Abstract
Through analysis of a reported microarray-based high-throughput examination, we found that miR-1275 was significantly down-regulated in nasopharyngeal carcinoma (NPC). While its role and mechanism participated in NPC progression are still little known. Here, we explored the effect of miR-1275 on the progression of NPC. Results demonstrated that miR-1275 was markedly down-regulated in NPC tissues and cell lines. MiR-1275 markedly repressed cell growth as confirmed by CCK8 and colony formation assay, via inhibition of HOXB5 in NPC cell lines. Moreover, miR-1275 suppressed G1/S transition via inhibition of HOXB5. Further, oncogene HOXB5 was evidenced to be a potential target of miR-1275, and its expression was conversely correlated with miR-1275 expression in NPC. Collectively, our study indicated that miR-1275, a tumor suppressor, played a critical effect on NPC progression via inhibition of cell growth, and suppression of G1/S transition by targeting oncogenic HOXB5.
Electronic supplementary material
The online version of this article (doi:10.1007/s12079-016-0351-9) contains supplementary material, which is available to authorized users.
Keywords: microRNA-1275 (miR-1275), HOXB5, Nasopharyngeal carcinoma (NPC), Proliferation
Introduction
MicroRNAs (miRNAs), are a class of single-stranded, small, and non-coding RNAs. They mediate post-transcriptional regulation of target genes via combination with 3′-untranslated regions (UTRs) at some sequence-specific sites, which contribute to suppression of these genes expression (Liu et al. 2014; Belgardt et al. 2015). miRNAs being critical regulatory factors in numerous of cancers have been confirmed by various of publications (Wulczyn et al. 2007; Chen et al. 2014; Dinami et al. 2014; Jin et al. 2014; Li et al. 2015). Recent studies reveal that miRNAs lead to tumor cell growth, metastasis, apoptosis, stem cell maintenance, cell identity, and senescence (Diaz-Martin et al. 2014; Hsu et al. 2014; Huang et al. 2015; Mei et al. 2015; Sun et al. 2015a). Although recent studies on miRNAs have broadened our horizon in human tumors, there are still numerous of unknown details which require to be further elaborated.
Nasopharyngeal carcinoma (NPC) is a kind of tumor derived from the epithelial cells located in nasopharynx. It is frequently occurring on some regions in Africa and East Asia, and common with diet or genetic factors involved in its nosetiology, as well as Epstein-Barr virus (EBV) exposure, (Chang and Adami 2006; Cai et al. 2015a, b). NPC also occupies 1/3 of childhood nasopharyngeal neoplasms in the USA (Young and Miller 1975). It is reported that dysregulation of miRNAs has been widely involved in NPC development. For instance, both EBV-miR-BART7-3p, and EBV-miR-BART1 could contribute to tumor EMT and metastasis through suppressing the PTEN-dependent pathways (Cai et al. 2015a, b). Zhao et al. found that miR-3188 regulated NPC cell growth and chemosensitivity via regulation of mTOR–p-PI3K/AKT-c-JUN by targeting oncogenic FOXO1 (Zhao et al. 2016). Another study indicated miR-93 facilitated cell proliferation and metastasis by targeting 3′-UTR of disabled homolog-2 mRNA in NPC (Xu et al. 2015). Additionally, PDCD4, a tumor suppressor, could modulate miR-184-induced inhibition of survival, and cell growth in NPC (Zhen et al. 2013).
MiR-1275 (MI0006415), a confirmed tumor-repressing miRNA, plays an important role in several diseases. Including obese (Pang et al. 2016), hepatocellular carcinoma (Fawzy et al. 2015), and oral squamous cell carcinoma (Manikandan et al. 2016). Katsushima et al. also found that miR-1275 exerted a tumor-suppressing role in human glioma stem-like cells by regulation ofClaudin11 protein suppression (Katsushima et al. 2012). Recently, Plieskatt et al. have reported that miR-1275 was markedly down-regulated in NPC (Plieskatt et al. 2014), indicating the tumor-suppressing functions of miR-1275 on NPC, but up to now this suggestion has not been rigorously elaborated.
In the present study, we explore the biological efficiency of miR-1275 in NPC, and investigate the potential mechanisms of its action. We discover that HOXB5 is a direct target of miR-1275, and is up-regulated in several tumors, including gastric carcinoma (Hong et al. 2015), breast cancer (Lee et al. 2015), bladder cancer (Luo et al. 2012), and oral squamous cell carcinoma (Tucci et al. 2011). While whether HOXB5 is up-regulated in NPC is not be explored. In this study, we show that miR-1275 is indeed repressed in NPC tissues and cell lines, and find human HOXB5 is indeed a direct target of miR-1275. In conclusion, we reveal that miR-1275 represses NPC cell growth, and suppresses G1/S transition through directly targeting 3′-UTR of HOXB5.
Materials and methods
Ethical statement
For the analyzed tissue specimens, all patients gave informed consent to use excess pathological specimens for research purposes. The protocols employed in this Subjects Committee. The use of human tissues was approved by the institutional review board of the Wuhan University and conformed to the Helsinki Declaration and to the local legislation. Patients offering samples for the study signed informed consent forms.
Tissue collection
Fresh and formalin-fixed, paraffin-embedded, NPC tumor tissue samples were obtained from patients who were diagnosed with primary NPC. Elective surgery was carried out on these patients at People’s Hospital of Wuhan University (Wuhan, China). In total, 114 cases of fresh NPC tissues were freshly frozen in liquid nitrogen and stored at −80 °C until further use. 114 cases of archived, formalin-fixed, paraffin-embedded NPC tissue samples were collected and used in clinicopathological and prognostic investigation of miR-1275. A comprehensive set of clinicopathological data were recorded, including age, gender, size of primary tumor, tumor differentiation, T stage, lymph node metastasis, and distant metastasis. The stage of disease was determined according to the tumor size, lymph node, and metastasis (pTNM) classification system. The use of tissues for this study has been approved by the ethics committee of People’s Hospital of Wuhan University. Before using these clinical materials for research purposes, all the patients signed the informed consent. None of these patients received any pre-operative chemotherapy or radiotherapy.
Cell culture and transfection
The human NPC cell lines, namely, SUNE-1, CNE-1, HNE-1, CNE-2, C666–1 and HONE-1 were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) supplemented with 10 %. fetal bovine serum (FBS). The human immortalized nasopharyngeal epithelial cell line NP69 was cultured in keratinocyte/serum-free medium (Invitrogen) supplemented with bovine pituitary extract. MiR-1275 mimic and mimic negative control, miR-1275 inhibitor and inhibitor negative control were purchased from GenePharma Co.,Ltd. (Shanghai, China). Complete medium without antibiotics was used to culture the cells at least twenty-four hours prior to transfection. The cells were washed with 1× PBS (pH 7.4) and then transiently transfected with 50 nM NC or miR-1275, 100 nM ASO-1275 or si-HOXB5, using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Western blot analysis
Western blot was performed using the protocol described previously (Sun et al. 2015a, b, c, d; Sun et al. 2016a). The following primary antibodies were used: rabbit anti-HOXB5 (Santa Cruz, USA), rabbit anti-GAPDH (Santa Cruz, USA).
RNA isolation and quantitative reverse transcription poly-merase chain reaction (qRT-PCR)
RNA isolation and qRT-PCR was carried out using the protocol described previously (Sun et al. 2015a). GADPH and U6 were used as endogenous controls. In addition, melting curves were used to evaluate non-specific amplification. The relative expression level was calculated using the 2-ΔΔCt method. The primer sequences used in this study are as follows: human HOXB5: sense: 5′- TCAGCCATGATATGACCGGG-3′, antisense: 5′- AGATCTTGATCTGGCGCTCG-3′; human GAPDH: sense: 5′-CTCTGCTCCTCCTGTTCGAC-3′, antisense: 5′-ACCAAATCCGTTGACTCCGA −3′. The formula and its derivations were obtained from the ABI Prism 7300 sequence detection system user guide. Statistical analysis was performed on the fold change.
Colony formation assay
Colony formation assay was carried out using the protocol described previously (Sun et al. 2015a).
Luciferase reporter assays
Luciferase reporter assays was carried out using the protocol described previously (Sun et al. 2015a).
BrdU immunofluorescence assay
BrdU immunofluorescence assay was carried out using the protocol described previously (Sun et al. 2015a).
CCK8 assay
Cell growth was measured using the cell proliferation reagent WST-8 (Roche Biochemicals, Mannheim, Germany). After plating cells in 96-well microtiter plates (Corning Costar, Corning, NY) at 1.0× 10 3 /well, 10 μL of CCK8 was added to each well at the time of harvest, according to the manufacturer’s instructions. One hour after adding CCK8, cellular viability was determined by measuring the absorbance of the converted dye at 450 nm.
Cell-cycle analysis
Transfected cells were harvested forty-eight hours after transfection. The cells were fixed in 70 % ethanol, washed once with PBS, and then labeled with propidium iodide (Sigma-Aldrich) in the presence of RNase A (Sigma-Aldrich) for 30 min in the dark (50 g/mL). Samples were run on a FACSalibur flow cytometer (Becton-Dickinson, FL, NJ, USA), and the percentages of cells within each phase of the cell cycle were analyzed using Cell Quest software.
Statistical analysis
All experiments were repeated for three times independently. Results were shown as the means ± standard error mean (SEM). Two independent sample t-test or One-Way Analysis of Variance (ANOVA) was performed using SPSS 20.0 software to assess significant differences in measured variables among groups. A value of P < 0.05 was considered to indicate a statistically significant difference.
Results
MiR-1275 is down-regulated in NPC tissues and cell lines, and is a favorable factor for prognosis
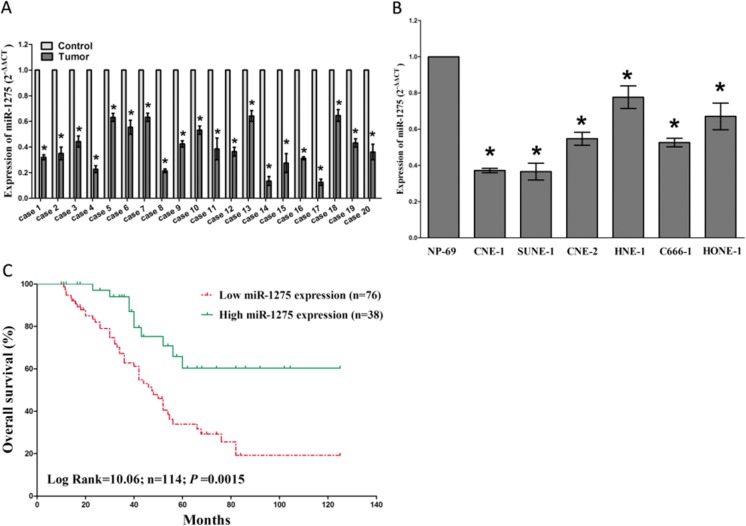
To validate the roles of miR-1275 in NPC, we firstly examined the miR-1275 expression level in human NPC tissues and their pair-matched adjacent NP tissues by qRT-PCR. The results demonstrated miR-1275 expression in the 114 NPC tumors tissues was markedly (P < 0.05) reduced (mean = 58 % of decrease) than their matched NP controls (Fig. 1a). Then, we tested miR-1275 levels in NPC cell lines, and results revealed a lower expression of miR-1275 in CNE-1, SUNE-1, CNE-2, C666–1, HNE-1, and HONE-1 cell lines, in comparison to that of in NP69 (a normal NP cell lines) (Fig. 1b). Moreover, patients with low miR-1275 levels (≤58 % of decrease, n = 78) had a shorter overall survival than that of inpatients with high miR-1275 expression levels (>58 % of decrease, n = 36)(Fig. 1c), as confirmed by Kaplan–Meier survival analysis. Additionally, we also evaluated the association of miR-1275 expression with clinic-pathological parameters. Results revealed that miR-1275 expression levels in NPC were significantly corrected with clinical stages (P = 0.0023). While miR-1275 expression was not correlated with histological type (P = 0. 3962), gender (P = 0.9111), or age (P = 0.3248) in NPC (Table 1 ).
Fig. 1.
MiR-1275 is down-regulated in primary human NPC and NPC cell lines, and benefits for prognosis. a. miR-1275 is significantly decreased in primary human NPC tissues in comparison to adjacent-normal NP tissues. n = 20 for each group. b. The RNA expression level of miR-1275 in six NPC cell lines and normal NP-69 cells. Assays were performed in triplicate. c. Kaplan-Meier survival analysis reveals that down-regulated miR-1275 is associated with poor prognosis in patients with NPC. *P < 0.05, Means ± SEM was shown. Statistical analysis was conducted using student t-test
Table 1.
Correlation between miR-1275 expression and clinipathological parameters of NPC patients (n = 114)
| Parameter | n | MiR-1275 | ||
|---|---|---|---|---|
| Age/years | High | Low | P-value [a] | |
| ≤40 | 37 | 18 | 19 | 0.3248 |
| >40 | 77 | 45 | 32 | |
| Gender | ||||
| Male | 81 | 50 | 31 | 0.9111 |
| Female | 33 | 20 | 13 | |
| Histological type | ||||
| DNC | 16 | 8 | 8 | 0.3962 |
| UDNC | 98 | 60 | 38 | |
| Clinical stages | ||||
| Stage I | 14 | 5 | 9 | 0.0023* |
| Stage II | 33 | 13 | 20 | |
| Stage III | 39 | 27 | 12 | |
| Stage IV | 28 | 22 | 6 | |
aChi-square test
*P < 0.05
HOXB5 is up-regulated in NPC tissues and its expression is negatively related to miR-1275
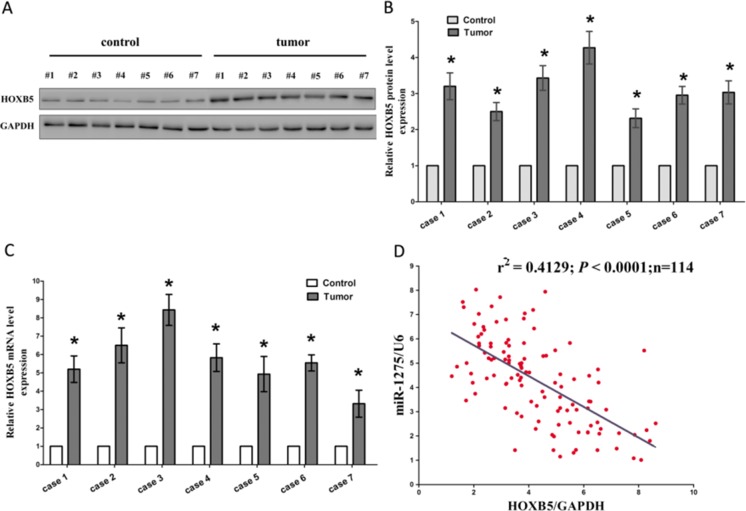
HOXB5 is a well-known oncogene, which is up-regulated in several tumors. Such as, HOXB5 is overexpressed in human gastric carcinoma and induces invasion and migration through direct transcriptional up-regulation of β-catenin (Hong et al. 2015), and HOXB5 is aberrantly overexpressed in breast cancer tissues and cell lines and promotes proliferation and invasion of breast cancer cells (Lee et al. 2015). Moreover, Luo et al. reported that HOXB5 was frequently over-expressed both in bladder cancer tissues and cell lines and inhibition of HOXB5 suppressed the oncogenic function of cancer cells (Luo et al. 2012). Hence, we then tested HOXB5 expression in NPC tissues and their pair-matched adjacent NP tissues, and results of western blot showed that HOXB5 protein levels were aggrandized in NPC tissues in comparison to NP tissues (Fig. 2a-b), which was verified by qRT-PCR (Fig. 2c). On account of the fact that HOXB5 is the critical factor in supervision of cell growth, cell cycle, and cell metastasis, aberration of its protein expression might result in the initial and development of human NPC. Additionally, we also assessed the association between HOXB5 mRNA and miR-1275 levels in 114 NPC tissues, and Pearson correlation analysis demonstrated that the expression of HOXB5 mRNA showed a significantly reverse correlation with miR-1275 levels(r2 = 0.4129, P < 0.0001) (Fig. 2d).
Fig. 2.
Expression of HOXB5 is up-regulated in primary human NPC tissues and negatively expressed related to miR-1275. a-c. Western-blot of HOXB5 protein and qRT-PCR of HOXB5 mRNA in NPC and adjacent-normal NP tissues. n = 20 for each group. d. Scatter plots show the inverse association between miR-1275 level and HOXB5 mRNA expression. *P < 0.05, Means ± SEM was shown. Statistical analysis was conducted using student t-test
MiR-1275 targets human HOXB5
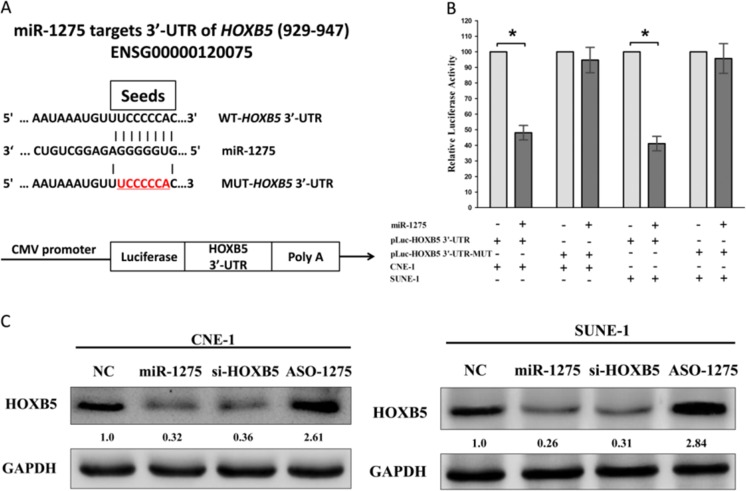
Having known miR-1275 is down-regulated in NPC, and expression of HOXB5 mRNA and miR-1275 exhibits inverse correlation. We next investigated the potential molecular mechanism of miR-1275’s anticancer property in NPC cells lines. Actually, we seek several data bases, including PicTar, microRNA.org and TargetScan, for its underlying onco-targets. And we selected HOXB5 that harbors a conserved miR-1275 cognate sites at its 929–947 of HOXB5 3′-UTR) (Fig. 3a), as an underlying target of miR-1275. To validate whether HOXB5 expression is indeed a direct target of miR-1275, luciferase reporter assays were performed. Results demonstrated that miR-1275 inhibited luciferase activity in CNE-1 cells and SUNE-1 cells with the WT HOXB5 3′-UTR reporter plasmid carried (Fig. 3b), while no marked inhibition was shown with the MUT HOXB5 3′-UTR reporter plasmid carried. Indicating that miR-1275 directly binds to HOXB5 3′-UTR at the predicted binding sites. To examine the efficiency of miR-1275 on the expression of HOXB5, we treated CNE-1 and SUNE-1 cells with NC, miR-1275, si-HOXB5 or ASO-1275 for indicated time. And results of western blot demonstrated that both miR-1275 and si-HOXB5 treatment repressed the protein level of HOXB5 in CNE-1 and SUNE-1 cells, but ASO-1275 treatment increased the HOXB5 protein expression (Fig. 3c).
Fig. 3.
HOXB5 proto-oncogene is a target of miR-1275 at specific 3′-UTR sites. a. Upper, the 3′-UTR of HOXB5 harbors one miR-1275 cognate sites. Lower, pmiR-RB-REPORT™ dual-luciferase reporter vector. b. Relative luciferase activity of reporter plasmids carrying wild-type or mutant HOXB5 3′-UTR in CNE-1 and SUNE-1 cells co-transfected with negative control (NC) or miR-1275 mimic. c. Protein expression of HOXB5 in CNE-1 and SUNE-1 cells after transfected with NC, miR-1275, si-HOXB5 or ASO-1275. Assays were performed in triplicate. *P < 0.05, Means ± SEM was shown. Statistical analysis was conducted using student t-test
MiR-1275 retards cell growth, and suppresses G1/S transition through down-regulation of HOXB5 in NPC
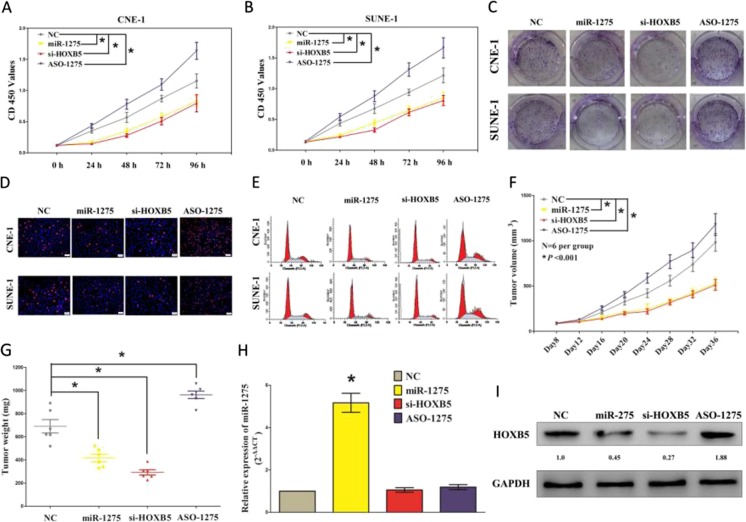
To further validate the tumor suppressive efficiency of miR-1275 in NPC, we treated CNE-1 and SUNE-1 cells with NC, miR-1275, si-HOXB5 or ASO-1275 for indicated time. First, we examined its role on NPC cell (CNE-1 and SUNE-1) growth. Both miR-1275 and si-HOXB5 treatment significantly attenuated CNE-1 and SUNE-1 cells vitality, while ASO-1275 significantly promoted CNE-1 and SUNE-1 cells vitality than NC treated CNE-1 and SUNE-1 cells, as demonstrated by CCK8 assay (Fig. 4a-b). These results were verified using colony formation assay, a method which was frequency used to assess the efficiency of miRNAs on tumor cell clonogenic survival (Fig. 4c, Fig. S1 ). Moreover, BrdU staining demonstrated that both miR-1275 and si-HOXB5 treatment suppressed CNE-1 and SUNE-1 cell DNA synthesis than that of in NC treated CNE-1 and SUNE-1 cells, respectively. However, ASO-1275 treatment increased CNE-1 and SUNE-1 cell DNA synthesis compared with that of in NC treated CNE-1 and SUNE-1 cells, separately (Fig. 4d, Fig. S2 ). Furthermore, the growth inhibitory efficiency of miR-1275 and si-HOXB5 on CNE-1 and SUNE-1 cells were accompanied by a corresponding increase in the ratio of cells in G1 and a decrease in the ratio of cells in the S phase (Fig. 4e, Fig. S3 ).
Fig. 4.
MiR-1275 inhibits NPC cell growth, and suppresses G1/S transition through down-regulation of HOXB5. a-b. CCK8 assays of CNE-1 and SUNE-1 cells after transfection. c. Shown are representative photos of colony formation assay after transfection for fourteen days. d. Shown are representative photos of BrdU immunofluorescence assay after transfection. Bar = 50 μm. e. Cell-cycle analysis was performed following the treatment CNE-1 and SUNE-1 cells with NC, miR-1275, si-HOXB5 and ASO-1275 for forty eight hours. The DNA content was quantified by flow cytometric analysis. f. Tumor volume in nude mice. g. Tumor weight in nude mice. Each group contained six mice (n = 6); the data are presented as the mean ± SEM; *p < 0.001, compared with the NC group. h. The miR-1275 expression in nude mice. i. The expression of HOXB5 protein in nude mice. Assays were performed in triplicate. *P < 0.05. Means ± SEM was shown. Statistical analysis was conducted using ANOVA
To verify the anticancer efficiency of miR-1275 in vivo, we performed tumor formation in nude mice using CNE-1 cells. Results indicated that the tumor volume and weight of mice treated with miR-1275 or si-HOXB5 were significantly reduced relative to that of treated with NC (Fig. 4f-g). While ASO-1275 treatment significantly increased tumor growth compared with that of in NC treatment in vivo (Fig. 4f-g). These results demonstrated that miR-1275 significantly repressed the tumorigenicity of CNE-1 cells in the nude mouse xenograft model. In addition, our results of western-blot demonstrated decreased expression of HOXB5 in tumors derived from miR-1275- or si-HOXB5-treated nude mice relative to that of in NC treatment (Fig. 4i). Thus, miR-1275 reduces the growth of established NPC xenografts.
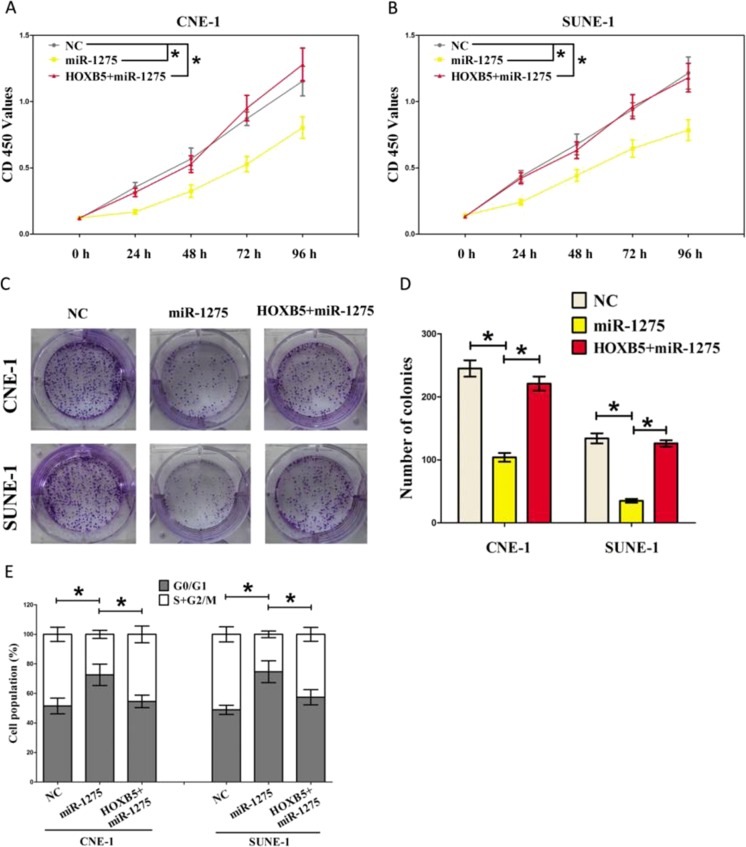
In addition, we also treated CNE-1 and SUNE-1 cells with NC, miR-1275 + HOXB5 for indicated time to verify miR-1275 inhibiting the tumorigenic potential of NPC cells by down-regulating oncogenic HOXB5 gene. CCK8 and colony formation assays demonstrated that miR-1275 suppressed cell growth in CNE-1 and SUNE-1 cells, while overexpression of HOXB5 in miR-1275 treated cells reversed the growth-inhibitory role of miR-1275 in both CNE-1 and SUNE-1 cells, and normalized to that of in NC levels (Fig. 5a-d, Fig. S4 ). Overexpression of HOXB5 also reversed the role of miR-1275 on cell arrest in both CNE-1 and SUNE-1 cells, as demonstrated by cell cycle analysis (Fig. 5e).
Fig. 5.
MiR-1275 exerts its tumor suppressive role in NPC cells through down-regulation of HOXB5. a-b. CCK8 assays of CNE-1 and SUNE-1 cells after transfection. c. Shown are representative photos of colony formation assay after transfection for fourteen days. d. Statistics of colony formation assay. e. Cell-cycle analysis was performed following the treatment CNE-1 and SUNE-1 cells with NC, miR-1275, HOXB5 + miR-1275 for forty eight hours. The DNA content was quantified by flow cytometric analysis. Assays were performed in triplicate. *P < 0.05. Means ± SEM was shown. Statistical analysis was conducted using ANOVA
Collectively, miR-1275 retards cell growth, and suppresses G1/S transition through down-regulation of HOXB5 in NPC.
Discussion
The critical role of miRNAs in tumorigenesis via the modulation of various targets has promoted intensive research into miRNAs as biomarkers and miRNA-based therapeutic strategies for the treating cancers. In the present study, we identified that miR-1275 was frequently down-regulated in human NPC tissues and cell lines. Subsequent analysis indicated that miR-1275 was significantly correlated with advanced clinical stage. We also found that miR-1275 exhibited its tumor suppressive role on NPC cells by down-regulation of oncogenic HOXB5 gene. In NPC tissues, expression of miR-1275 was also found to be correlated inversely with HOXB5 expression. This suggested that the miR-1275 in NPC was a strong tumor suppressor by regulating NPC cell growth and cell cycle. Suggesting that miR-1275 has a critical role on suppression of the initiation and progression of NPC.
Until now, the detail molecular mechanisms on progression of NPC have not been well elaborated. Though aberrant of miRNAs has been demonstrated in numerous of human cancers (Sun et al. 2015b, 2016a, b, c, d, e), the dysregulation and underlying efficiency of miRNAs in NPC were still little known. Decreased expression of miR-1275 has been revealed by several studies on obese (Pang et al. 2016), hepatocellular carcinoma (Fawzy et al. 2015), and oral squamous cell carcinoma (Manikandan et al. 2016). Our results also showed that miR-1275 was down-regulated in NPC tissues and cell lines, which suggested the deregulation of miR-1275 was an early event of NPC tumorigenesis. It has been reported that miR-1275 could control a number of target genes simultaneously. Such as, miR-1275 inhibited adipogenesis via ELK1 and its expression decreases in obese subjects (Pang et al. 2016), and miR-1275 could hamper tumor growth by targeting three IGF2-mRNA-binding proteins in hepatocellular carcinoma (Fawzy et al. 2015). Moreover, miR-1275 regulated stem-like cells of human glioma by suppression of Claudin11 protein expression (Katsushima et al. 2012). While among all of the predicted target genes of miR-1275, we selected that HOXB5 functions as a crucial effector of miR-1275. And results of luciferase reporter assays demonstrated miR-1275 could markedly suppress the luciferase activity of Luc-HOXB5–3′-UTR by targeting the 3′-UTR of HOXB5 mRNA. Hence, we selected HOXB5 for further analysis.
Next, we explored the potential anticancer role of miR-1275 on human NPC cell lines. We first investigated its efficiency on NPC cell growth and cell cycle, and found that miR-1275 markedly suppressed cell growth in the NPC cell lines CNE-1 and SUNE-1. We also found that the growth-inhibitory efficiency of miR-1275 in NPC might be attributed to that miR-1275 targeting 3′-UTR of HOXB5 mRNA, and suppressing the protein expression of HOXB5 in NPC cells. We identified HOXB5 as a direct target of miR-1275 in NPC cells, and discovered that the tumor-suppressive activity of miR-1275 was induced by the modulation of HOXB5 expression. Moreover, miR-1275 could also retard G1/S transition in CNE-1 and SUNE-1 cells by directly targeting 3′-UTR of HOXB5 mRNA.
Collectively, our present study showed that miR-1275 was down-regulated in NPC cells and tissues, and its inhibitory effects on tumor progression were mediated by the down-regulation of its target HOXB5. The present results elucidate a potential mechanism underlying the tumor-suppressor role of miR-1275, and indicate that miR-1275 could be a useful marker and potential therapeutic target in NPC.
Electronic supplementary material
(DOCX 2076 kb)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Funding
The Fundamental Research Funds for the Central Universities (No. 2,015,305,020,202) to Cheng-Cao Sun, and The Fund of the Health and Family Planning of Hubei province, China (NO: WJ2015MB036).
References
- Belgardt BF, Ahmed K, Spranger M, Latreille M, Denzler R, Kondratiuk N, von Meyenn F, Villena FN, Herrmanns K, Bosco D, Kerr-Conte J, Pattou F, Rulicke T, Stoffel M. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. NAT MED. 2015;21(6):619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- Cai LM, Lyu XM, Luo WR, Cui XF, Ye YF, Yuan CC, Peng QX, Wu DH, Liu TF, Wang E, Marincola FM, Yao KT, Fang WY, Cai HB, Li X. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. ONCOGENE. 2015;34(17):2156–2166. doi: 10.1038/onc.2014.341. [DOI] [PubMed] [Google Scholar]
- Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J, Wang S, Liu T, Cai H, Yao K, Li JL, Li X. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. NAT COMMUN. 2015;6:7353. doi: 10.1038/ncomms8353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomark Prev. 2006;15(10):1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- Chen H, Li L, Wang S, Lei Y, Ge Q, Lv N, Zhou X, Chen C. Reduced miR-126 expression facilitates angiogenesis of gastric cancer through its regulation on VEGF-A. ONCOTARGET. 2014;5(23):11873–11885. doi: 10.18632/oncotarget.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Martin J, Diaz-Lopez A, Moreno-Bueno G, castilla MA, Rosa-Rosa JM, Cano A, Palacios J. A core microRNA signature associated with inducers of the epithelial-to-mesenchymal transition. J PATHOL. 2014;232(3):319–329. doi: 10.1002/path.4289. [DOI] [PubMed] [Google Scholar]
- Dinami R, Ercolani C, Petti E, Piazza S, Ciani Y, Sestito R, Sacconi A, Biagioni F, le Sage C, Agami R, Benetti R, Mottolese M, Schneider C, Blandino G, Schoeftner S. miR-155 drives telomere fragility in human breast cancer by targeting TRF1. CANCER RES. 2014;74(15):4145–4156. doi: 10.1158/0008-5472.CAN-13-2038. [DOI] [PubMed] [Google Scholar]
- Fawzy IO, Hamza MT, Hosny KA, Esmat G, El TH, Abdelaziz AI. miR-1275: A single microRNA that targets the three IGF2-mRNA-binding proteins hindering tumor growth in hepatocellular carcinoma. FEBS LETT. 2015;589(17):2257–2265. doi: 10.1016/j.febslet.2015.06.038. [DOI] [PubMed] [Google Scholar]
- Hong CS, Jeong O, Piao Z, Guo C, Jung MR, Choi C, Park YK. HOXB5 induces invasion and migration through direct transcriptional up-regulation of beta-catenin in human gastric carcinoma. BIOCHEM J. 2015;472(3):393–403. doi: 10.1042/BJ20150213. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Hsieh TH, Tsai CF, Tsai HP, Chen HS, Chang Y, Chuang HY, Lee JN, Hsu YL, Tsai EM. miRNA-199a-5p regulates VEGFA in endometrial mesenchymal stem cells and contributes to the pathogenesis of endometriosis. J PATHOL. 2014;232(3):330–343. doi: 10.1002/path.4295. [DOI] [PubMed] [Google Scholar]
- Huang CF, Sun CC, Zhao F, Zhang YD, Li DJ. miR-33a levels in hepatic and serum after chronic HBV-induced fibrosis. J GASTROENTEROL. 2015;50(4):480–490. doi: 10.1007/s00535-014-0986-3. [DOI] [PubMed] [Google Scholar]
- Jin M, Zhang T, Liu C, Badeaux MA, Liu B, Liu R, Jeter C, Chen X, Vlassov AV, Tang DG. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. CANCER RES. 2014;74(15):4183–4195. doi: 10.1158/0008-5472.CAN-14-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsushima K, Shinjo K, Natsume A, Ohka F, Fujii M, Osada H, Sekido Y, Kondo Y. Contribution of microRNA-1275 to Claudin11 protein suppression via a polycomb-mediated silencing mechanism in human glioma stem-like cells. J BIOL CHEM. 2012;287(33):27396–27406. doi: 10.1074/jbc.M112.359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Hur H, Yun HJ, Kim Y, Yang S, Kim SI, Kim MH. HOXB5 Promotes the Proliferation and Invasion of Breast Cancer Cells. INT J BIOL SCI. 2015;11(6):701–711. doi: 10.7150/ijbs.11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kuscu C, Banach A, Zhang Q, Pulkoski-Gross A, Kim D, Liu J, Roth E, Li E, Shroyer KR, Denoya PI, Zhu X, Chen L, Cao J. miR-181a-5p Inhibits Cancer Cell Migration and Angiogenesis via Downregulation of Matrix Metalloproteinase-14. CANCER RES. 2015;75(13):2674–2685. doi: 10.1158/0008-5472.CAN-14-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zheng Q, Vrettos N, Maragkakis M, Alexiou P, Gregory BD, Mourelatos Z. A MicroRNA precursor surveillance system in quality control of MicroRNA synthesis. MOL CELL. 2014;55(6):868–879. doi: 10.1016/j.molcel.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Cai Q, Wang W, Huang H, Zeng H, He W, Deng W, Yu H, Chan E, Ng CF, Huang J, Lin T. A microRNA-7 binding site polymorphism in HOXB5 leads to differential gene expression in bladder cancer. PLOS ONE. 2012;7(6) doi: 10.1371/journal.pone.0040127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikandan M, deva MRA, Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R, Munirajan AK. Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. MOL CANCER. 2016;15:28. doi: 10.1186/s12943-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Q, Xue G, Li X, Wu Z, Li X, Yan H, Guo M, Sun S, Han W. Methylation-induced loss of miR-484 in microsatellite-unstable colorectal cancer promotes both viability and IL-8 production via CD137L. J PATHOL. 2015;236(2):165–174. doi: 10.1002/path.4525. [DOI] [PubMed] [Google Scholar]
- Pang L, You L, Ji CB, Shi C, Chen L, Yang L, Huang F, Zhou Y, Zhang J, Chen X, Guo X. MiR-1275 inhibits adipogenesis via ELK1 and its expression decreases in obese subjects. 2016. [DOI] [PubMed] [Google Scholar]
- Plieskatt JL, Rinaldi G, Feng Y, Levine PH, Easley S, Martinez E, Hashmi S, Sadeghi N, Brindley PJ, Bethony JM, Mulvenna JP. Methods and matrices: approaches to identifying miRNAs for nasopharyngeal carcinoma. J TRANSL MED. 2014;12:3. doi: 10.1186/1479-5876-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Yang C, Xue R, Li S, Zhang T, Pan L, Ma X, Wang L, Li D. Sulforaphane alleviates muscular dystrophy in mdx mice by activation of Nrf2. J Appl Physiol (1985) 2015;118(2):224–237. doi: 10.1152/japplphysiol.00744.2014. [DOI] [PubMed] [Google Scholar]
- Sun CC, Li SJ, Yang CL, Xue RL, Xi YY, Wang L, Zhao QL, Li DJ. Sulforaphane Attenuates Muscle Inflammation in Dystrophin-deficient mdx Mice via NF-E2-related Factor 2 (Nrf2)-mediated Inhibition of NF-kappaB Signaling Pathway. J BIOL CHEM. 2015;290(29):17784–17795. doi: 10.1074/jbc.M115.655019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sun C, Liu Z, Li S, Yang C, Xue R, Xi Y, Wang L, Wang S, He Q, Huang J, Xie S, Jiang W, Li D. Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. ONCOTARGET. 2015;6(28):25533–25574. doi: 10.18632/oncotarget.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Sang M, Li S, Sun X, Yang C, Xi Y, Wang L, Zhang F, Bi Y, Fu Y, Li D. Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-Met. ONCOTARGET. 2015;6(37):39756–39792. doi: 10.18632/oncotarget.5476. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sun C, Huang C, Li S, Yang C, Xi Y, Wang L, Zhang F, Fu Y, Li D. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. ONCOTARGET. 2016;7(7):8341–8359. doi: 10.18632/oncotarget.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Li S, Yang C, Xi Y, Wang L, Zhang F, Li D. MicroRNA-187-3p mitigates non-small cell lung cancer (NSCLC) development through down-regulation of BCL6. Biochem Biophys Res Commun. 2016;471(1):82–88. doi: 10.1016/j.bbrc.2016.01.175. [DOI] [PubMed] [Google Scholar]
- Sun CC, Li SJ, Zhang F, Pan JY, Wang L, Yang CL, Xi YY, Li DJ (2016c) Hsa-miR-329 exerts tumor suppressor function through down-regulation of MET in non-small cell lung cancer. ONCOTARGET 7(16):21510–21526 [DOI] [PMC free article] [PubMed]
- Sun C, Li S, Li D. Sulforaphane mitigates muscle fibrosis in mdx mice via Nrf2-mediated inhibition of TGF-beta/Smad signaling. J Appl Physiol (1985) 2016;120(4):377–390. doi: 10.1152/japplphysiol.00721.2015. [DOI] [PubMed] [Google Scholar]
- Sun C, Li S, Li D (2016e) Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. ONCOTARGET 7(24):35960–35978 [DOI] [PMC free article] [PubMed]
- Tucci R, Campos MS, Matizonkas-Antonio LF, Durazzo M, Pinto JDS, Nunes FD. HOXB5 expression in oral squamous cell carcinoma. J APPL ORAL SCI. 2011;19(2):125–129. doi: 10.1590/S1678-77572011000200008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21(2):415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Xu YF, Mao YP, Li YQ, Ren XY, He QM, Tang XR, Sun Y, Liu N, Ma J. MicroRNA-93 promotes cell growth and invasion in nasopharyngeal carcinoma by targeting disabled homolog-2. CANCER LETT. 2015;363(2):146–155. doi: 10.1016/j.canlet.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Young JJ, Miller RW. Incidence of malignant tumors in U. S. children. J Pediatr. 1975;86(2):254–258. doi: 10.1016/S0022-3476(75)80484-7. [DOI] [PubMed] [Google Scholar]
- Zhao M, Luo R, Liu Y, Gao L, Fu Z, Fu Q, Luo X, Chen Y, Deng X, Liang Z, Li X, Cheng C, Liu Z, Fang W. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. NAT COMMUN. 2016;7:11309. doi: 10.1038/ncomms11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Liu Z, Yang H, Yu X, Wu Q, Hua S, Long X, Jiang Q, Song Y, Cheng C, Wang H, Zhao M, Fu Q, Lyu X, Chen Y, Fan Y, Liu Y, Li X, Fang W. Tumor suppressor PDCD4 modulates miR-184-mediated direct suppression of C-MYC and BCL2 blocking cell growth and survival in nasopharyngeal carcinoma. CELL DEATH DIS. 2013;4 doi: 10.1038/cddis.2013.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 2076 kb)