Abstract
Background
A major obstacle to the development of broadly protective M protein-based group A streptococcal (GAS) vaccines is the variability within the N-terminal epitopes that evoke potent bactericidal antibodies. The concept of M type-specific protective immune responses has recently been challenged based on the observation that multivalent M protein vaccines elicited cross-reactive bactericidal antibodies against a number of non-vaccine M types of GAS. Additionally, a new “cluster-based” typing system of 175 M proteins identified a limited number of clusters containing closely related M proteins. In the current study, we used the emm cluster typing system, in combination with computational structure-based peptide modeling, as a novel approach to the design of potentially broadly protective M protein-based vaccines.
Methods
M protein sequences (AA 16–50) from the E4 cluster containing 17 emm types of GAS were analyzed using de novo 3-D structure prediction tools and the resulting structures subjected to chemical diversity analysis to identify sequences that were the most representative of the 3-D physicochemical properties of the M peptides in the cluster. Five peptides that spanned the range of physicochemical attributes of all 17 peptides were used to formulate synthetic and recombinant vaccines. Rabbit antisera were assayed for antibodies that cross-reacted with E4 peptides and whole bacteria by ELISA and for bactericidal activity against all E4 GAS.
Results
The synthetic vaccine rabbit antisera reacted with all 17 E4 M peptides and demonstrated bactericidal activity against 15/17 E4 GAS. A recombinant hybrid vaccine containing the same E4 peptides also elicited antibodies that cross-reacted with all E4 M peptides.
Conclusions
Comprehensive studies using structure-based design may result in a broadly protective M peptide vaccine that will elicit cluster-specific and emm type-specific antibody responses against the majority of clinically relevant emm types of GAS.
Keywords: group A streptococcal vaccine, M protein, structure-based design, broadly neutralizing antibodies
1. Introduction
Despite the impressive advances in the development and deployment of microbial vaccines, infectious diseases remain a major cause of morbidity and mortality throughout the world [1]. Many of the infectious diseases prevalent today, for which there are no licensed vaccines, are caused by organisms that maintain a stealthy appearance by displaying a variable array of protective antigens among members of the same species or that undergo mutation over time, thus thwarting the acquired immune responses within a population. These scenarios, among others, together with the knowledge that 3-D structure varies less than sequence, have prompted a new field of “structural vaccinology”, which uses protein structure to identify and, in some cases, engineer vaccine antigens that stimulate broadly neutralizing immune responses potentially capable of protecting against microbes that otherwise elicit strain-specific or no immunity following natural infection [2].
A classic example of the heterogeneity of protective antigens within a species is the M protein of group A streptococci (GAS). This is now perceived as a major obstacle to the development of effective M protein-based vaccines. The M protein is an alpha-helical dimer that extends from the surface of the organism and functions as a major virulence determinant by conferring resistance to opsonization and phagocytic killing in the non-immune host [3]. N-terminal M protein sequences are variable among the >200 emm types of GAS, evoke antibodies with the greatest opsonic activity [4, 5], and protect animals against lethal challenge infections with GAS [6]. These observations have served as the basis for the design and development of recombinant multivalent vaccines containing as many as 30 different N-terminal peptides of M proteins linked in tandem [7]. We have previously shown that multivalent M protein-based vaccines were immunogenic in animals [5, 7, 8] and humans [9, 10]. The assumption has been that protection against GAS infection is “type-specific”, which has led to the conclusion that covering infections by all emm types would be impractical, if not impossible. However, recent data suggest that protection may not be as type-specific as previously thought. A 30-valent M protein based vaccine stimulated antibodies that opsonized 30 emm types that were not represented in the vaccine [7, 11].
The observation that a number of non-vaccine emm types were cross-opsonized by vaccine antisera was later combined with a comprehensive analysis of the sequences and function of M proteins from a global collection of 175 emm types of GAS [12]. The results provided a new emm cluster-typing system that classified 96.2% (P.R.S., personal communication) of all contemporary GAS isolates into 48 emm clusters containing structurally and functionally related M proteins. Furthermore, 117 emm types, contained in only 16 clusters, accounted for 94.4% of GAS infections in the world. The remaining 32 clusters contained single M proteins that were structurally distinct. Cross-opsonization data combined with extensive structure/function data suggested that there is a need for a paradigm shift away from the concept of “type-specific” immunity against GAS infections to one that is “type-specific” and “cluster-specific” to account for cross-opsonization of different emm types within the same clusters [12]. The results also suggested that the number of emm types may not be an insurmountable obstacle to the development of broadly protective M protein-based vaccines.
In the current study, we have used the emm cluster typing system, in combination with computational structure-based peptide modeling, as a novel approach to the design of potentially broadly protective M protein-based vaccines. We focused on one emm cluster, E4, which contains 17 GAS emm types that, in aggregate, are more prevalent in both the US and elsewhere in the world when compared to any one of the remaining emm clusters [13] [P.R.S., personal communication]. The overall goal was to identify a small number of N-terminal M peptides from the E4 cluster that evoked antibodies that cross-reacted with all other E4 peptides and promoted functional bactericidal activity against all emm types of GAS in the cluster. Our results, coupled with an iterative bioinformatics process that utilized data from peptide cross-reactive antibody binding, provide an experimental framework that can be applied to all clusters of M proteins, potentially leading to the development of a broadly efficacious vaccine.
2. Materials and Methods
Group A streptococcal strains
The bacterial strains used in this study were obtained from our laboratory collection, the North America Group A Streptococcal Pharyngitis Serotype Surveillance Study [13], and Bernard Beall at the Centers for Disease Control and Prevention.
Synthetic peptides
Peptides copying AA 16–50 of each of the mature M proteins expressed by the GAS in the E4 cluster were synthesized (Biosyn, Lewisville, TX) to contain a C-terminal cysteine to facilitate coupling to KLH. Control peptides from non-E4 clusters were synthesized for a previous study [7].
Computational modeling
De novo structure prediction tools [14–16] were used to generate peptide structural ensembles that were subjected to chemical diversity analysis to identify sequences that are the most representative of the structural physicochemical properties of the peptides in the E4 cluster. The de novo approach was implemented using PEP-FOLD [17], which has been shown to generate near-native or native structures for 95% of a set of 56 structurally diverse peptides with 25–52 residues. PEP-FOLD was used to generate thermodynamically-allowed structures for all 17 E4 peptides, AA 1–35 and 16–50. The resulting structures were analyzed using a set of 22 “protein” Quantitative Structure-Activity Relationship (QSAR) descriptors in the MOEv2014 program (Chemical Computing Group, Montreal, Canada). The descriptors quantify the physicochemical properties of a peptide with a given structure and sequence.
Vaccine formulations and immunization of animals
Five E4 peptides copying AA 15–50 of M2, M8, M22, M89, and M112, were selected as vaccine components based on the structural predictions described above. Each of the five peptides was conjugated individually to KLH using a bifunctional cross-linker, as described [18]. The peptide-KLH conjugates (100 μg each) were formulated with complete Freund’s adjuvant (FA) for the initial injection and incomplete FA for subsequent injections, and used to immunize three rabbits at time 0, 3 weeks and 6 weeks. Serum was obtained 3 weeks following the final injection.
A recombinant pentavalent E4 hybrid protein vaccine containing the same M peptides was produced from extracts of E. coli containing pUC57 into which was inserted a chemically synthesized hybrid gene (Genscript, Piscataway, NJ), using methods previously described [7]. The synthetic gene was also designed to encode an upstream T7 promoter, and a 3′ poly-histidine motif followed by a stop codon.
ELISA and peptide inhibition assays
ELISA was performed using the synthetic peptides or whole bacteria as solid-phase antigens, as previously described [19]. Endpoint titers were determined using 2-fold dilutions of pre-immune or immune sera, starting with a dilution of 1:200. Titer was defined as the reciprocal of the last dilution of antiserum resulting in an O.D. of >0.1. Non-specific immunoglobulin binding to the surface of whole streptococci was blocked by the addition of a mixture of 3% pig and 2% goat serum prior to the addition of the test antiserum. Peptide inhibition assays were performed by pre-incubating the E4 vaccine rabbit antisera with each E4 synthetic peptide (10 μg/ml) prior to the addition of the serum to ELISA wells containing solid-phase peptide antigens. Percent inhibition was calculated using PBS as a negative control.
Bactericidal assays
Bactericidal assays were performed in quadruplicate as previously described [20]. Briefly, 0.01 ml of Todd Hewitt broth containing log-phase bacteria was added to 0.05 ml of test serum or normal rabbit serum (NRS). Then 0.140 ml of lightly heparinized non-immune human blood was added and the complete mixture was rotated for 2 hrs at 37°C. Aliquots of this mixture were plated onto blood agar plates, and incubated overnight at 37°C. Colony forming units (CFU) were counted, averaged, and the results were expressed as percent killing, calculated using the formula: [(1- (total CFU with test serum/total CFU with NRS)) x 100]. Only those assays that resulted in growth of the test strain to at least four generations in the presence of NRS were considered valid tests. Data were reported as the average percent killing in quadruplicate assays +/− the standard deviation calculated after averaging the CFU in the four control samples.
Bivariate contingency analysis
The results of ELISA peptide-inhibition assays were used to determine the antigenic relatedness among all E4 peptides using antiserum against the E4 peptide vaccine. A bivariate contingency statistical analysis was used in the program MOE where the correlation between the 22 protein descriptors and percent inhibition were calculated, as well as the dependency between the variables following a Chi distribution and the corresponding uncertainties. The contingency coefficients, entropic uncertainty, linear correlations and Cramer’s coefficients were calculated and the principal component analysis (PCA) descriptors that were found to have significant statistical significance in representing the percent inhibition of cross-reactive antibodies were used to re-analyze the structural relatedness of all E4 peptides in diversity space.
Study approvals
All protocols using human blood samples were approved by the University of Tennessee Health Science Center Institutional Review Board. Animal protocols were approved by the University of Tennessee Health Science Center Institutional Animal Care and Use Committee.
3. Results
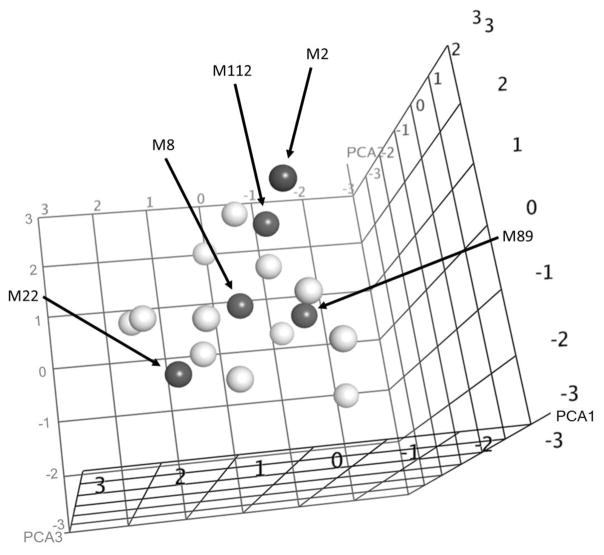
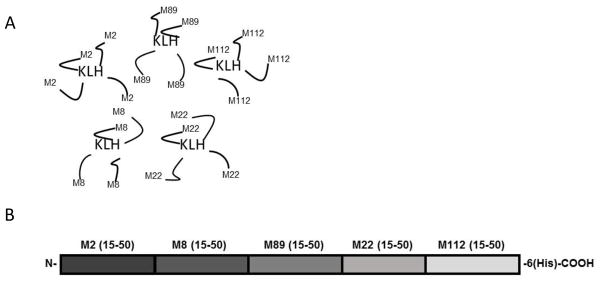
To determine the feasibility of using the emm cluster typing system as the basis for structure-based vaccine design, we focused on the E4 emm cluster, which contains 17 GAS emm types that are the most prevalent, compared to any one of the other emm clusters, in both the US and elsewhere in the world [12, 13]. Our goal was to find a small number of peptides that would evoke antibodies that cross-reacted with all other E4 peptides and promoted functional bactericidal activity against all emm types in the cluster. The first step was to perform computational de novo modeling of 3-D peptide structures, for which we used the PEP-FOLD software [17]. This software was limited to peptides of ~35 amino acid (AA) residues. Therefore, we initially analyzed the relatedness of the 17 E4 N-terminal M peptides spanning the sequences AA 1–35 and AA 16–50. A comparison of the 3-D structures of both sets of predictions revealed greater structural similarities among the 17 peptides copying the 16–50 AA, and these were subsequently analyzed using a set of 22 “protein” QSAR descriptors in the MOEv2014 program (Chemical Computing Group, Montreal, Canada). The descriptors quantify the physicochemical properties of a peptide with a given 3-D structure and sequence. The first three components in a principal component analysis (PCA) of the descriptor space represented 74% of the overall variance (Fig. 1). Five peptides, emm2, 8, 22, 89, and 112, were selected to represent the E4 cluster by identifying sequences that spanned the physicochemical descriptor range of the 17 peptides in PCA space. These peptide sequences were used to formulate the synthetic and recombinant pentavalent E4 vaccines (Fig. 2).
Fig 1.
Relatedness of physicochemical properties of E4 peptides calculated from structures derived using de novo 3-D peptide structure prediction and represented in principal component space. Peptides selected to represent all E4 peptides are indicated by solid symbols.
Fig. 2.
Schematic representation of the pentavalent E4 synthetic (A) and recombinant hybrid (B) vaccines.
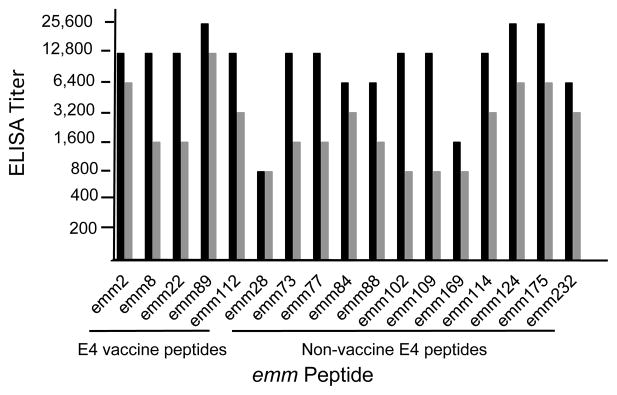
Vaccine-specific and cross-reactive peptide antibodies elicited by the pentavalent E4 synthetic peptide vaccine
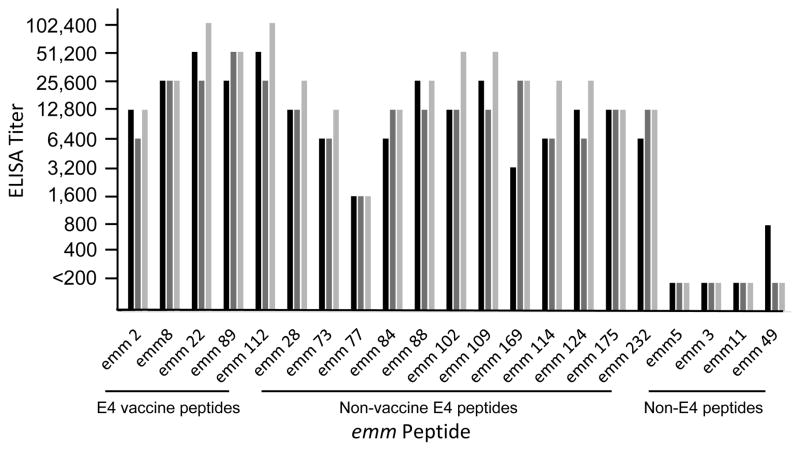
The pentavalent E4 synthetic peptide vaccine evoked high levels of antibodies in three rabbits against all 5 vaccine peptides, as well as significant levels (≥8-fold) of antibodies against the 12 non-vaccine E4 peptides (Fig. 3). The antisera did not react at all with three of the non-E4 peptides tested (emm5, clade Y; emm 3, cluster A-C5; and emm11, cluster E6). Antiserum from one rabbit showed low levels of antibody binding to the emm49 peptide, which is located in the adjacent E3 cluster [12]. Antiserum from rabbit #3, which had the highest titers against all peptides, was selected for detailed studies of antibody binding and functional bactericidal assays.
Fig. 3.
E4 peptide-specific and cross-reactive antibodies evoked by pentavalent E4 synthetic peptide vaccine. Each of the three bars represents the titer obtained with serum from one of the three rabbits.
Bactericidal activity of E4 pentavalent peptide vaccine antisera
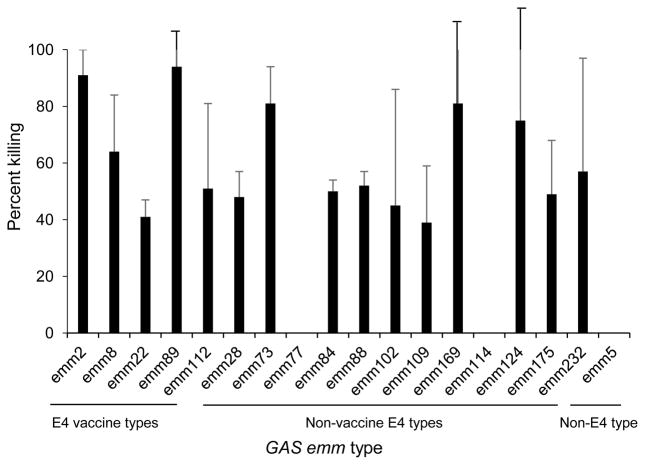
Each of the E4 emm types of GAS was used in assays to assess opsonic activity of the immune sera, as determined by bactericidal killing mediated by neutrophils in whole human blood (Fig. 4). Bactericidal activity ranging from 40–90% was demonstrated against 15 of the 17 E4 emm types. None of the antisera showed bactericidal activity against emm77 or emm114 GAS in repeated assays. We concluded that the level (or avidity) of antibodies against emm77 in the E4 vaccine antisera (titer of 1,600) was not sufficient to promote opsonization. A more interesting observation was that emm114 was not opsonized by the E4 vaccine immune sera, although the antibodies cross-reacted with the emm114 peptide with a titer of 25,600 (Fig. 3).
Fig. 4.
Bactericidal activity of pentavalent E4 peptide vaccine antisera against vaccine emm types and non-vaccine emm types of GAS in the E4 cluster. Each data point represents the mean bactericidal killing observed in four independent experiments (+/− S.D.).
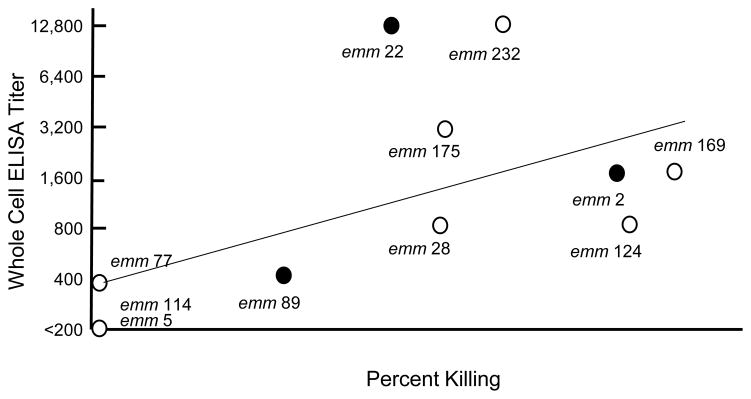
Binding of peptide antibodies to the native M protein on the bacterial surface
The dissociation between antibody binding to the emm114 peptide and the functional bactericidal activity has at least two potential explanations. First, the antibody may bind in high titer to the peptide but not to the epitopes in their native conformation on the surface of the organism. Second, the epitopes on the surface of the organism may be blocked by the specific interaction of human plasma proteins known to participate in the resistance of GAS to opsonization [3]. To differentiate between these two possibilities, we performed ELISA using ten E4 GAS as solid-phase antigens and the E4 peptide vaccine antisera and compared antibody binding to bactericidal activity (Fig. 5). In general, there is a correlation between antibody binding to the bacterial cell surface and functional activity. The results indicate that the emm types that resulted in no bactericidal activity (emm77, 114, and 5, which is the non-E4 emm type) also showed little to no antibody binding in the absence of plasma proteins. This suggests that the peptide antisera did not promote bactericidal killing of emm77 and emm114 because the antibodies did not bind to the epitopes on the bacterial surface and not because the binding was blocked by host proteins in the whole blood bactericidal assay. While the result with emm77 (titer of 400 with whole bacteria) supports our hypothesis that the titer (or avidity) of cross-reactive antibodies was insufficient to promote opsonization, the absence of bactericidal activity against emm114 indicates that the antibodies that cross-reacted with the emm114 peptide did not recognize or were unable to access the epitopes in the native protein on the surface of the organism, due possibly to a change in conformation of the epitope region arising from interactions with the remainder of the M114 protein.
Fig. 5.
Association between E4 synthetic peptide vaccine antibody binding to the M protein on the surface of whole streptococci and functional bactericidal activity in human blood. Vaccine emm types are indicated by solid symbols. (R=0.6, p=0.05, Pearson’s correlation coefficient).
Peptide specificity of antibodies evoked by the E4 vaccine
To directly assess the antigenic relatedness among the E4 peptides, we performed ELISA inhibition assays using the E4 vaccine peptides as soluble inhibitors of E4 vaccine antibodies binding to all peptides from the E4 cluster (Table 1). As expected, the homologous vaccine peptide resulted in significant antibody inhibition. Interestingly, in most cases a single vaccine peptide resulted in significant inhibition of antibodies cross-reactive with multiple non-vaccine peptides (range 2–4).
Table 1.
Peptide specificity of E4 vaccine antibodies that cross-reacted with non-vaccine E4 peptides
| ELISA Peptide | % Inhibition of Antibody Binding by Vaccine Peptide:
|
||||
|---|---|---|---|---|---|
| emm2 | emm8 | emm22 | emm89 | emm112 | |
| emm 2 | 89 | 0 | 0 | 0 | 0 |
| emm 8 | 0 | 90 | 0 | 2 | 0 |
| emm 22 | 0 | 4 | 76 | 0 | 7 |
| emm 89 | 7 | 2 | 5 | 76 | 0 |
| emm 112 | 0 | 6 | 0 | 0 | 85 |
| emm 73 | 76 | 0 | 0 | 1 | 0 |
| emm 77 | 18 | 0 | 13 | 1 | 40 |
| emm 84 | 1 | 1 | 0 | 88 | 0 |
| emm 88 | 0 | 32 | 0 | 0 | 100 |
| emm 102 | 10 | 0 | 0 | 12 | 59 |
| emm 109 | 6 | 3 | 81 | 0 | 11 |
| emm 114 | 84 | 0 | 9 | 1 | 0 |
| emm 28 | 4 | 0 | 78 | 0 | 14 |
| emm 124 | 80 | 0 | 7 | 0 | 0 |
| emm 169 | 2 | 40 | 0 | 0 | 5 |
| emm 175 | 63 | 0 | 13 | 18 | 0 |
| emm 232 | 0 | 2 | 0 | 82 | 0 |
In addition to testing the initial structural predictions (Fig. 1), the results from peptide-inhibition studies also provided quantitative data to inform the computational models used to predict structural and functional relatedness. We applied these empirical data to modify the physicochemical descriptors used to predict the structural relatedness of peptides within the E4 cluster. Of 22 descriptors that were used in the PCA calculations to represent the physicochemical and structural properties of the peptides, only the peptide masses, isoelectric points, volumes, and hydrophobicity were found to have significant statistical significance in representing the percent inhibition of cross-reactive antibodies. The peptide sequences with axes corresponding to values of the three most statistically important protein descriptors, volume, mass and isoelectric potential, were then displayed in regions of diversity space that correlated with percentage antibody inhibition that defines the structural and antigenic relatedness within a cluster (results not shown), thus representing an iterative process to inform the computational modeling leading to the identification of a different set of five peptides (M2, 84, 102, 124, and 175) that could then be formulated into a new E4 vaccine predicted to evoke high titer functional antibodies against all E4 emm types of GAS.
Prototype recombinant hybrid pentavalent E4 vaccine
Although synthetic peptides are becoming more attractive and affordable as vaccine antigens, the most cost-effective approach to vaccine formulation remains recombinant proteins. Therefore, as a proof-of-principle we constructed a recombinant fusion protein containing the same E4 peptides that were incorporated in the synthetic peptide vaccine described above (Fig. 2B). Two rabbits received three 200μg doses of the unconjugated protein on alum at 4-week intervals. The fusion protein evoked antibodies against each of the vaccine peptides and cross-reactive antibodies against the remaining E4 peptides (Fig. 6), albeit with slightly lower titers than those observed with the synthetic peptide vaccine formulated with Freund’s adjuvant.
Fig. 6.
E4 peptide-specific and cross-reactive antibodies evoked by recombinant pentavalent E4 hybrid peptide vaccine. Each of the two bars represents the titer obtained with serum from one of two immunized rabbits.
4. Discussion
Structure-based design of vaccines is a relatively new approach to identify and/or engineer vaccine antigens that elicit broadly neutralizing antibodies (bNAb) against human pathogens that display highly variable protective epitopes [2]. In some cases, structural vaccinology has utilized crystal structures of potential protective antigens in complex with bNAbs to define peptide structures that elicit broadly protective immunity [2]. Another approach has involved identifying protective epitopes within larger structurally defined proteins and excluding non-protective regions, thus minimizing the total amount of protein required to elicit protective immunity. A minimal number of protective epitopes may be combined in ways that retain antigenic structures that elicit an array of bNAbs against multiple different strains of the same organism. This strategy has been used successfully in the design of a novel chimeric vaccine against group B streptococcus that contained immunogenic domains of six structurally related but antigenically different pilus subunits [21].
A major obstacle to the development of M protein-based GAS vaccines has been the number of different M proteins expressed by these organisms (>200), each of which by definition contains different N-terminal sequences that evoke antibodies with the greatest bactericidal activity [4]. The assumption that protection against infection in humans is M type-specific, coupled with the complexity of the global epidemiology of GAS infections [22], has created the perception that this is an insurmountable hurdle for the development of a broadly efficacious vaccine that could be deployed to all regions of the world. There is an especially pressing need for a safe and effective vaccine in in low- and middle-income countries where populations are at high risk for developing acute rheumatic fever and rheumatic heart disease, which are associated with significant morbidity and mortality [23].
The convergence of our observations of cross-opsonic immune responses among multiple emm types [7] and the new emm cluster typing system [12] has led to a working hypothesis that immunity to GAS infections is a combination of “cluster-specific” and “type-specific” immune responses that depend on the conformational properties of the epitopes concerned. Our overall hypothesis is that broadly protective vaccines can be developed using computational design that takes full advantage of the structural relatedness of M proteins. It is well-known that three-dimensional peptide structure is more conserved than sequence [24]. Hence, determination of the structural properties of the M protein peptides with the greatest potential to evoke broadly cross-protective immune responses should permit a reduction in the number of peptides required for a highly efficacious vaccine.
In this study, we have demonstrated the feasibility of using de novo peptide structure predictions to determine antigenic relatedness among 17 different emm peptides in one M cluster and to predict those that evoke antibodies that cross-react with other emm types in the cluster. Unlike some previous reports of structure based vaccine design, we have focused only on the array of native peptide structures expressed by GAS. The feasibility of this approach is partly supported by the observation that the structures of the N-terminal regions of M proteins are constrained by their function(s) in virulence, including binding of the complement regulatory proteins C4BP [25, 26], factor H (and factor H-like protein) [27] and protection from proteolytic digestion [28]. Thus, the new cluster-based system predicts groups of related peptide structures whose diversity is limited by functional constraints. The ability to predict closely related 3-dimensional peptide structures allowed the selection of potentially cross-protective peptides. Indeed, in the current study the rabbit antisera against the E4 pentavalent vaccine reacted with all vaccine peptides and cross-reacted with all twelve non-vaccine E4 peptides. The synthetic E4 vaccine was formulated with Freund’s adjuvant to elicit high levels of antibodies for evaluation of cross-reactive functional activity. The lower levels of antibodies evoked by the recombinant vaccine formulated on alum were likely the result of reduced adjuvant activity. Eventual formulations for human trials may take advantage of different adjuvants, such as mono-phosphoryl lipid A or QS-21 [29], to enhance the potential of eliciting high titer cross-opsonic antibodies.
Our finding that not all emm types within the cluster were opsonized equally by the E4 vaccine antisera resulted in the development of an informatics-based assessment of physical-chemical descriptors combined with empiric data on antibody binding that defined a new set of peptides within the cluster that should result in broader cross-protective immunity. Finally, we demonstrated the feasibility of using recombinant hybrid proteins to formulate multivalent vaccines containing peptides representing the emm clusters. We believe that these preliminary results set the stage for comprehensive studies to design, develop, and test a broadly protective vaccine that will elicit cluster-specific and emm type-specific antibody responses against the majority of the 175 clinically relevant emm types of GAS.
Highlights.
emm clusters were used as the basis for structure-based design of vaccines
3-D structures and physicochemical properties identified five M peptides representing 17 emm types
Pentavalent synthetic peptide vaccine antibodies reacted with all 17 M peptides in the cluster
Vaccine antibodies promoted bactericidal activity against 15/17 streptococcal emm types
Structure-based design may result in broadly protective group A streptococcal vaccines
Acknowledgments
This work was supported by research funds from the U.S.P.H.S. National Institutes of Health AI-010085 (J.B.D.).
Footnotes
Author Contributions
J.B.D. conceived of and designed the study, analyzed the results, and wrote the manuscript; P.R.S. contributed to the design of the study, analyzed data, and edited the manuscript; H.S.C. contributed to the experimental design, interpretation of results, and wrote the manuscript; T.A.P. conducted experiments, acquired data, and analyzed results; C.M.H. conducted experiments, acquired data, and analyzed results; J.C.S. contributed to the design of the study, analyzed the results, and edited the manuscript; J.Y.B. contributed to the design of the study, conducted experiments, acquired data, analyzed results, and edited the manuscript.
Conflict of Interest
JBD is the inventor of certain technologies related to the development of group A streptococcal vaccines. The University of Tennessee Research Foundation has licensed the technology to Vaxent, LLC, of which JBD is the Chief Scientific Officer and a member. All other authors have no conflicts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulp DW, Schief WR. Advances in structure-based vaccine design. Curr Opin Virol. 2013;3:322–31. doi: 10.1016/j.coviro.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, et al. Disease manifestations and pathogenic mechanisms of group a streptococcus. Clin Microbiol Rev. 2014;27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones KF, Fischetti VA. The importance of the location of antibody binding on the M6 protein for opsonization and phagocytosis of group A M6 streptococci. J Exp Med. 1988;167:1114–23. doi: 10.1084/jem.167.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu MC, Walls MA, Stroop SD, Reddish MA, Beall B, Dale JB. Immunogenicity of a 26-valent group A streptococcal vaccine. Infect Immun. 2002;70:2171–7. doi: 10.1128/IAI.70.4.2171-2177.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penfound Ta, Chiang EY, Ahmed Ea, Dale JB. Protective efficacy of group A streptococcal vaccines containing type-specific and conserved M protein epitopes. Vaccine. 2010;28:5017–22. doi: 10.1016/j.vaccine.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale JB, Penfound TA, Chiang EY, Walton WJ. New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci. Vaccine. 2011;29:8175–8. doi: 10.1016/j.vaccine.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dale JB. Multivalent group A streptococcal vaccine designed to optimize the immunogenicity of six tandem M protein fragments. Vaccine. 1999;17:193–200. doi: 10.1016/s0264-410x(98)00150-9. [DOI] [PubMed] [Google Scholar]
- 9.Kotloff KL, Corretti M, Palmer K, Campbell JD, Reddish MA, Hu MC, et al. Safety and immunogenicity of a recombinant multivalent group a streptococcal vaccine in healthy adults: phase 1 trial. JAMA. 2004;292:709–15. doi: 10.1001/jama.292.6.709. [DOI] [PubMed] [Google Scholar]
- 10.McNeil SA, Halperin SA, Langley JM, Smith B, Warren A, Sharratt GP, et al. Safety and immunogenicity of 26-valent group A streptococcus vaccine in healthy adult volunteers. Clin Infect Dis. 2005;41:1114–22. doi: 10.1086/444458. [DOI] [PubMed] [Google Scholar]
- 11.Tapia MD, Sow SO, Tamboura B, Keita MM, Berthe A, Samake M, et al. Streptococcal Pharyngitis in Schoolchildren in Bamako. Mali Pediatr Infect Dis J. 2014 doi: 10.1097/INF.0000000000000608. [DOI] [PMC free article] [PubMed]
- 12.Sanderson-Smith M, De Oliveira DMP, Guglielmini J, McMillan DJ, Vu T, Holien JK, et al. A Systematic and Functional Classification of Streptococcus pyogenes That Serves as a New Tool for Molecular Typing and Vaccine Development. J Infect Dis. 2014;210:1325–38. doi: 10.1093/infdis/jiu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shulman ST, Tanz RR, Dale JB, Beall B, Kabat W, Kabat K, et al. Seven-year surveillance of north american pediatric group a streptococcal pharyngitis isolates. Clin Infect Dis. 2009;49:78–84. doi: 10.1086/599344. [DOI] [PubMed] [Google Scholar]
- 14.Daidone I, Ulmschneider MB, Di Nola A, Amadei A, Smith JC. Dehydration-driven solvent exposure of hydrophobic surfaces as a driving force in peptide folding. Proc Natl Acad Sci U S A. 2007;104:15230–5. doi: 10.1073/pnas.0701401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thukral L, Daidone I, Smith JC. Structured pathway across the transition state for peptide folding revealed by molecular dynamics simulations. PLoS Comput Biol. 2011;7:e1002137. doi: 10.1371/journal.pcbi.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maupetit J, Derreumaux P, Tuffery P. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009;37:W498–503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Y, Maupetit J, Derreumaux P, Tuffery P. Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J Chem Theory Comput. 2014;10:4745–58. doi: 10.1021/ct500592m. [DOI] [PubMed] [Google Scholar]
- 18.Dale JB, Beachey EH. Localization of protective epitopes of the amino terminus of type 5 streptococcal M protein. J Exp Med. 1986;163:1191–202. doi: 10.1084/jem.163.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courtney HS, Hasty DL, Dale JB. Anti-phagocytic mechanisms of Streptococcus pyogenes: binding of fibrinogen to M-related protein. Mol Microbiol. 2006;59:936–47. doi: 10.1111/j.1365-2958.2005.04977.x. [DOI] [PubMed] [Google Scholar]
- 20.Dale JB, Niedermeyer SE, Agbaosi T, Hysmith ND, Penfound TA, Hohn CM, et al. Protective immunogenicity of group A streptococcal M-related proteins. Clin Vaccine Immunol. 2015;22:344–50. doi: 10.1128/CVI.00795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuccitelli A, Cozzi R, Gourlay LJ, Donnarumma D, Necchi F, Norais N, et al. Structure-based approach to rationally design a chimeric protein for an effective vaccine against Group B Streptococcus infections. Proc Natl Acad Sci U S A. 2011;108:10278–83. doi: 10.1073/pnas.1106590108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9:611–6. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 23.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–94. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 24.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–6. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persson J, Beall B, Linse S, Lindahl G. Extreme sequence divergence but conserved ligand-binding specificity in Streptococcus pyogenes M protein. PLoS Pathog. 2006;2:e47. doi: 10.1371/journal.ppat.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buffalo CZ, Bahn-Suh AJ, Hirakis SP, Biswas T, Amaro RE, Nizet V, et al. Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein. Nat Microbiol. 2016;1:16155. doi: 10.1038/nmicrobiol.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gustafsson MC, Lannergard J, Nilsson OR, Kristensen BM, Olsen JE, Harris CL, et al. Factor H binds to the hypervariable region of many Streptococcus pyogenes M proteins but does not promote phagocytosis resistance or acute virulence. PLoS Pathog. 2013;9:e1003323. doi: 10.1371/journal.ppat.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penfound TA, Ofek I, Courtney HS, Hasty DL, Dale JB. The NH(2)-terminal region of Streptococcus pyogenes M5 protein confers protection against degradation by proteases and enhances mucosal colonization of mice. J Infect Dis. 2010;201:1580–8. doi: 10.1086/652005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunningham AL, Garcon N, Leo O, Friedland LR, Strugnell R, Laupeze B, et al. Vaccine development: From concept to early clinical testing. Vaccine. 2016 doi: 10.1016/j.vaccine.2016.10.016. [DOI] [PubMed] [Google Scholar]