Abstract
The Notch signaling pathway has a critical role in cell fate determination and tissue homeostasis in a variety of different lineages. In the context of normal Notch signaling, the Notch receptor of the “signal-receiving” cell is activated in trans by a Notch ligand from a neighboring “signal-sending” cell. Genetic studies in several model organisms have established that ubiquitination of the Notch ligand, and its regulated endocytosis, is essential for transmission of this activation signal. In mammals, this ubiquitination step is dependent on the protein Mind bomb 1 (Mib1), a large multi-domain RING-type E3 ligase, and its direct interaction with the intracellular tails of Notch ligand molecules. Here, we discuss our current understanding of Mind bomb structure and mechanism in the context of Notch signaling and beyond.
Introduction to Notch signaling
Notch signaling is a juxtamembrane cell-cell communication pathway conserved in all metazoans [1]. These signals play critical roles in cell-fate determination, cellular proliferation and apoptosis [2]. The four mammalian homologs of Drosophila Notch and their five Delta-like and Jagged ligands (Delta-like 1,3 and 4, and Jagged 1,2) are expressed at the cell surface as single-pass transmembrane proteins. In canonical activation, the distal portion of the Notch extracellular region binds to the headpiece of one of these ligands expressed on an adjacent cell (Figure 1a), leading to receptor proteolysis and delivery of the NICD to the nucleus where it acts as an accessory transcription factor [1].
Figure 1.
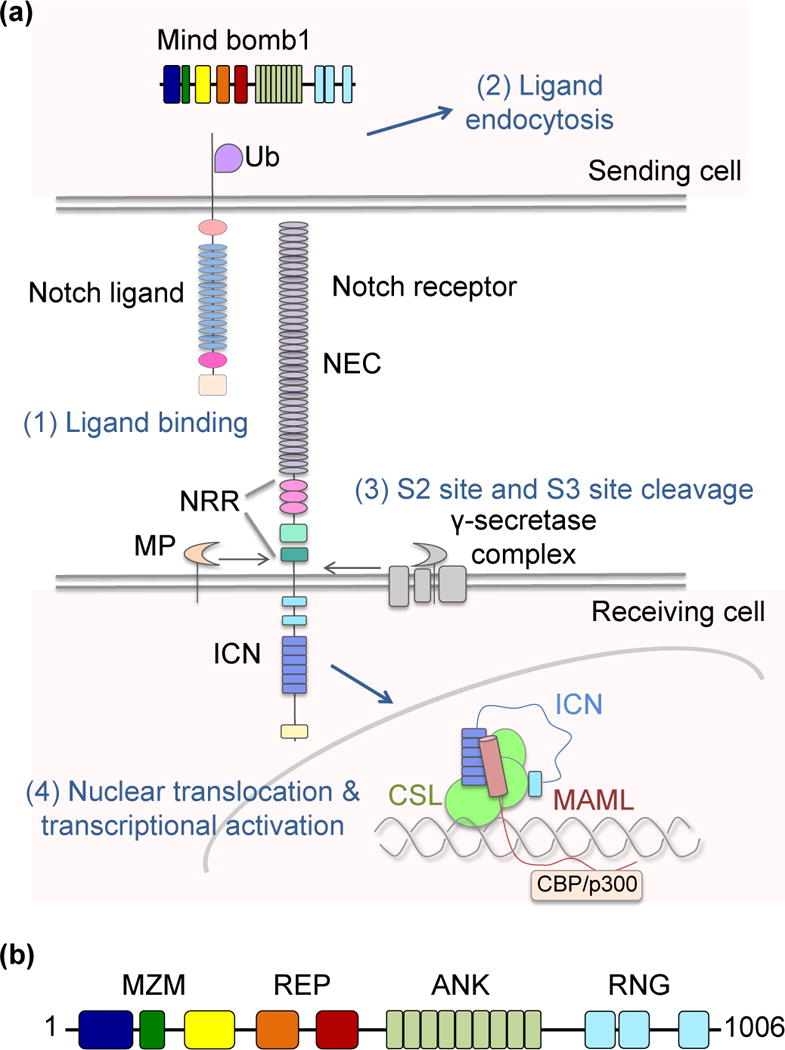
Schematic of major components in the Notch signaling pathway and Mib domain organization. (a) Notch signaling initiates upon transcellular engagement of a Notch ligand with a Notch receptor, and relies on ubiquitin-dependent internalization of the ligand. In vertebrates, ubiquitination is dependent on the E3 ligase Mind bomb 1 (Mib1). Transcellular delivery of force is thought to expose a site (S2) within the Notch negative regulatory region (NRR) for cleavage by ADAM-family metalloproteases. S2 cleavage is followed by cleavage at site S3 by gamma secretase, leading to release and entry of of the intracellular Notch (ICN) domain into the nucleus. Within the nucleus, ICN forms a ternary transcriptional complex with CSL and MAML to regulate target gene expression. (b) Domain organization of Mib1 highlighting the MZM, REP, ANK, and RNG regions of the protein. The MZM domain contains two Mib-Herc2 domains (blue and yellow) flanking a ZZ Zinc finger (green). The REP domain contains two tandem “Mib repeat” elements (orange and red). The ANK domain is composed of nine ankyrin-type repeats (light green), and the C-terminal RNG domain is composed of three RING elements (cyan).
The structural basis for binding of Notch1 to an evolved version of its highest affinity ligand, Delta-like 4, has recently been reported [3]. This transcellular handshake, however, is not sufficient for activation of a Notch signal [4]. Work from many groups has shown that endocytosis of the Notch ligand is required for receptor activation and strongly suggests that this event is intimately associated with the application of mechanical force to the Notch molecule on the signal-receiving cell [5,6]. The transmitted force appears to relieve autoinhibition of the receptor by unmasking the ligand-dependent metalloprotease cleavage site. Proteolytic cleavage at this site by members of the ADAM metalloprotease family, followed by gamma secretase cleavage and release of the intracellular Notch (ICN) domain produces the intracellular effector that forms a ternary nuclear complex with the transcription factor CSL (short for CBF1, Su(H) and LAG-1) and a Mastermind (MAML) coactivator to regulate target gene expression (Figure 1a).
Role of E3 ligases in Notch signaling
Ubiquitination of the intracellular tail of a Notch ligand is a key event that licenses these molecules for subsequent endocytosis and productive signaling. Genetic studies initially performed in flies and zebrafish identified two families of E3 ubiquitin ligase capable of ligand ubiquitination: Mind bomb (Mib) and Neuralized (Neur) proteins [7–9•]. In the fly, both Neuralized and Mind bomb have essential and non-redundant roles in Notch pathway activation, but can functionally substitute for each other when ectopically expressed [10•]. Suprisingly, these two E3 ligase families share virtually no similarities in structure or domain architecture [11], and their respective Notch ligand epitopes are non-overlapping [12••].
In vertebrates, the Neuralized ligases do not perform an essential role in Notch signaling, and accordingly the epitope recognized by Neur is no longer evident in the ligand intracellular tail [12••]. A series of germline and conditional knockout mice of the Mind bomb (paralogs 1 and 2) and Neuralized (paralogs 1 and 2) have shown that loss of Mind bomb 1 alone is sufficient for wide-spread Notch loss-of-function phenotypes across numerous tissues [13,14]. Loss of Mib2 or either Neuralized protein, however, is tolerated without major Notch phenotypes, indicating that Mib1 has a primary, and perhaps exclusive, role in Notch ligand ubiquitination and trafficking in mammals [15]. In humans, single-allele familial mutations of Mib1 correlate with left ventricular non-compaction cardiomyopathy (LVNC) and aberrant Notch signaling [16••].
Mib1 functions outside the Notch pathway
Mind bomb proteins have been implicated in a number of cellular processes in addition to Notch ligand ubiquitination. Unbiased proteomic studies using immunoprecipitation followed by mass spectrometry and yeast two-hybrid screens have been performed to elucidate a more complete Mib1 interactome. These studies Mib1 associates with diverse targets including membrane trafficking proteins, cell adhesion components, replication/transcription/translation factors and signal transduction proteins [17,18].
More detailed functional studies demonstrate a role for Mib1 in the regulation of seemingly unconnected areas of cell biology. For example, Mib1 positively regulates Wnt/β-catenin signaling through ubiquitination and degradation of receptor-like tyrosine kinase (RYK) [19]. Multiple regulators of apoptosis are modulated by Mib1 ubiquitination, including death-associated protein kinase (DAPK) and cellular FLICE-like inhibitory protein (cFLIP) [20,21]. An important role for Mib1 in centriolar biology is emerging, and suggests direct interaction and regulation of multiple factors including PLK4 (polo-like kinase 4), AZI1 (5-azacytidine induced 1), and PCM1 (pericentriolar material 1) [22,23]. In neuronal cells, Mib1 directly modulates cyclin-dependent kinase 5 (CDK5) and survival of motor neuron (SMN) protein levels independent of its well-studied role in Notch-mediated neurogenesis [17,24,25]. The structural mechanism for these non-canonical interactions is currently unclear, but may utilize binding modes similar to that elucidated for its engagement of Notch ligand substrates [26••].
Mind bomb architecture
Mind bomb proteins are large multi-domain E3 ligases with a distinct modular architecture (Figure 1b). At their N-termini, Mib proteins possess two independent substrate recognition regions, referred to below as MZM and REP. At the C-terminus, each protein contains multiple RING domains, one or more of which bind to E2~Ubiquitin (E2~Ub) conjugates. Separating the substrate recognition modules from the RING domains are a series of ankyrin repeats (Figure 1b). It remains unclear how this particular architecture facilitates Mind bomb-catalyzed ubiquitin transfer. The overall layout, however, resembles the organization of the multi-protein Cullin scaffolded ligases, with all subunits instead contained within a single polypeptide (Figure 2). Similar to the Cullin component of the Cullin-RING ligases (CRLs), reviewed in [27], the ankyrin repeats may act as a spacer to hold the substrate and E2~Ub at the proper distance and correct orientation for optimal catalysis of ubiquitin transfer.
Figure 2.
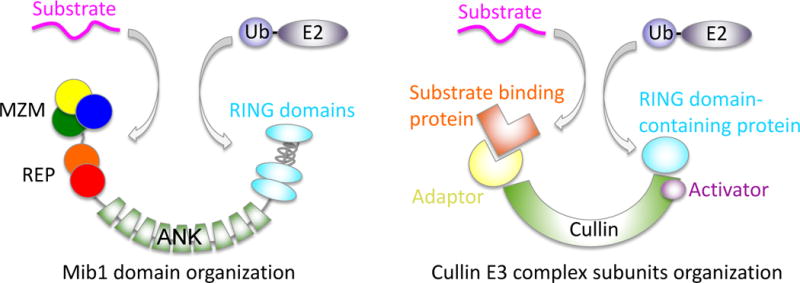
Potential architecture of Mib1 and comparison to a typical Cullin E3 ligase.
Structure of the substrate recognition domains
Past structural efforts have focused on the N-terminal portion of the protein, which is necessary and sufficient for this ligand recognition [9•,26••,28]. The structure of this fragment, determined by X-ray crystallography, revealed that the Mind bomb N-terminus is actually composed of two separate elements that independently recognize discrete and independent epitopes on the ligand cytoplasmic tail (Figure 1b, 3a). The N-terminal domain, referred to as the MZM element (short for Mib-Herc2/ZZ Zinc Finger/Mib-Herc2), adopts a tricorn overall shape. The two Mib-Herc2 subdomains sandwich the ZZ Zinc finger, with extensive interdomain interactions that are further stabilized by a two-stranded anti-parallel β-sheet that bridges all three sub-domains (Figure 3a) [26••]. Such an intimate organization between the two Mib-Herc2 modules and ZZ Zinc finger was unexpected, as other Mib-Herc2 containing proteins, including the E3 ubiquitin ligases Herc2 and Hectd1, have only a single Mib-Herc2 domain and no neighboring ZZ Zinc finger.
Figure 3.
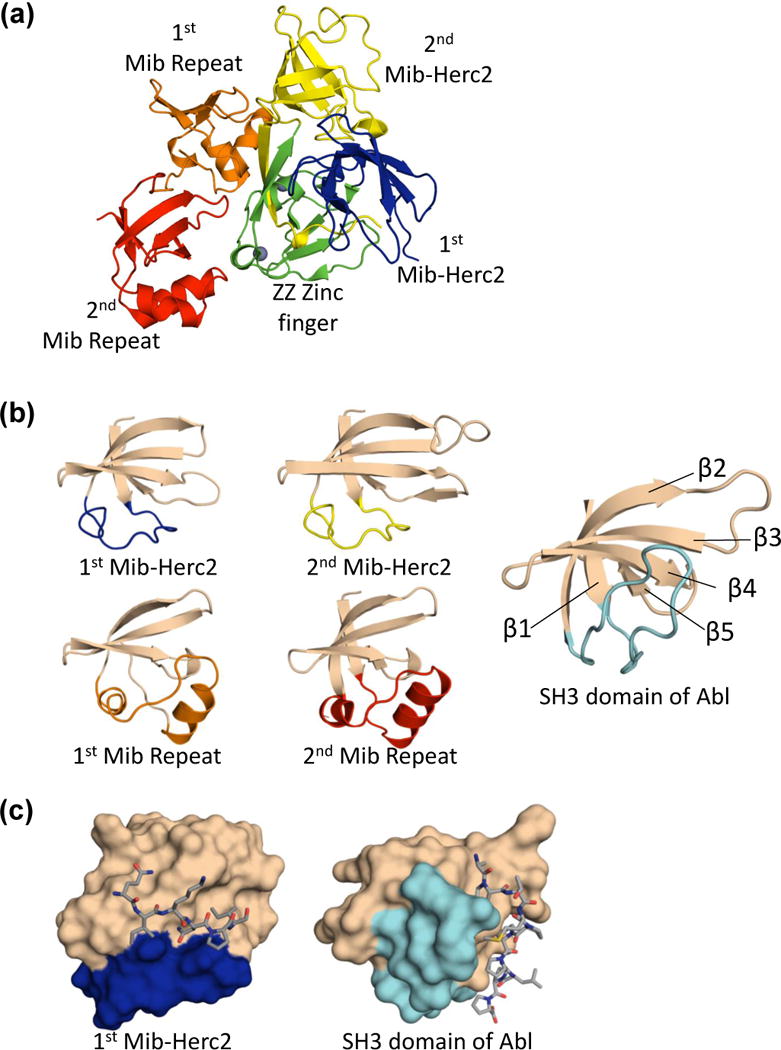
Structural features of the Mib1 MZM-REP region. (a) Ribbon representation of the MZM-REP structure, colored as in Figure 1. (b) Comparison of the Mib-Herc2 and Mib repeat domain folds to the SH3 domain of Abl. The variable linker connecting β strands 1 and 2 is highlighted in the color of the analogous domain within the larger MZM-REP fragment shown in panel (a). (c) Peptide binding mode for the Mib-Herc2 and SH3 domains. The Mib-Herc2 and SH3 domains are represented as a surface, and colored as in (b). Respective peptide epitopes are shown as sticks.
The second substrate recognition domain of Mind bomb, referred to as the REP domain, consists of two imperfect tandem repeats (Figure 1b, 3a). These modules have no apparent amino acid sequence identity with other protein domains and are not detected outside the Mind bomb family. The crystal structure of the isolated REP region revealed these tandem repeats to be independently folded with a flexible tether between them. Although the REP domain has substantial contacts with the MZM domain in the structure of the complete MZM-RAP fragment (Figure 3a), it is unclear whether these interdomain contacts are functionally important or are a consequence of a crystal packing requirement: small angle x-ray scattering measurements suggest that the “closed” MZM-REP crystal conformation is not well populated under the strongent high-salt conditions required for the analysis [26••].
Unexpectedly, both the Mib-Herc2 and Mib repeats bear topological similarity to a src homology-3 (SH3) domains despite sharing virtually no sequence identity with them (Figure 3b). All three domains adopt the same topological arrangement, and share the core β-sheet topology. The Mib-Herc2 and Mib repeats diverge, however, from typical SH3 domains in the linker between the first and second β-strands. This linker varies substantially in length, orientation and secondary structure among the SH3, Mib-Herc2 and Mib repeat folds, and plays a critical role in substrate recognition for all three domain types (Figure 3b). For each module type, the binding pocket is formed at the interface of the variable linker and the β-sheet. The structural mechanism for these binding events has been clearly elucidated for SH3 domains bound to proline-rich peptides [29], and for the binding of the first Mib-Herc2 module to its cognate epitope on the intracellular tails of Notch ligands (Figure 3c and 4a). The role of this interface in the binding of the Mib repeats to their cognate motif has instead been inferred by peptide-like crystal contacts and high sequence conservation, with accompanying testing by point mutagenesis in biochemical and cellular assays [26••].
Figure 4.
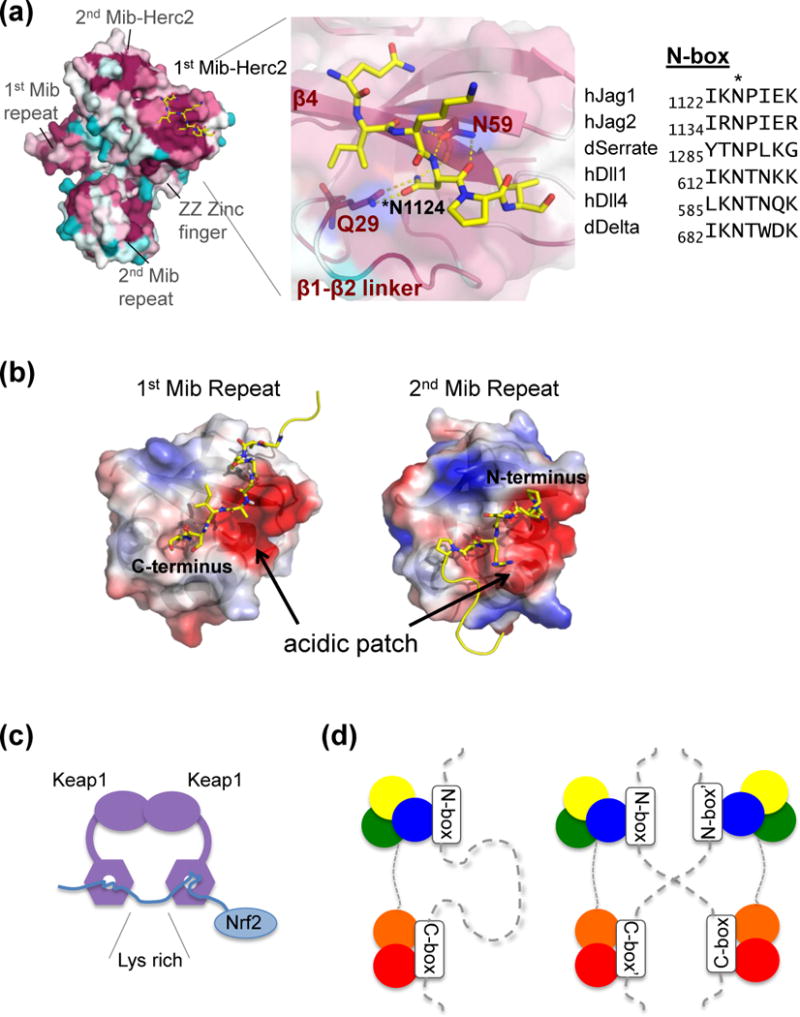
Bipartite ligand recognition of Mib1. (a) Co-crystal structure of MZM-REP and a Jag1 N-box peptide. Left panel: Mib1 represented as a surface and colored by conservation on a sliding scale from crimson (most conserved), to cyan (least conserved). The N-box peptide is represented as sticks (yellow, colored by atom type). Middle panel: close-up view of the Mib-Herc2:peptide binding interface. Right panel: sequence alignment of N-box epitopes from human (h) and Drosophila (d) Notch ligands. (b) Structure of the isolated Mib repeat region and intermolecular crystal contacts. Mib repeats 1 and 2 are shown in the same orientation and colored by surface charge (red: acidic, blue: basic). Peptides from the N-terminus and C-terminus of neighboring molecules are shown as sticks (yellow, and colored by atom). (c) Model of Nrf2 recognition by a Keap1 homodimer, and (d) schematic of ligand recognition by Mib1 in cis (left) and in trans (right).
Structural basis for epitope binding
Detailed biochemical and functional studies have shown that the MZM and REP domains function synergistically in recognition of the Notch ligand Jagged 1 (Jag1) [26••]. Although each domain is independently capable of a low-affinity interaction with the intracellular tail, the full MZM-REP domain exhibits a higher affinity for the ligand tail and bivalent ligand engagement results in enhanced ubiquitination rates. Two non-overlapping epitopes on Jag1 were identified as important for binding to Mib1. The first, near the juxtamembrane N-terminal portion of the intracellular tail, is termed the N-box (sequence IKNPIEK), and the second, near the C-terminus of the ligand, is termed the C-box (sequence KQDNRD). The N-box and C-box epitopes are independently recognized by the MZM and REP regions, respectively. The N-box epitope is conserved in both the Delta and Serrate/Jagged ligand families, and co-crystal structures of MZM-REP with peptides from human Jag1 or from fly Delta suggest these various interactions to be similar across all ligands (Figure 4a). The C-box epitope, however, is not apparent in the Delta family of Notch ligands, and a functional role for a direction interaction between REP and Delta-like ligands has not yet been demonstrated [26••].
High resolution atomic structures of the MZM-REP fragment in complex with N-box peptides have revealed the binding mode for engagement of N-box peptides with the MZM region. The N-box epitope contains a central asparagine residue, and the crystal structure shows that this side-chain forms key hydrogen bonds to Q29 of the Mib-Herc2 repeat that anchor the bound peptide (Figure 4a). Although the residues around this asparagine in the N-box are more variable, the co-crystal structures show packing of hydrophobic sidechains at positions −2 and +2 relative to the central asparagine. The Mib1 Q29 side chain, which hydrogen bonds with the epitope asparagine, and N59, which hydrogen bonds with the epitope mainchain, are both critical for biochemical binding and cellular function (Figure 4a) [26••].
The unliganded REP domain, crystallized in the absence of MZM, identified potential features for peptide recognition. Within the crystal, intermolecular contacts are formed at the α-β interface with N- and C-terminal extensions of neighboring molecules (Figure 4b). These sites, which are negatively charged and conserved among Mib repeats, are analogous to the binding grooves in SH3 and Mib-Herc2 domains. Neutralization of this charged patch disrupts the interaction with C-box epitopes, but the structural basis for C-box binding remains unresolved [26••].
Bipartite binding and role in ubiquitin transfer
The use of two independent binding epitopes for ligand recognition has potential implications for the assembly of ligand-Mib1 complexes at the membrane and the mechanism for transfer of ubiquitin to the ligand tails. The presence of two independent recognition epitopes on a single substrate is unusual, but not unprecedented in E3 biology.
One instructive example, with both similarities and differences when compared to the Mib1-ligand complex, is the Keap1-Cul3-RBX1 E3 ligase complex bound to its substrate Nrf2 [30,31•]. Nrf2 contains two discontinuous epitopes and uses this bipartite binding interface to position an intervening lysine-rich region for ubiquitination (Figure 4c). Similarly, lysine residues between the Jag1 N- and C-box epitopes are essential for the function of ligand molecules in signaling assays [32•], suggesting that bipartite binding by Mib might simultaneously play a role in lysine presentation to the ubiquitin-charged E2 subunit.
A notable distinction between Keap1 and Mib1 is that the stoichiometries of their complexes with substrate differ. Keap1 forms a dimer, in which each Keap1 subunit binds a different Nrf2 epitope on a single polypeptide chain, resulting in a Keap1:Nrf2 stoichiometry of 2:1 (Figure 4c). In contrast, the stoichiometry of Mib1-ligand complexes is 1:1, in which structurally distinct MZM and REP domains within a single Mib1 protein engage two different epitopes from a single ligand tail (Figure 4d). Intriguingly, modeling and cell-based studies by Luxán et al [16••] as well as unpublished work (BG, BJM and SCB) suggest that Mind bomb can dimerize, likely through the C-terminal RING region (see below). Thus, the potential exists for epitope engagement by Mib1 as 2:2 clusters with N- and C-box epitopes in trans, as well as for much higher order complexes assembled by “daisy chaining.” Whether Mib1 exhibits bipartite binding of other molecular partners outside of Notch signaling remains unresolved. The potential also exists for MZM and REP to bind different molecules within a larger protein complex concurrently, so that ubiquitination occurs only in the context of an assembled complex.
RING domains and E2~ubiquitin binding
RING domains, which are common to many E3 ligases, typically bind to ubiquitin loaded E2 proteins, catalyzing Ub transfer to substrate proteins [33]. All Mib1 orthologs possess three RING domains near their C-termini. Mind bomb, however, does not function as a RING-inbetween-RING (RBR)-type ligase as it does not share the characteristic C6HC zinc coordination motif of the RBR family [34,35] or its cysteine~Ub intermediate. The most C-terminal RING domain has been shown to be critical for several endpoints of Mib function, including neurogenesis, auto-ubiquitination, and ubiquitination of RYK [9•,19,21]. The question of why Mib1 has three RING domains remains unclear: no clear role for the first or second RING domain has been elucidated, and Mib2, which is a close homolog of Mib1, has only two RING domains.
Between the second and third RING domains of Mib1 is a predicted coiled coil region and a familial mutation within this sequence, V943F, causes left ventricular non-compaction cardiomyopathy [16••]. Intriguingly, the BIRC7 and IDOL E3 ligases contain a short coiled-coil segment immediately preceding their own RING domains and this helical interface helps drive the dimerization of these proteins [36•,37]. The V943F mutation could potentially disrupt Mind bomb function by interfering with an analogous coiled-coil dimerization motif.
Conclusions
The importance of Mib1 in the Notch pathway has been firmly established, and roles for Mib1 beyond Notch signal transduction are emerging from cellular and proteomic studies. Recent structural studies of N-terminal fragments of Mib1 in isolation and in complex with the cytoplasmic tail of Jag1 has identified a bipartite mode of recognition, but it remains to be seen whether a similar mechanism exists for recognition of other substrates. Moreover, the functional implications of bipartite binding, for example, in the formation of 2:2 or larger homo- or hetero-oligomeric complexes, are not yet clear. Structural elucidation of full-length Mib1 in complex with substrate and E2~Ub will fill current gaps in understanding the molecular logic of this E3 family and how its architecture promotes ubiquitin transfer to substrate proteins.
Highlights.
N-terminal region of Mind bomb1 recognizes two separate epitopes on Notch ligands
An architecture resembling cullin-E3 ligases is proposed for full-length Mind bomb
Bipartite tail recognition mecanism suggests potential for creating oligomeric assemblies
Acknowledgments
We thank Thomas Klein and members of the Blacklow laboratory for helpful discussions. This work is supported by NIH grants R01 CA092433 and P01 CA119070, Project 3 (to SCB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- 3.Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S, Garcia KC. Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science. 2015;347:847–853. doi: 10.1126/science.1261093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun X, Artavanis-Tsakonas S. The intracellular deletions of Delta and Serrate define dominant negative forms of the Drosophila Notch ligands. Dev Camb Engl. 1996;122:2465–2474. doi: 10.1242/dev.122.8.2465. [DOI] [PubMed] [Google Scholar]
- 5.Musse AA, Meloty-Kapella L, Weinmaster G. Notch ligand endocytosis: mechanistic basis of signaling activity. Semin Cell Dev Biol. 2012;23:429–436. doi: 10.1016/j.semcdb.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, et al. Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Dev Cell. 2015;33:729–736. doi: 10.1016/j.devcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haddon C, Jiang YJ, Smithers L, Lewis J. Delta-Notch signalling and the patterning of sensory cell differentiation in the zebrafish ear: evidence from the mind bomb mutant. Development. 1998;125:4637–4644. doi: 10.1242/dev.125.23.4637. [DOI] [PubMed] [Google Scholar]
- 8.Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- 9•.Itoh M, Kim C-H, Palardy G, Oda T, Jiang Y-J, Maust D, Yeo S-Y, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Mind Bomb Is a Ubiquitin Ligase that Is Essential for Efficient Activation of Notch Signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. Using a zebrafish model system, this study established the role of Mind bomb in Notch ligand endocytosis and receptor activation. [DOI] [PubMed] [Google Scholar]
- 10•.Le Borgne R, Remaud S, Hamel S, Schweisguth F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 2005;3:e96. doi: 10.1371/journal.pbio.0030096. Demonstrated that Neuralized and Mind bomb regulate distinct subsets of Notch-mediated development events in the fly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He F, Saito K, Kobayashi N, Harada T, Watanabe S, Kigawa T, Güntert P, Ohara O, Tanaka A, Unzai S, et al. Structural and functional characterization of the NHR1 domain of the Drosophila neuralized E3 ligase in the notch signaling pathway. J Mol Biol. 2009;393:478–495. doi: 10.1016/j.jmb.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 12••.Daskalaki A, Shalaby NA, Kux K, Tsoumpekos G, Tsibidis GD, Muskavitch MAT, Delidakis C. Distinct intracellular motifs of Delta mediate its ubiquitylation and activation by Mindbomb1 and Neuralized. J Cell Biol. 2011;195:1017–1031. doi: 10.1083/jcb.201105166. Identified non-overlapping sequence motifs in the fly ligand Delta, which are recognized separately by Mind bomb and Neuralized ligases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barsi JC, Rajendra R, Wu JI, Artzt K. Mind bomb1 is a ubiquitin ligase essential for mouse embryonic development and Notch signaling. Mech Dev. 2005;122:1106–1117. doi: 10.1016/j.mod.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Koo B-K, Lim H-S, Song R, Yoon M-J, Yoon K-J, Moon J-S, Kim Y-W, Kwon M, Yoo K-W, Kong M-P, et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- 15.Koo B-K, Yoon M-J, Yoon K-J, Im S-K, Kim Y-Y, Kim C-H, Suh P-G, Jan YN, Kong Y-Y. An obligatory role of mind bomb-1 in notch signaling of mammalian development. PloS One. 2007;2:e1221. doi: 10.1371/journal.pone.0001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Luxán G, Casanova JC, Martínez-Poveda B, Prados B, D’Amato G, MacGrogan D, Gonzalez-Rajal A, Dobarro D, Torroja C, Martinez F, et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med. 2013;19:193–201. doi: 10.1038/nm.3046. First paper reporting a heritable developmental defect resulting from mutation in a Mind bomb protein. Human germline mutations in Mind bomb 1 were shown to result in autosomal dominant left ventricular non-compaction cardiomyopathy. [DOI] [PubMed] [Google Scholar]
- 17.Mertz J, Tan H, Pagala V, Bai B, Chen P-C, Li Y, Cho J-H, Shaw T, Wang X, Peng J. Sequential Elution Interactome Analysis of the Mind Bomb 1 Ubiquitin Ligase Reveals a Novel Role in Dendritic Spine Outgrowth. Mol Cell Proteomics MCP. 2015;14:1898–1910. doi: 10.1074/mcp.M114.045898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tseng L-C, Zhang C, Cheng C-M, Xu H, Hsu C-H, Jiang Y-J. New Classes of Mind Bomb-Interacting Proteins Identified from Yeast Two-Hybrid Screens. PLOS ONE. 2014;9:e93394. doi: 10.1371/journal.pone.0093394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berndt JD, Aoyagi A, Yang P, Anastas JN, Tang L, Moon RT. Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/β-catenin signaling. J Cell Biol. 2011;194:737–750. doi: 10.1083/jcb.201107021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin Y, Blue EK, Dixon S, Shao Z, Gallagher PJ. A Death-associated Protein Kinase (DAPK)-interacting Protein, DIP-1, Is an E3 Ubiquitin Ligase That Promotes Tumor Necrosis Factor-induced Apoptosis and Regulates the Cellular Levels of DAPK. J Biol Chem. 2002;277:46980–46986. doi: 10.1074/jbc.M208585200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Gallagher PJ. Mind bomb 1 regulation of cFLIP interactions. Am J Physiol Cell Physiol. 2009;297:C1275–1283. doi: 10.1152/ajpcell.00214.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villumsen BH, Danielsen JR, Povlsen L, Sylvestersen KB, Merdes A, Beli P, Yang Y-G, Choudhary C, Nielsen ML, Mailand N, et al. A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J. 2013;32:3029–3040. doi: 10.1038/emboj.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Čajánek L, Glatter T, Nigg EA. The E3 ubiquitin ligase Mib1 regulates Plk4 and centriole biogenesis. J Cell Sci. 2015;128:1674–1682. doi: 10.1242/jcs.166496. [DOI] [PubMed] [Google Scholar]
- 24.Choe E-A, Liao L, Zhou J-Y, Cheng D, Duong DM, Jin P, Tsai L-H, Peng J. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J Neurosci Off J Soc Neurosci. 2007;27:9503–9512. doi: 10.1523/JNEUROSCI.1408-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon DY, Dimitriadi M, Terzic B, Cable C, Hart AC, Chitnis A, Fischbeck KH, Burnett BG. The E3 ubiquitin ligase mind bomb 1 ubiquitinates and promotes the degradation of survival of motor neuron protein. Mol Biol Cell. 2013;24:1863–1871. doi: 10.1091/mbc.E13-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.McMillan BJ, Schnute B, Ohlenhard N, Zimmerman B, Miles L, Beglova N, Klein T, Blacklow SC. A Tail of Two Sites: A Bipartite Mechanism for Recognition of Notch Ligands by Mind Bomb E3 Ligases. Mol Cell. 2015;57:912–924. doi: 10.1016/j.molcel.2015.01.019. Elucidated the structure of the Mib1 substrate recognition domains, and identified a bipartite mechanism for binding of the two recognition modules to different peptide epitopes on the Notch ligand Jag1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N, Schulman BA. Structural regulation of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2011;21:257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen W, Casey Corliss D. Three modules of zebrafish Mind bomb work cooperatively to promote Delta ubiquitination and endocytosis. Dev Biol. 2004;267:361–373. doi: 10.1016/j.ydbio.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 30.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 Recruits Neh2 through Binding to ETGE and DLG Motifs: Characterization of the Two-Site Molecular Recognition Model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88(Part B):101–107. doi: 10.1016/j.freeradbiomed.2015.05.034. Cogent and concise structure/function review of the bipartite Keap1:Nrf2 interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Glittenberg M, Pitsouli C, Garvey C, Delidakis C, Bray S. Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. EMBO J. 2006;25:4697–4706. doi: 10.1038/sj.emboj.7601337. This work distinguished between bulk endocytosis of the ligand Serrate dependent on a di-leucine motif from a second motif in the intracellular tail required for trans-activation of Notch signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budhidarmo R, Nakatani Y, Day CL. RINGs hold the key to ubiquitin transfer. Trends Biochem Sci. 2012;37:58–65. doi: 10.1016/j.tibs.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Lechtenberg BC, Rajput A, Sanishvili R, Dobaczewska MK, Ware CF, Mace PD, Riedl SJ. Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature. 2016;529:546–550. doi: 10.1038/nature16511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenhaber B, Chumak N, Eisenhaber F, Hauser M-T. The ring between ring fingers (RBR) protein family. Genome Biol. 2007;8:209. doi: 10.1186/gb-2007-8-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Zhang L, Fairall L, Goult BT, Calkin AC, Hong C, Millard CJ, Tontonoz P, Schwabe JWR. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 2011;25:1262–1274. doi: 10.1101/gad.2056211. This work demonstrated functional dimerization of the IDOL E3 ligase driven by a coiled-coil/RING portion of the protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7–E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]


