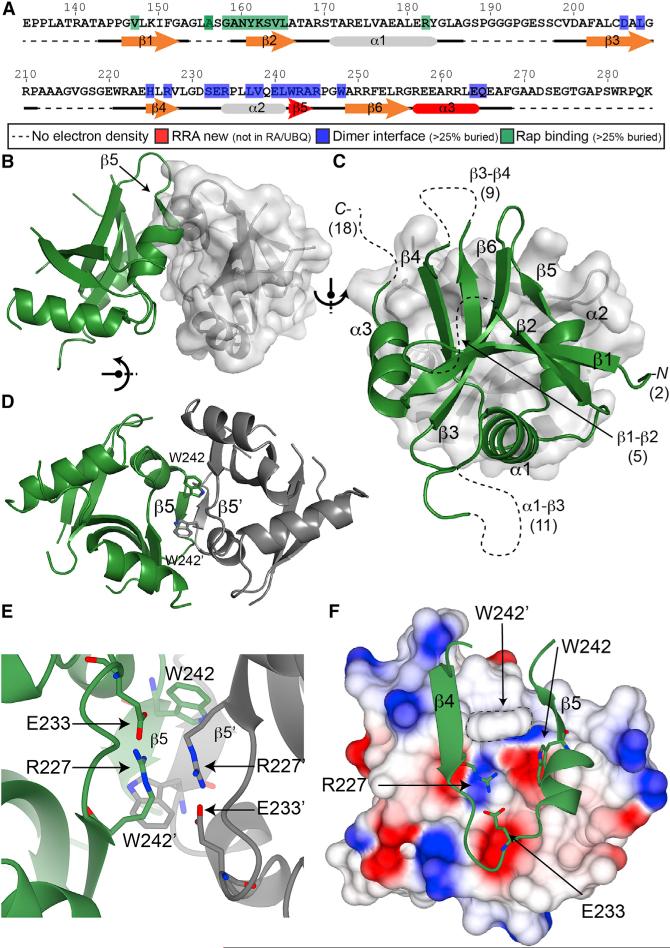
The human Rasip1 RA domain was purified after trypsin cleavage and crystallized. The structure (PDB: 5KHQ) was solved and refined to 2.8 Å resolution with an Rwork of 23.4% and Rfree of 26.2% (Table 1). The asymmetric unit contained two RA domains forming an atypical dimer not found in most other RA/UBQ domains (Figure 2B) and each monomer was almost identical (root-mean-square deviation [RMSD] = 0.36 Å2; Figure 3A). The RRA had a UBQ fold (Figure 3B) similar to other Ras-association domains such as RalGDS (Figure 3C), with two unusual features: (1) it had a short three-residue β5 strand inserted at the C terminus of the α2 helix important for dimer formation, and (2) an extra C-terminal α3 helix after the β6 strand or β5 strand in the RA/UBQ fold. The RRA amphipathic α3 helix packed against a small hydrophobic interface on the β4 strand was composed of Leu226 and Val228, which explained why the domain had to be extended at the C terminus. The current RRA model excludes many residues; 141–142 (N terminus), 154–158 (loop β1-β2), 187–197 (loop α1-β3), 212–220 (loop β3-β4), and 268–285 (C terminus, not part of the RRA) as highlighted in Figures 2A and 2C, and many side chains were also poorly defined in the electron density map. Of the 144 amino acids, 45 loop residues were not observed in the electron density map. This observation was in agreement with the NMR spectrum, which showed at the least 30 sharp peaks. Thus, the Rasip1 RA domain contained atypical features not found in other RA domains: (1) a β5 strand inserted after the α2 helix, (2) an additional C-terminal α3 helix, and (iii) many unstructured loops (Figure 2A).
Table 1.
Data Collection and Refinement Statistics for the Rasip1 RA Domain Alone and Bound to Rap1
| Rasip1 RA (PDB: 5KHQ) | Rasip1 RA-Rap1 (PDB: 5KHO) | |
|---|---|---|
| Data Collection | ||
| Space group | P6122 | P21212 |
| Cell dimensions | ||
| a, b, c (Å) | 89.2, 89.2, 178.0 | 101.6, 154.7, 38.9 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0–2.8 (3.0–2.8)a | 50–2.78 (2.99–2.78)a |
| CC1/2b | 100.0 (95.1)a | 99.9 (73.0)a |
| R meas | 0.109 (2.871)a | 0.075 (1.533)a |
| I/σI | 26.4 (2.1)a | 19.3 (1.5)a |
| Completeness (%) | 99.5 (97.6)a | 99.5 (98.5)a |
| Redundancy | 24.8 (25.7)a | 8.3 (8.3)a |
| Refinement | ||
| Resolution (Å) | 47.1–2.8 | 48.2–2.78 |
| No. reflections | 10,332 | 15,232 |
| Rwork/Rfree | 23.4/26.2 | 20.5/28.6 |
| No. atoms | 1,573 | 4,132 |
| Protein | 1,558 | 4,048 |
| Ligand/ion | 6 | 72 |
| Water | 9 | 14 |
| B factors | 105.2 | 91.4 |
| Protein | 107.6 | 96.8 |
| Ligand/ion | 97.1 | 89.3 |
| Water | 91.8 | 79.8 |
| RMSD | ||
| Bond lengths (Å) | 0.009 | 0.009 |
| Bond angles (°) | 1.485 | 1.397 |
| Ramachandran (%) | ||
| Favored, allowed, outliers | 98.4, 1.6, 0 | 93.7, 5.5, 0.8 |
Highest resolution shell is shown in parentheses.
As defined in XDS.
Figure 2. Crystal structure of the Rasip1 RA Domain Dimer.
(A) Sequence of human Rasip1 RA domain with secondary and tertiary structure elements highlighted.
(B–F) Cartoon representation of the Rasip1 dimer with one monomer shown in green and the other in gray.
(B) Side view of the dimer with a surface representation of one of the monomers.
(C) View of one monomer on top of the other one. The secondary structure elements of a monomer are labeled. The missing loop and terminus residues in the crystal structure are shown in brackets (total of 45).
(D) View of the dimer interface with a focus on the β5 strands of each monomer forming a short three-residue antiparallel β sheet. The side chains of Trp242 are also making a small hydrophobic interface.
(E) Different view of the dimer interface with a focus on the side chains of Glu233 and Arg227 making a salt bridge within the same molecule. They also pack against the same amino acid pair from the opposite monomer in an antiparallel way.
(F) Top view of the dimer interface as shown in (C) with a surface charge representation of one of the monomers to highlight the relatively hydrophilic dimer interface.
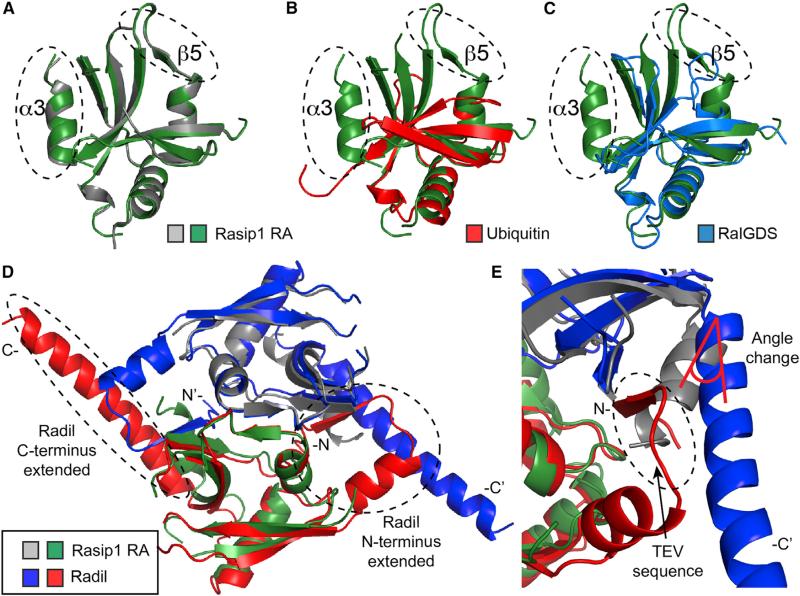
Figure 3. Proteins Containing RA and UBQ Domains.
(A–C) Superimposition of the Rasip1 RA domain monomer, as a green ribbon diagram, with (A) the second Rasip1 RA domain in the dimer (gray); (B) ubiquitin (PDB: 1AAR; red); (C) RalGDS (PDB: 1LFD; blue).
(D and E) Superimposition of the RA domain dimer of Rasip1 with Radil (PDB: 3EC8; red and blue). (D) Radil has extended N and C termini in comparison with Rasip1. (E) The Radil crystal structure contains the N-terminal TEV cleavage sequence forming an artificial extra β strand. The β strand alters the angle of the C-terminal helix.
The dimer was stabilized by an extensive interface burying 15.9% (975 Å2) of the total surface area of each monomer (Figure 2C). A short three-residue antiparallel β sheet was formed by the β5 strand from each molecule, involving backbone hydrogen bonds between Trp242, Arg243, and Ala244 (Figure 2D). Furthermore, the Trp242 side chain was found near Trp242 from the other monomer, creating a small hydrophobic interface (Figure 2D). Interestingly, the dimer interface included the side chains of Arg227 and Glu233, forming a salt bridge within the same RRA monomer, not across the dimer, and they packed against the same amino acid pair from the opposite monomer in an antiparallel fashion (Figure 2E). The surface charge representation of the dimer interface highlighted the presence of a relatively hydrophilic dimer interface (Figure 2F). Thus, the crystal structure of the Rasip1 RA domain shows a dimer that is stabilized by (1) the two Trp242 side chains forming a small hydrophobic interface, (2) a short antiparallel β sheet formed by β5 strands, and (3) a salt bridge between Arg227 and Glu233, allowing tighter packing of the dimer interface.
Rasip1 RA Domain Dimer Binds Two Rap1 Molecules
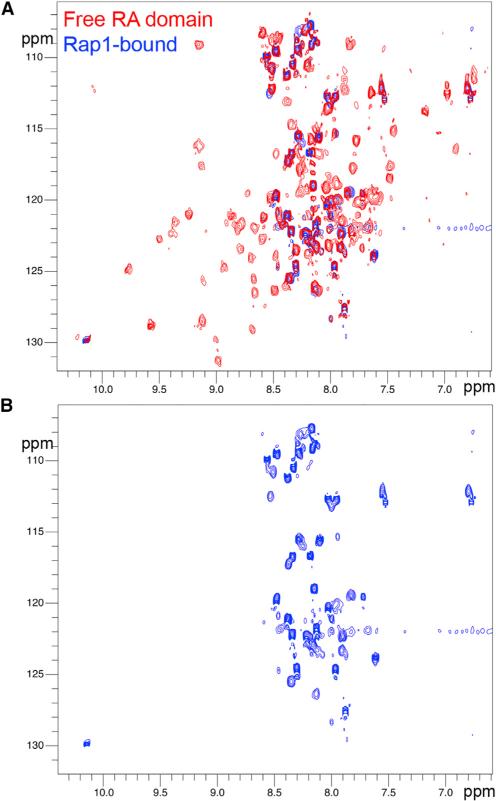
Analysis of the RRA crystal structure dimer showed that the two RA-binding motifs, located in the β2 strand of Rasip1, were available for binding and positioned at each end of the dimer, suggesting it could bind two Rap1 molecules. We previously showed using SEC that RRA formed a complex with GMP-PNP (a GTP analog)-bound Rap1 at equimolar concentration (de Kreuk et al., 2016). Isothermal titration calorimetry (ITC) measured KD = 0.77 ± 0.09 μM for the interaction with a stoichiometry constant of N = 1.22, in agreement with a stoichiometry of 1, showing that each Rasip1 RA domain monomer could bind a Rap1 molecule. NMR titrations using 15N-labeled Rasip1 showed that, upon Rap1 binding, all the well-dispersed peaks disappeared from the spectrum and only the sharp ones remained (Figure 4A). This suggested that the core of the protein was in complex with Rap1 and that most of the loop residues were not involved in the interaction and remained unstructured (~37 sharp peaks) (Figure 4B) and therefore available for other interactions. Thus, a RRA dimer binds to two Rap1 molecules and contains numerous unstructured regions that remain unstructured after Rap1 binding.
Figure 4. Most Rasip1 RA Domain Unfolded Loops Are Not Involved in the Rap1 Interaction.
(A) 1H,15N-sfHMQC spectra (298K, 600 MHz) of 160 μM Rasip1 RA domain in the free form (red) and in the presence of 250 μM Rap1 (~1.5-fold excess, blue). Almost all the broad peaks with good dispersion disappeared upon complex formation.
(B) Focus on the ~37 sharp peaks with poor dispersion left from the Rasip1 spectrum after Rap1 binding.
Crystal Structure of the Rasip1 RA Domain Dimer Bound to Two Rap1 Molecules
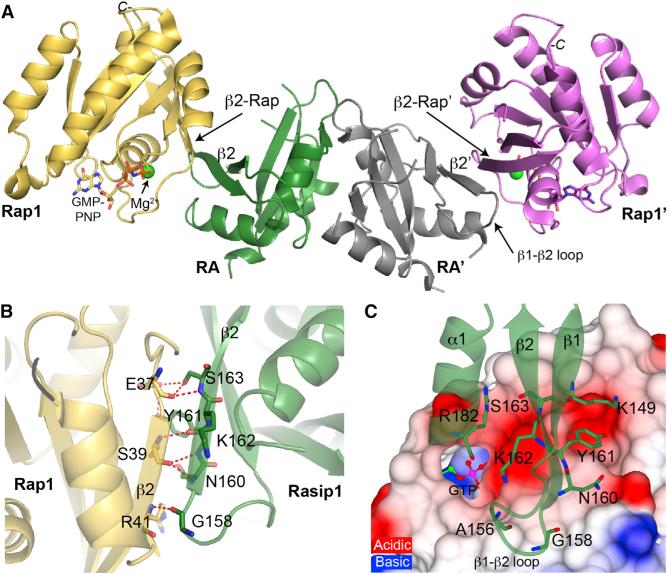
The human Rasip1 RA domain after trypsin cleavage in complex with human Rap1B (residues 1–167) was purified by SEC and crystallized. We determined the Rasip1-Rap1 complex structure to 2.78 Å resolution (PDB: 5KHO) with an Rwork of 20.5% and Rfree of 28.6% (Table 1). The asymmetric unit contained one Rasip1 RA domain dimer bound to two Rap1 molecules (Figure 5A), and the two individual RA-Rap1 were almost identical (RMSD = 0.51 Å). Similar to the free RA domain structure, the current Rasip1 RA domain model excludes many residues, 141–142 (N terminus), 186–195 (loop α1-β3), 212–220 (loop β3-β4), and 268–285 (C terminus), and many side chains were also poorly defined in the electron density map. This observation was in agreement with the NMR spectrum of the complex, which showed that most of the sharp peaks remained sharp when in complex with Rap1; this suggested that the unstructured loops remained unstructured in the complex. The asymmetric unit contained two Rap1 molecules for which we observed good electron density, with the exception of residues 62 and 63 in the switch II region: Rasip1 did not interact with switch II. In the crystal structure, the Rasip1-Rap1 complex was stabilized by an extensive interface, burying 533.5 Å2 of the total surface area of each molecule. Thus, the Rasip1 RA domain dimer binds two Rap1 molecules, and the majority of Rasip1 loops remained unstructured.
Figure 5. Crystal Structure of the Rasip1 RA Domain Bound to Two Rap1 Molecules.
Cartoon representation of the Rasip1 dimer (green and gray monomers) bound to two Rap1 molecules (yellow and magenta).
(A) Side view of the Rasip1 RA domain dimer bound to two Rap1. A new extended antiparallel β sheet across the β2 strand of Rasip1 and the β2 strand of Rap1 is formed.
(B) View of the key hydrogen bonds at the Rasip1-Rap1 binding interface, with a focus on the interactions across the newly formed β sheet.
(C) Top view of the binding interface on the surface charge representation of Rap1. The Rasip1 residues making hydrogen bonds with Rap1 are labeled. The Rasip1 Arg182 side chain is sitting into an acidic pocket on the surface of Rap1.
Rap1 Binding to Rasip1 RA Domain Stabilizes the RRA β2 Strand and the β1-β2 Loop
Because we had solved the structure of the Rasip1 RA domain dimer alone, we had the opportunity to examine the effect of Rap1 binding on the structure of the RA domain. The major binding determinant was mediated by main-chain and side-chain interactions across a newly formed antiparallel β sheet composed of β2 strand from Rap1, also called switch I, and β2 strand from the Rasip1 RA domain (Figure 5B). Interestingly, Asn160 made a hydrogen bond with Rap1-Ser39, extending the β2 strand from the Rasip1 by a few residues at the N terminus, in comparison with the free protein structure. Also the Rasip1 β1-β2 loop was stabilized by Ala156 and Gly158, making hydrogen bonds with Rap1 (Figure 5C), and an electron density was observed for residues 154–158. As summarized in Figure 6A, most of the residues involved in the interaction were centered around Rasip1 β2 strand and Rap1 switch I. Rasip1 Arg182 is located at the bottom of α1 helix and makes a hydrogen bond with the backbone carbonyl group of Rap1 Pro34, but most importantly the side chain is sitting into an acidic pocket on the surface of Rap1 (Figure 5C). Thus, Rasip1 binding to Rap1 stabilized the Rasip1 β2 strand and the β1-β2 loop, otherwise inducing no other structural changes.
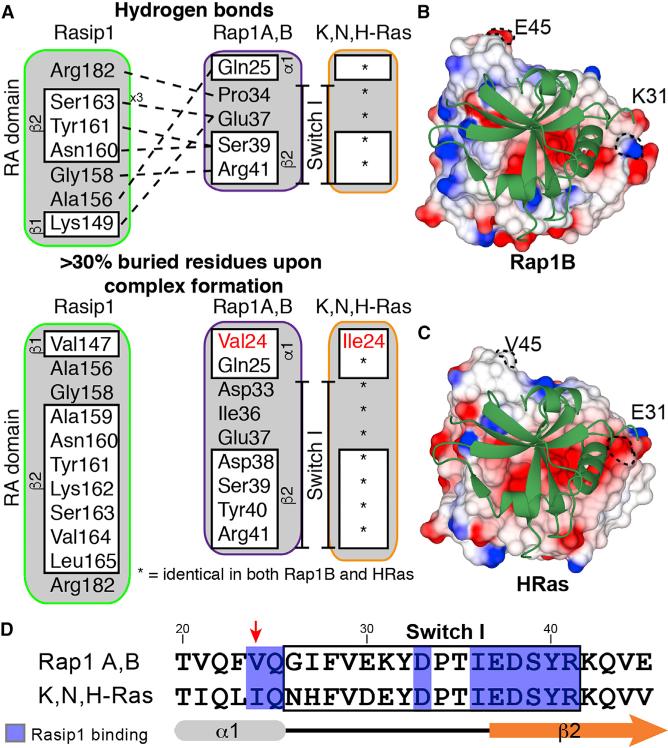
Figure 6. The Rap1 Binding Interface for Rasip1 Is Highly Homologous to HRas.
(A) Summary of the Rasip1-Rap1 interactions as identified using COCOMAPS. The upper section shows the hydrogen bonds between the two proteins and the lower section shows the residues buried by formation of the complex. HRas is very similar to Rap1, and the residues that are different are highlighted in red.
(B) View of the Rasip1 RA domain on the surface electrostatic potential representation of Rap1.
(C) View of the Rasip1 RA domain on the surface electrostatic potential representation of HRas.
(D) Sequence alignment of the effector region between proteins of the Ras subfamily of GTP-binding proteins. The residues that come in contact with Rasip1 are highlighted in blue and the red arrow points to the amino acid that is different between the two sequences.
Rasip1 True Rap1 or Ras Binding Domain? Structural Analysis
Mitin et al. (2004) showed that Rasip1 RA binds Rap1A, HRas, and KRas, but not RhoA, and suggested that Rasip1 may not be an exclusive effector for Ras-mediated signaling in cells. The structure described in this study created the opportunity to examine the Rasip1 RA domain specificity for Rap1 versus HRas; the amino acid sequences of the two GTPases are very similar, exhibiting 57.5% identity for residues 1 to 167. A first look at the surface charge of both Rap1 and HRas, as seen from the Rasip1 binding interface (Figures 6B and 6C), shows very little difference. Analysis of the Rasip1-Rap1 interface using COCOMAPS (Vangone et al., 2011) identifies many hydrogen bonds between the two proteins, and many residues are buried by formation of the complex as summarized in Figure 6A. The difference between Rap1 and HRas within the Rasip1 binding interface is limited to one residue as highlighted in Figure 6A, i.e., Rap1-Val24 and HRas-Ile24. Furthermore, this is the only difference found in the Rasip1 binding interface for most of the Ras subfamily members; Rap1A,B and K,N,H-Ras (Figure 6D). We noted that this small difference would not predict specific binding to either Rap1 or HRas.
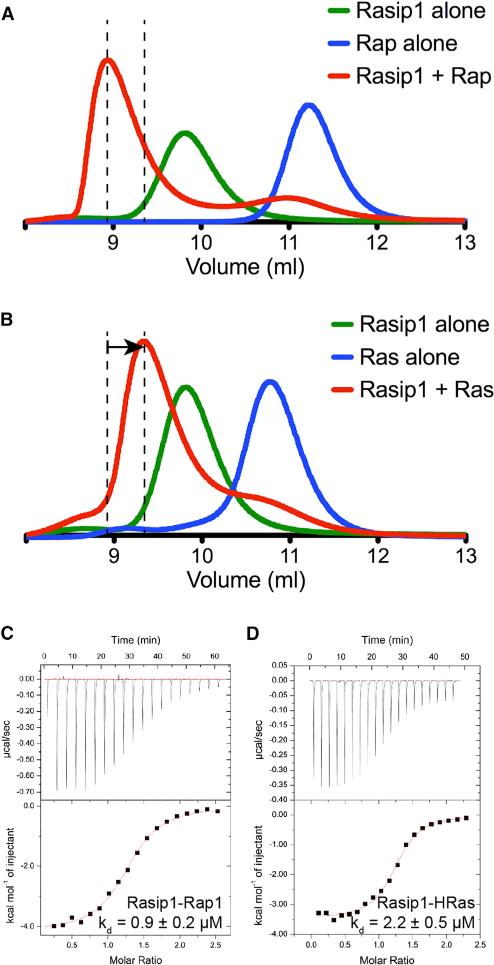
Since there was no obvious structural feature in the Rasip1 RA domain pointing to Rap1 or Ras binding specificity, we tested binding of the Rasip1 RA domain to both Rap1B and HRas using SEC. Both proteins formed a complex with the Rasip1 RA domain; Rap1 binding induced a clear shift toward a smaller volume of elution (Ve), indicating the formation of the complex (Figure 7A), while HRas showed a smaller shift toward an increased Ve (Figure 7B). Persistence of a complex through the SEC column implies that the proteins physically interact and equilibrate quickly enough to associate on a scale of minutes to hours. SEC is sensitive to both on and off rates; a differentiated early peak corresponds to high-nanomolar binding, and an early shoulder usually corresponds to low- to mid-micromolar affinity of binding (Mayer et al., 2009). This suggested that Rasip1 bound to both Rap1 and HRas, but that it has higher affinity for Rap1.
Figure 7. Rasip1 RA Domain Binds Rap1 and HRas with Comparable Affinity.
(A and B) Binding of the Rasip1 RA domain to (A) Rap1 or (B) HRas was analyzed on a Superdex-75 (10/300) GL gel filtration column at room temperature. Incubation of both GTPases with Rasip1 resulted in the formation of a complex with an increased Ve (50 μM each). The arrow in (B) indicates the difference in elution between the Rap1 and HRas complexes.
(C and D) Calorimetric titration of the Rasip1 RA domain in the cell with (C) Rap1 bound to GMP-PNP or (D) HRas bound to GMP-PNP. We performed each experiment two times and obtained similar results.
We then quantified the binding affinity of the interactions by ITC to determine whether the binding affinities are the same or different. The Rasip1 RA domain bound Rap1 with KD = 0.9 ± 0.2 μM (Figure 7C); and also HRas with KD = 2.2 ± 0.5 μM (Figure 7D) (Table 2). Furthermore, the stoichiometry constant measured for both complexes (Table 2) was in agreement with a stoichiometry of 1 and showed that each Rasip1 monomer could bind one ligand, or that a dimer can bind two simultaneously. The ITC data were in agreement with the SEC data, showing that the Rasip1 RA domain binds both Rap1 and HRas with very similar affinity. Interestingly, our structural analysis suggested that Rasip1 would not show specificity for either protein and the biochemical data showed that there was a minor difference. The difference in ΔG between Rap1 and HRas binding was of 0.4 kcal/mol, which was consistent with a maximum one hydrogen bond difference or less between the two complexes. Depending on geometry and environment, the hydrogen bond free-energy content is between 1 and 5 kcal/mol. Thus, our structural and biochemical analyses of RRA suggested that it might be an effector for both Rap and Ras-mediated signaling in cells.
Table 2.
Thermodynamic Parameters Obtained from ITC Titrations
| KD (μM) | N | ΔH (cal/mol) | ΔS (cal/mol K) | -TΔS (cal/mol) | ΔG (cal/mol) | |
|---|---|---|---|---|---|---|
| Rasip1 versus Rap1 | 0.9 ± 0.2 | 1.22 ± 0.01 | −3,400 ± 100 | 16.2 ± 0.5 | −4,800 ± 200 | −8,200 ± 300 |
| Rasip1 versus Hras | 2.2 ± 0.5 | 1.20 ± 0.06 | −3,700 ± 500 | 14 ± 2 | −4,100 ± 600 | −7,800 ± 1,100 |
| Radi1 versus Rap1 | 1.2 ± 0.1 | 1.04 ± 0.05 | −6,500 ± 100 | 5.3 ± 0.2 | −1,600 ± 100 | −8,100 ± 500 |
| Radi1 versus Hras | 3.9 ± 0.9 | 1.21 ± 0.05 | −5,500 ± 100 | 6.8 ± 0.1 | −2000 ± 100 | −7,500 ± 400 |
The SE reported is based on two different titrations using the same protein samples.
Radil RA Domain: Structure Comparison
We previously noted that Rasip1 and Radil had a highly conserved sequence (31% identity) (de Kreuk et al., 2016). Indeed, a Radil crystal structure also showed a dimer (PDB: 3EC8) and the two structures could be superimposed (Figure 3D; RMSD 1.85 Å2 for the dimer or 1.14 Å2 for monomers). Two main differences were observed with the Rasip1 structure at the N and C termini (Figure 3D). First, the Radil N terminus contained the tobacco etch virus (TEV) cleavage sequence, forming an artificial extra β strand next to the β4 stand, extending the β sheet (Figure 3E). Second, the C-terminal α helix in the Radil structure is much longer and stacked against the altered N terminus of the other molecule in the dimer, also altering the angle (Figure 3E). However, the Rap1 binding site located in the β2 strand of both structures looked almost identical, and only two amino acids were different; Rasip1 Ala159, Asn160 equivalent in Radil are Thr76, His77. Thus, both Rasip1 and Radil RA domain structures are dimeric, and their Rap1 binding sites were almost identical.
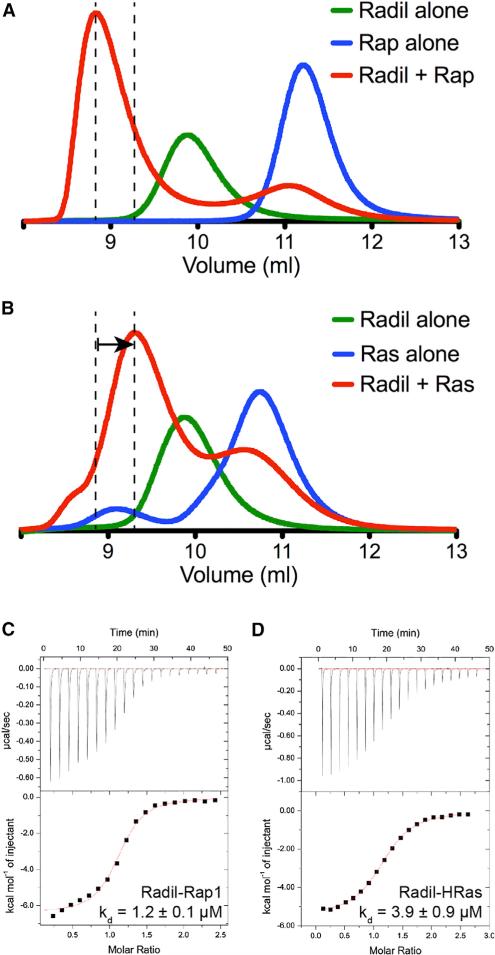
Radil RA Domain Also Binds Rap1 and HRas with Comparable Affinity
Radil is a paralog of Rasip1. Since there was no obvious structural feature in the Rasip1 RA domain pointing to Rap1 or Ras binding specificity, we also tested binding of the Radil RA domain to both Rap1B and HRas using SEC. Similar to Rasip1, both proteins formed a complex with the Radil RA domain; Rap1 binding showed a prominent shift toward a smaller Ve due to the formation of the complex (Figure 8A), while HRas showed a smaller shift toward an increased Ve (Figure 8B). This datum suggested that Radil could bind both Rap1 and HRas. Using ITC, we found that the Radil RA domain bound Rap1 with KD = 1.2 ± 0.1 μM (Figure 8C) and HRas with KD = 3.9 ± 0.9 μM (Figure 8D) (Table 2). Again, the binding stoichiometry constant observed for both complexes (Table 2) was in agreement with a stoichiometry of 1 and showed that each Radil monomer could bind one ligand, or that a dimer can bind two simultaneously. The ITC data were in agreement with the SEC data, showing that the Radil RA domain binds both HRas and Rap1 with comparable affinity. As for Rasip1, our structural analysis suggested that Radil would not show high specificity for either protein, and the biochemical data showed that there was only a minor difference. The difference in ΔG between Rap1 and HRas binding was 0.6 kcal/mol, which was consistent with a maximum one hydrogen bond difference or less between the two GTPases (Sheu et al., 2003). Thus, our structural and biochemical analysis of Radil suggested that, like Rasip1, it could be an effector for both Rap- and Ras-mediated signaling in cells.
Figure 8. Radil RA Domain Also Binds Rap1 and HRas with Comparable Affinity.
(A and B) Binding of the Radil RA domain to (A) Rap1 or (B) HRas was analyzed on a Superdex-75 (10/300) GL gel filtration column at room temperature. Incubation of both GTPases with Radil resulted in the formation of a complex with an increased Ve (50 μM each). The arrow in (B) indicates the difference in elution between the Rap1 and HRas complexes.
(C and D) Calorimetric titration of the Radil RA domain in the cell with (C) Rap1 bound to GMP-PNP or (D) HRas bound to GMP-PNP. We performed each experiment two times and obtained similar results.
DISCUSSION
The capacity of Rasip1 to bind Rap1 is important for the recruitment of Rasip1 to EC cell-cell junctions and to stabilize them. Here, we report the first crystal structure of Rasip1 in the form of the isolated RA domain, which adopts an atypical ubiquitin-like (UBQ) fold different to other RA-domain-containing proteins with a unique feature of being a dimer. The dimer is stabilized by the β5 strand, not found in other RA/UBQ domains, which mediates an antiparallel β sheet with a β5-β5′ topology. The dimer is stable in solution as determined by SEC and NMR linewidth. The Rasip1 RA domain contains many unstructured loops as demonstrated by both the presence of many sharp peaks in the NMR sfHMQC and the absence of electron density in the crystal structure. It is possible that those unstructured loops are binding sites for protein-protein interactions. The dimer is stabilized by (1) a short antiparallel β sheet formed by the β5 strand of each molecule, (2) the side chain of Trp242 of both chains forming a small hydrophobic interface, and (3) an intra-chain salt bridge between Arg227 and Glu223 that allows tighter packing of the dimer interface. An early study on Rasip1, called Rain at the time, using residues 120–233 reported that the protein was unstable and no affinity for Rap1 and HRas could be measured (Wohlgemuth et al., 2005). This can now be explained due to the premature stop of the domain exposing a hydrophobic surface on the β4 strand hindering correct folding; an additional amphipathic C-terminal α3 helix not found in other RA/UBQ domains is necessary to mask the hydrophobic interface. Taken together, our results show that Rasip1 contains an atypical β5 strand and α3 helix, not found in other RA/UBQ domains, necessary for correct folding and dimer formation.
Our biochemical data show that the Rasip1 RA domain dimer can bind two Rap1 molecules: (1) we observe a shift by SEC until an equimolar concentration of monomers is achieved, and (2) ITC titrations show a stoichiometry of 1 for the interaction. We purified the Rasip1-Rap1 complex, and we report, to our knowledge, the first crystal structure of a dimeric RA domain in complex with two GTP-binding proteins of the Ras subfamily, one bound at each end of the dimer. The major binding determinant is mediated by a newly formed antiparallel β sheet involving main-chain and side-chain interactions between the Rasip1 β2 strand and Rap1 switch I β2-strand. Comparison of the Rasip1 RA domain structures shows that Rap1 binding induces only a few changes to the overall structure of the RA domain. Overall, the β2 strand and the β1-β2 loop of Rasip1 are stabilized in the complex by hydrogen bonds with Rap1. Thus, Rasip1 RA domain folds as a dimer and binds two Rap1 GTP-binding proteins, and these data define the structure of a protein complex that regulates vascular integrity and development.
As for the free protein, no electron density can be observed for most of the Rasip1 loops, suggesting that they remain unfolded in the complex. To support this observation, analysis of the NMR sfHMQC linewidth of Rasip1 in the complex shows that the core of the Rasip1 dimer is in complex with Rap1 and that most of the loop residues remain free and flexible. Those flexible loops are likely binding sites for protein-protein interactions, and Rap1-binding to the RA domain could regulate this binding, for example, by restricting access to certain loops such as the β1-β2 loop. Moreover, there is a predicted forkhead-associated (FHA) and a dilute (DIL) domain at the C terminus of the RA domain in both Rasip1 and Radil proteins, so it is possible that the FHA domain folds back onto the RA domain, interacting with one of those loops. Upon Rap1 or Ras binding, a conformation change in the protein could expose a binding site in the FHA domain leading to, for example, HEG1 receptor binding and membrane localization of Rasip1 (de Kreuk et al., 2016). Considering that Rasip1 is a homodimer, the affinity of Rasip1 to certain ligands, such as transmembrane proteins like HEG1, should be increased by the multivalence of the dimer and possibly induce clustering of those receptors.
The structure described in this study gives us the opportunity to identify residues on the surface of Rasip1 and Rap1 that establish the effector specificity in the Ras subfamily of GTP-binding proteins, as we did previously for the KRIT1 FERM domain (Gingras et al., 2013) and others for Raf (Nassar et al., 1996). However, to our surprise, using the published crystal structures of different Rap and Ras proteins, we could not find a salt bridge or a hydrogen bond that would favor one member of the family. Mitin et al. (2004) suggested that Rasip1 may not be an exclusive effector for Ras-mediated signaling in cells; they observed binding to Rap1A, HRas, and KRas, but not RhoA. Using SEC and ITC, we find that the Rasip1 RA domain can bind both Rap1 and HRas (KD = 0.9 and 2.2 μM, respectively) with comparable affinity. An extensive in vitro biochemical study by Wohlgemuth et al. (2005) with many RA-domain-containing proteins showed that, as a general rule, a 10-fold difference in affinity between Rap1 and HRas is observed, and that MRas behaves more or less similarly to HRas. Furthermore, an average ΔG difference of ~1.5 kcal/mol is sufficient to provide this specificity, which is approximately one hydrogen bond difference. Depending on the geometry and environment, the hydrogen bond free-energy content is estimated to be between 0.5 and 8 kcal/mol (Sheu et al., 2003). The difference in ΔG between Rap1 and HRas for Rasip1 binding was low (0.4 kcal/mol), which is consistent with a maximum one hydrogen bond difference or less between the two complexes. Thus, our structural and biochemical analysis of Rasip1 suggested that it can be an effector for both Rap- and Ras-mediated signaling in cells.
It is arguable whether or not the relative affinities of the isolated domains for Ras or Rap, measured in vitro, reflect the specificity of that interaction in vivo. In the case of Rasip1, there is strong evidence that it acts together with Rap in vascular development (Post et al., 2013; Wilson et al., 2013; Xu et al., 2011), consistent with our affinity measurements. Also, although both Ras and Rap interact with full-length Rasip1, as shown by co-precipitation studies (Mitin et al., 2004), Rap recruits and co-localizes with Rasip1 at the plasma membrane (Post et al., 2015). Less work has been done on the Ras interaction with Rasip1, but interestingly, both KRas and Rasip1 have been shown to play an important role in endothelial tubulogenesis (Norden et al., 2016); more studies will need to investigate this potential connection. Rasip1 shows no specificity toward Rap and Ras in vitro, and we could not create a structure-guided mutant of Rasip1 that would selectively bind to either Rap or Ras to help these investigations. We previously showed Arg182 to be important by mutagenesis where Rasip1(R182E) exhibited a markedly reduced affinity for Rap1 (de Kreuk et al., 2016). This mutation creates repulsion between the negatively charged glutamate side chain of the mutant and the Rap1 surface. However, this mutant also blocks binding to Ras, thus it can be used to study the downstream effects of Rasip1 binding to Rap and/or Ras in vivo.
In our search for an explanation for the low specificity of Rasip1 toward members of the Ras subfamily of GTP-binding proteins, we investigated Radil for comparison. The Radil RA domain has a high sequence identity with the Rasip1 RA domain and was shown to bind Rap and not H-, N-, or K-Ras by co-precipitation studies (Smolen et al., 2007). To our surprise, we also found that the dimeric Radil RA domain binds both Rap and Ras in vitro (KD = 1.2 and 3.9 μM, respectively) with similar affinity as observed with Rasip1. Similar to Rasip1, there is strong evidence that Radil is an effector of Rap (Post et al., 2013, 2015), but little evidence on whether it can also function as a Ras effector in vivo. Interestingly, Radil was described as an AF6-like protein (also called afadin) and AF6, which contains two N-terminal RA domains, also interacts with both Ras and Rap (Boettner et al., 2000; Kuriyama et al., 1996; Zhang et al., 2005).
Our data on both Rasip1 and Radil point to a dimeric RA domain subfamily that can bind both Rap and Ras in vitro. The fact that the RA domain is a dimer is important, since the Ras subfamily member are attached to the membrane by prenylation and palmitoylation on their CAAX box. This can provide a membrane anchor for Rasip1 and, since the dimeric RA domain can bind two molecules, the affinity for two ligands attached to the membrane will cooperatively augment. Furthermore, it is also possible that Rasip1 will bind one Rap and one Ras simultaneously to create membrane nanoclusters of Ras subfamily GTP-binding proteins. Interestingly, Ras-GTP forms dimers at the membrane to activate the MAPK pathway (Nan et al., 2015), so it is possible that Rasip1 binding to two molecules promotes Ras and/or Rap dimerization and possible activation of specific pathways in vivo.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human RA domains of Rasip1 (residues 134–285) and Radil (residues 51–193) were cloned into the pETM-11 (His-tagged, EMBL) vector and were expressed in Escherichia coli BL21 Star (DE3) cultured in lysogeny broth medium or in minimal medium for 15N-labeled samples for NMR. Recombinant His-tagged RA domain polypeptides were purified by nickel-affinity chromatography following standard procedures. The His-tag was removed by cleavage with TEV protease overnight, and the protein was further purified by SEC using a Superdex-75 (16/600) column (GE Healthcare). The protein concentration for RA domain monomers was assessed using a molar absorption coefficient of E280 = 24,980 M−1 for Rasip1 and 15,470 M−1 for Radil.
Human Rap1 isoform Rap1B (residues 1–167) and HRas (residues 1–189) cloned into pTAC vector in the E. coli strain CK600K was the generous gift of Professor Alfred Wittinghofer (Max Planck Institute of Molecular Physiology, Germany). Cultures were grown at 37°C until they reached an A600 of 0.8, transferred to an 18°C shaker for 1 hr, and then induced with 1 mM isopropyl β-D-1-thiogalactopyranoside overnight. Untagged GTPases were purified by ion exchange, followed by Superdex-75 (26/600) gel filtration. The column was pre-equilibrated and run with 20 mM Tris, 150 mM NaCl, 3 mM MgCl2, and 2 mM DTT (pH 7.4; TBS-MD). Nucleotide exchange for GMP-PNP (guanosine 5’-[β,γ-imido]triphosphate) was achieved as described by John et al. (1990) using the non-EDTA-containing approach. The final sample was buffer exchanged using a PD-10 column in TBS-MD buffer before freezing. The protein concentration was assessed using a molar absorption coefficient of E280 = 18,450 M−1 for HRas-GTP and 19,480 M−1 for Rap1B-GTP as reported by Tucker et al. (1986).
RA Domains Binding to Rap1 and HRas by Size Exclusion Chromatography
SEC of recombinant RA domains with Rap1B and HRas was performed using a Superdex-75 (10/300) GL at room temperature. The proteins, 50 μM each, were mixed in a volume of 100 μL and incubated for 30 min at room temperature before loading onto the column, which was pre-equilibrated with and run with TBS-MD (pH 7.4).
Purification of Rasip1 after Trypsin Treatment
Recombinant His-tagged Rasip1 RA domain was purified by nickel-affinity chromatography as described above. The protein was exchanged into 20 mM Tris, 150 mM NaCl, 2 mM DTT (pH 7.4; TBS-D) using a HiPrep (26/10) desalting column (GE Healthcare). A total of 48 mg of Rasip1 in TBS-D buffer supplemented with 2 mM CaCl2 was digested with 20 μg of trypsin for 5 hr at 30° C (ratio of 1:2,500; Promega V511A). The solution was then incubated in 50 μL of trypsin inhibitor agarose (Sigma T0637) for 15 min at room temperature. The beads were pelleted by centrifugation and the suspension loaded onto a Superdex-75 (16/600) column equilibrated in TBS-D buffer. The final protein concentration was determined using a molar absorption coefficient of E280 = 24,980 M−1 for Rasip1, which was partially accurate since the protein lost ~2 kDa as determined by SDS-PAGE, but no tryptophan residue based on NMR.
Purification of the Trypsin-Treated Rasip1-Rap1 Complex
Equal molar concentrations of trypsin-treated Rasip1 RA domain and GMP-PNP loaded Rap1B were mixed and loaded on a Superdex-75 (16/600). The column was pre-equilibrated and run with 20 mM Tris, 50 mM NaCl, 3 mM MgCl2, and 2 mM DTT (pH 8). The final complex concentration was determined using a molar absorption coefficient of E280 = 39,420 M−1 for the Rasip1-Rap1B complex.
Isothermal Titration Calorimetry
ITC experiments were performed using an ITC200 microcalorimeter (MicroCal) at 23°C. All proteins were dialyzed into 20 mM sodium phosphate, 200 mM NaCl, 3 mM MgCl2, 1 mM tris(2-carboxyethyl)phosphine, 0.1 mM GMP-PNP (pH 6.5) before performing the experiment. During each titration, 19 injections of 2 μL of HRas or Rap1 (between 0.34 and 1 mM) were made to cells containing Rasip1 or Radil (between 30 and 80 μM). ITC data were analyzed by fitting to a single-site binding equation using MicroCal Origin software. The SE reported is based on two different titrations using the same protein samples.
Crystallization of the Trypsin-Treated Rasip1 RA Domain and Rasip1-Rap1 Complex
The purified trypsin-treated Rasip1 RA domain and trypsin-treated Rasip1-Rap1 complex were concentrated to 4.67 and 3.63 mg/mL, respectively. Crystals were grown at room temperature using the sitting-drop method by mixing equal volumes of protein complex and reservoir solution (2 + 2 μL). The reservoir solution contained (Rasip1) 1.25–1.5 M ammonium sulfate, 100 mM 2-(N-morpholino)ethanesulfonic acid (MES; pH 6.0), and (Rasip1-Rap1) 14–19% PEG 8K, 200 mM calcium acetate, 100 mM MES (pH 6.5). The crystals were briefly transferred to reservoir solution containing 20% glycerol before freezing in liquid nitrogen.
Structure Determination
Diffraction data for the Rasip1 RA domain and RRA-Rap1 complex were collected at Stanford Synchrotron Radiation Lightsource experimental station 12-2. All crystals displayed strong to severe anisotropy of diffraction in two directions, where reflection intensities extend in one direction to higher resolution than in other directions, so we relied on the CC* to assess data quality (Karplus and Diederichs, 2012).
The data were processed with XDS (Kabsch, 2010). For the Rasip1-Rap1 complex, initial phases were determined using the structure of human Rap1B, excluding the nucleotide (PDB: 4HDQ, chain B) with Phaser; all the software used are part of the CCP4 software suite (Collaborative Computational ProjectNumber 4, 1994). Two copies of Rap1B were found, clear electron density was observed for the nonhydrolyzable GTP analog GMP-PNP, and it was very apparent that the Rap1 was bound to Rasip1 by the presence of a newly formed antiparallel β sheet formed by strand β2 from Rap1, also called switch I, and a strand from Rasip1. The GMP-PNP (GNP) was docked into the model, many residues of Rasp1 were built manually using Coot near Rap1 switch I, and cycles of maximum likelihood refinement in Refmac5 were performed. It became very apparent from that point that the Rap1 was bound to GMP-PNP and the quality of the map improved significantly. The model was then optimized using cycles of manual refinement with Coot and maximum likelihood refinement in Refmac5.
The structure of the Rasip1 was solved by molecular replacement using Phaser with the structure of the Rasip1 dimer from the Rasip1-Rap1 complex.
Highlights.
We solved the crystal structure of Raisp1 RA domain (RRA) and in complex with Rap1
In contrast to most RA domains, RRA has an extended C terminus and forms a dimer
The RRA dimer can bind two Rap1 or Ras molecules (KD = 0.9 and 2.2 μM, respectively)
Rap1 binding via its switch I domain induces few conformation changes in RRA
ACKNOWLEDGMENTS
The authors thank Alexander E. Aleshin, Robert C. Liddington, Robyn L. Stanfield, and Ian A. Wilson for data collection time on SSRL beamline 12-2. They also thank Bart-Jan de Kreuk and Igor L. Barsukov for useful discussions. A.R.G. is supported by the American Heart Association; Scientist Development Grant (12SDG11610043) and Grant-In-Aid (16GRNT29650005). M.H.G. is supported by the National Institute of Health (HL106489 and NS092521).
Footnotes
ACCESSION NUMBERS
The atomic coordinates and structure factors for the Rasip1 dimer and the Rasip1 in complex with Rap1B have been deposited in the PDB under the accession codes PDB: 5KHQ and 5KHO, respectively.
AUTHOR CONTRIBUTIONS
A.R.G. conducted the experiments with contributions from W.P.-M. A.A.B. conducted and analyzed the ITC experiments. A.R.G. designed the research, wrote the paper, and directed the project with contributions from M.H.G.
REFERENCES
- Boettner B, Govek EE, Cross J, Van Aelst L. The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc. Natl. Acad. Sci. USA. 2000;97:9064–9069. doi: 10.1073/pnas.97.16.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- de Kreuk BJ, Gingras AR, Knight JD, Liu JJ, Gingras AC, Ginsberg MH. Heart of glass anchors Rasip1 at endothelial cell-cell junctions to support vascular integrity. Elife. 2016;5:e11394. doi: 10.7554/eLife.11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AR, Puzon-McLaughlin W, Ginsberg MH. The structure of the ternary complex of Krev interaction trapped 1 (KRIT1) bound to both the Rap1 GTPase and the heart of glass (HEG1) cytoplasmic tail. J. Biol. Chem. 2013;288:23639–23649. doi: 10.1074/jbc.M113.462911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J, Sohmen R, Feuerstein J, Linke R, Wittinghofer A, Goody RS. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry. 1990;29:6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama M, Harada N, Kuroda S, Yamamoto T, Nakafuku M, Iwamatsu A, Yamamoto D, Prasad R, Croce C, Canaani E, et al. Identification of AF-6 and canoe as putative targets for Ras. J. Biol. Chem. 1996;271:607–610. doi: 10.1074/jbc.271.2.607. [DOI] [PubMed] [Google Scholar]
- Mayer CL, Snyder WK, Swietlicka MA, Vanschoiack AD, Austin CR, McFarland BJ. Size-exclusion chromatography can identify faster-associating protein complexes and evaluate design strategies. BMC Res. Notes. 2009;2:135. doi: 10.1186/1756-0500-2-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitin NY, Ramocki MB, Zullo AJ, Der CJ, Konieczny SF, Taparowsky EJ. Identification and characterization of rain, a novel Ras-interacting protein with a unique subcellular localization. J. Biol. Chem. 2004;279:22353–22361. doi: 10.1074/jbc.M312867200. [DOI] [PubMed] [Google Scholar]
- Nan X, Tamguney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, Lewis S, Gray JW, McCormick F, Chu S. Ras-GTP dimers activate the mitogen-activated protein Kinase (MAPK) pathway. Proc. Natl. Acad. Sci. USA. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar N, Horn G, Herrmann C, Block C, Janknecht R, Wittinghofer A. Ras/Rap effector specificity determined by charge reversal. Nat. Struct. Biol. 1996;3:723–729. doi: 10.1038/nsb0896-723. [DOI] [PubMed] [Google Scholar]
- Norden PR, Kim DJ, Barry DM, Cleaver OB, Davis GE. Cdc42 and k-ras control endothelial tubulogenesis through apical membrane and cytoskeletal polarization: novel stimulatory roles for GTPase effectors, the small GTPases, Rac2 and Rap1b, and inhibitory influence of Arhgap31 and Rasa1. PLoS One. 2016;11:e0147758. doi: 10.1371/journal.pone.0147758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post A, Pannekoek WJ, Ross SH, Verlaan I, Brouwer PM, Bos JL. Rasip1 mediates Rap1 regulation of Rho in endothelial barrier function through ArhGAP29. Proc. Natl. Acad. Sci. USA. 2013;110:11427–11432. doi: 10.1073/pnas.1306595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post A, Pannekoek WJ, Ponsioen B, Vliem MJ, Bos JL. Rap1 spatially controls ArhGAP29 to inhibit Rho signaling during endothelial barrier regulation. Mol. Cell. Biol. 2015;35:2495–2502. doi: 10.1128/MCB.01453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu SY, Yang DY, Selzle HL, Schlag EW. Energetics of hydrogen bonds in peptides. Proc. Natl. Acad. Sci. USA. 2003;100:12683–12687. doi: 10.1073/pnas.2133366100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen GA, Schott BJ, Stewart RA, Diederichs S, Muir B, Provencher HL, Look AT, Sgroi DC, Peterson RT, Haber DA. A Rap GTPase interactor, RADIL, mediates migration of neural crest precursors. Genes Dev. 2007;21:2131–2136. doi: 10.1101/gad.1561507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker J, Sczakiel G, Feuerstein J, John J, Goody RS, Wittinghofer A. Expression of p21 proteins in Escherichia coli and stereochemistry of the nucleotide-binding site. EMBO J. 1986;5:1351–1358. doi: 10.1002/j.1460-2075.1986.tb04366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangone A, Spinelli R, Scarano V, Cavallo L, Oliva R. COCOMAPS: a web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics. 2011;27:2915–2916. doi: 10.1093/bioinformatics/btr484. [DOI] [PubMed] [Google Scholar]
- Wilson CW, Parker LH, Hall CJ, Smyczek T, Mak J, Crow A, Posthuma G, De Maziere A, Sagolla M, Chalouni C, et al. Rasip1 regulates vertebrate vascular endothelial junction stability through Epac1-Rap1 signaling. Blood. 2013;122:3678–3690. doi: 10.1182/blood-2013-02-483156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth S, Kiel C, Kramer A, Serrano L, Wittinghofer F, Herrmann C. Recognizing and defining true Ras binding domains I: biochemical analysis. J. Mol. Biol. 2005;348:741–758. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- Xu K, Chong DC, Rankin SA, Zorn AM, Cleaver O. Rasip1 is required for endothelial cell motility, angiogenesis and vessel formation. Dev. Biol. 2009;329:269–279. doi: 10.1016/j.ydbio.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Sacharidou A, Fu S, Chong DC, Skaug B, Chen ZJ, Davis GE, Cleaver O. Blood vessel tubulogenesis requires Rasip1 regulation of GTPase signaling. Dev. Cell. 2011;20:526–539. doi: 10.1016/j.devcel.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Rehmann H, Price LS, Riedl J, Bos JL. AF6 negatively regulates Rap1-induced cell adhesion. J. Biol. Chem. 2005;280:33200–33205. doi: 10.1074/jbc.M505057200. [DOI] [PubMed] [Google Scholar]