Abstract
Oxytocin, a nine amino acid long neuropeptide hormone, is synthesized in the hypothalamus and stored and released from the neural lobe of the pituitary gland. Although commonly known for its central role in the regulation of parturition and lactation, oxytocin signaling also plays a key role in modulating social behavior, evoking contentment, initiating maternal behavior, inducing trust, generosity and bonding in humans and animals. Oxytocin signaling can prove to be of great importance in therapeutics and drug targeting because of its diverse range of actions. However, a well annotated map of oxytocin signaling pathway is currently lacking in the publicly available pathway resources. Therefore, we systematically curated the available signaling information of oxytocin from published literature and collated the data to develop a more complete map. We cataloged 66 molecules belonging to oxytocin signaling pathway, which included 9 protein-protein interactions, 39 post-translational modifications, 14 protein translocation events and 22 activation/inhibition events. Further, Oxytocin signaling network data is made freely available to academic fraternity by integrating this into NetPath (http://www.netpath.org/), a freely available human signaling pathway resource developed previously by our group.
Electronic supplementary material
The online version of this article (doi:10.1007/s12079-016-0353-7) contains supplementary material, which is available to authorized users.
Keywords: G-protein coupled receptor, Gene expression, Herring bodies, Nociception, Osteoclastogenesis, Prostaglandins
Introduction
The nine amino acid long cyclic peptide hormone oxytocin (OXT) is synthesized in the magnocellular neurosecretory cells of the supraoptic and paraventricular nuclei of the hypothalamus (Summar et al. 1990). It is expressed as an inactive precursor protein by the OXT gene located on chromosome number 20. The post-translational progressive hydrolysis of this inactive prohormone through the action of a series of enzymes results in the active form of OXT (Guillou et al. 1994). It is stored in the Herring bodies at the axon terminals of the posterior pituitary lobe before being released into circulation by exocytosis from the neurohypophysis nerve terminals (Jirikowski et al. 1990). OXT is also reportedly synthesized in several peripheral tissues such as uterus, placenta, amnion, corpus luteum, testis, thymus, kidney and pancreas (Gimpl and Fahrenholz 2001). OXT binds to oxytocin receptors (OXTR), which belongs to the G-protein coupled receptor (GPCR) superfamily (Arrowsmith and Wray 2014). The human OXTR gene is located on chromosome locus 3p25. The expression levels of oxytocin receptors in the myoepithelial cells surrounding the alveoli of mammary glands remain constantly elevated throughout the lactation period (Soloff et al. 1979).
OXT-OXTR signaling has been associated with various biological functions. However, its role in uterine contraction and labor induction was the most widely studied. OXT expression in the uterus is elevated during gestation and declines immediately after parturition. The cell specific difference in receptor expression levels aids oxytocin to switch targets between uteri during parturition, to mammary glands during the milk ejection reflex. OXT contributes to myometrial contractility and its receptor has been used as a target for tocolytic agents (Vrachnis et al. 2011). In mammals, OXT is released in a pulsatile manner by the neurohypophysis and is thought to have a key role in the peripartum period by stimulating smooth muscle contractility in the uterus and mammary gland (Smith et al. 2006). In uterus, OXT signaling causes increase in prostaglandins (PGs) after activation of OXTR, which are the mediators of the manifold actions of OXT. Several OXT agonists and antagonists are being used for therapeutic purposes, with differing effects, depending on the cellular context. The highly selective and orally active OXT antagonists are used for the prevention of preterm labor (Serradeil-Le Gal et al. 2004). Also, OXT is being used as an agonist to generate or enhance maternal labor, to prevent or treat postpartum uterine bleeding (Weeks 2015).
OXT-OXTR signaling also plays significant roles in heart and brain. It regulates hypothalamo-pituitary-adrenal axis, modulating behavioral response towards stress and social behavior (Neumann 2002). OXT plays an important role in potentiating a response to a range of pro-social behaviors and in evoking contentment, inducing trust, generosity, and bonding in humans and other animals (Grillon et al. 2013). Prostaglandins are mediators of OXT actions such as excitation in supraoptic nuclei of neurons released in response to OXTR agonists (Wang and Hatton 2006). Cardiac activity of OXT involves maintenance of blood pressure, increased angiogenesis and anti-inflammatory activity (Gutkowska and Jankowski 2012). Further, OXT has a significant role in bone development and is implicated in skeletal remodeling and osteoblast maturation (Majumder et al. 2013). In addition to this, it is involved in the pathophysiology of various disorders such as diabetes, osteoporosis and neuropsychiatric disorders (Elabd and Sabry 2015; Rozek et al. 2014). Considering the biomedical importance OXT signaling in the biological system with respect to various physiological and pathological actions, it is imperative to develop a well annotated and expanded signaling map for oxytocin. Towards this goal, we initiated this bioinformatics study to systematically bring together all the molecular reactions orchestrated by stimulation of OXTR by OXT.
Materials and methods
Literature searches were carried out using PubMed to compile the reactions induced by oxytocin-oxytocin receptor signaling using key search terms such as ‘oxytocin’, ‘oxytocin receptor’, ‘OXT signaling’ and ‘OXTR’. Experimental studies showing oxytocin receptor stimulation by oxytocin or oxytocin analogues (agonists and antagonists) were further selected for curation. All the signaling reactions were archived under the categories such as protein-protein interactions (PPIs), post translational modifications (PTMs), gene regulation, protein activation/inhibition and translocation. The reactions were manually curated and catalogued using PathBuilder, a web-based pathway curation tool developed in-house (Kandasamy et al. 2009). NetPath annotation pipeline was followed to develop OXT signaling, which has been described previously by other groups at our institute, to develop several signaling pathways including Leptin (Nanjappa et al. 2011);Thyroid stimulating hormone (Goel et al. 2011), corticotropin-releasing hormone (Subbannayya et al. 2013), brain-derived neurotrophic factor (Sandhya et al. 2013), Interleukin-17 (Sharma et al. 2015), and Interleukin-10 (Verma et al. 2015) signaling pathways.
Each molecular reaction has been hyperlinked to the research articles in the PubMed from which it was curated. Each reaction was annotated with information about the experimental conditions and cell types used and brief comments about the study. Sites and residues for post-translational modification have been mapped to a RefSeq sequence from the sequence information provided in the respective experiments. Pictorial representation of OXT pathway was generated using PathVisio (Version 3.2.2), freely available software for drawing and visualization (Wang et al. 2012). The reactions were then exported to a web-based pathway resource called NetPath (Kandasamy et al. 2010). Pictorial representation of OXT pathway map depicts all the four broad categories of curation such as molecular association, catalysis, transport, and gene expression at transcription and/or translational level. To validate the authenticity of curated molecular events pertaining to oxytocin pathway, these reactions were also reviewed by a pathway authority, who is a subject expert in the field.
Results and discussion
An extensive PubMed search of research articles pertaining to OXT-OXTR signaling fetched in a total of 1803 articles. These articles were screened for information pertaining to the molecular events induced by oxytocin, resulting in 83 articles from which the pathway map was curated. We documented a total of 66 unique proteins involved in either of 9 PPIs, 39 PTMs, 14 translocation and 22 activation-inhibition reactions. We also catalogued regulation of genes induced by oxytocin in different mammalian systems and identified 62 and 57 genes regulated at transcriptional and translational levels, respectively (Supplementary Table 1).
The OXT pathway data has been made freely available to scientific community through the NetPath resource (http://www.netpath.org/pathways?path_id=NetPath_169). It includes description of the OXT signaling pathway and statistics of total number of molecules and molecular reactions present in signaling network. The molecules involved in the pathway have been linked to molecule page of NetPath and HPRD (Prasad et al. 2009), which provides concise description about the molecule. Data is available to scientific community in standard data exchange format such as Biological Pathway Exchange (BioPAX level 3) (Demir et al. 2010), Proteomics Standards Initiative for Molecular Interaction (PSI-MI version2.5) (Orchard and Kerrien 2009) and Systems Biology Markup Language (SBML version 2.1) (Hucka et al. 2003). The data has been made accessible in tab delimited and Microsoft Excel formats. Figure 1 depicts the OXT-OXTR signaling map.
Fig. 1.
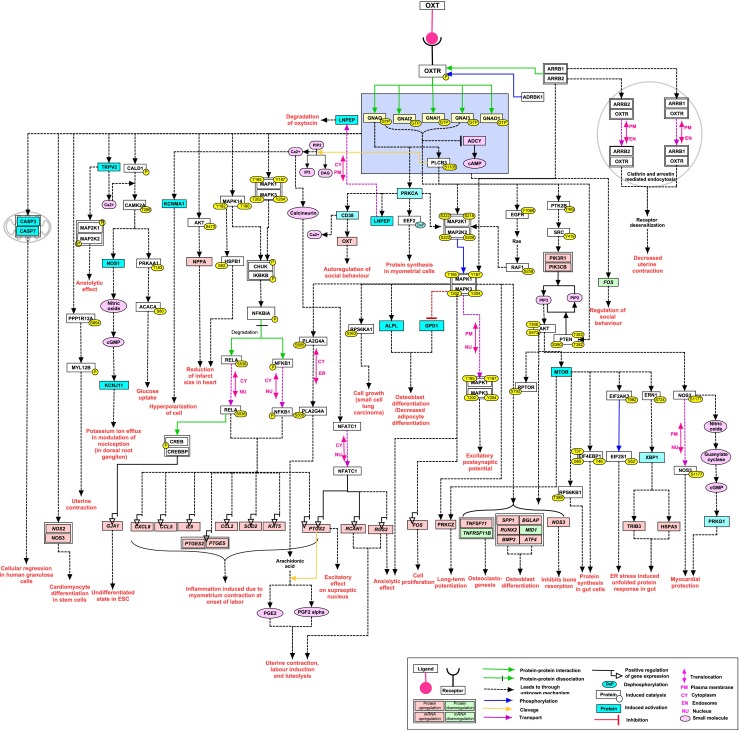
A schematic representation of reactions induced by oxytocin. The pathway map depicts reactions induced by stimulatory action of OXT through OXTR. Reactions represented in map are protein-protein interaction, post-translational modification, activation/inhibition and gene/protein expression. Sites and residues for post translational modifications are also shown. Legend is provided for identification of different pathway reactions
Molecular events induced by OXT-OXTR signaling are involved in various processes such as uterine contraction, labor induction, cell proliferation, cardiomyogenesis, bone formation and neuromodulation. OXTR stimulation is primarily mediated through Gαi/Gαq/Gαo protein activation. OXT induces pro-inflammatory cytokine overexpression through mitogen-activated protein kinase (MAPK)/Nuclear factor kappa B (NFκB) pathway, which in turn is associated with onset of labor (Kim et al. 2015). Prostaglandins (PGE2 and PGF2 alpha) production through arachidonic acid cleavage is one of the effective mediators in oxytocin induced processes especially to those related to onset of labor (Terzidou et al. 2011; Jeng et al. 2000). OXT also triggers osteoclastogenesis and osteoblast differentiation through MAPK1/3 activation thus it may play a significant role as a therapeutic agent in osteoporosis (Tamma et al. 2009). It also plays significant role in functions associated with the CNS such as regulation of nociception, which is mediated by activation of potassium ion channel (KCJN11) (Gong et al. 2015) and anxiolytic effect mediated through expression of regulator of G-protein signaling 2 (RGS2) (Okimoto et al. 2012). In the heart, OXT elicits cardioprotective function by reducing infarct size and post-ischemic recovery, which is mediated through PI3K-AKT/NOS/natriuretic peptide A (NPPA) expression and p38 MAPK/heat shock 27 kDa protein 1 (HSPB1) phosphorylation (Ondrejcakova et al. 2012).
Conclusions
Availability of an expanded and well curated map of OXT signaling reactions in a centralized resource will accelerate the understanding of the role of various molecules in the context of normal physiological or pathological conditions induced by OXT. The signaling data has been made available in multiple community exchange formats to ensure easy integration of data with multiple public repositories and pathway analysis software such as gene set enrichment and Gene Ontology analyses.
Electronic supplementary material
(XLSX 21 kb)
Acknowledgments
We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics (IOB), Bangalore. We thank the “Infosys Foundation” for research support to IOB. OC, PM and LG are recipients of INSPIRE Fellowship from the Department of Science and Technology (DST), Government of India. KP is a recipient of Junior Research Fellowship from Council of Scientific and Industrial Research (CSIR), Government of India. AS is a recipient of Senior Research Fellowship from the University Grants Commission (UGC), Government of India. JA is a recipient of Senior Research Fellowship from the Council of Scientific & Industrial Research (CSIR), Government of India.
Abbreviations
- OXT
Oxytocin
- OXTR
Oxytocin receptor
- CNS
Central nervous system
- GPCR
G-Protein coupled receptor
- PTMs
Post-translational modifications
- PPIs
Protein-protein interactions
- BioPAX
Biological pathway exchange
- SBML
Systems biology markup language
- PSI-MI
Proteomics standards initiative for molecular interaction
Compliance with ethical standards
Conflict of interest
The author(s) declare that they have no competing interests.
Footnotes
Oishi Chatterjee and Krutika Patil contributed equally to this work.
Contributor Information
Oishi Chatterjee, Email: oishi@ibioinformatics.org.
Krutika Patil, Email: krutika.2611@gmail.com.
Apeksha Sahu, Email: apeksha@ibioinformatics.org.
Lathika Gopalakrishnan, Email: lathika@ibioinformatics.org.
Praseeda Mol, Email: praseeda@ibioinformatics.org.
Jayshree Advani, Email: jayshree@ibioinformatics.org.
Srabani Mukherjee, Email: srabanimuk@yahoo.com.
Rita Christopher, Email: rita.nimhans@yahoo.com.
T. S. Keshava Prasad, Email: keshav@ibioinformatics.org.
References
- Arrowsmith S, Wray S. Oxytocin: its mechanism of action and receptor signalling in the myometrium. J Neuroendocrinol. 2014;26:356–369. doi: 10.1111/jne.12154. [DOI] [PubMed] [Google Scholar]
- Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D'Eustachio P, Schaefer C, Luciano J, Schacherer F, Martinez-Flores I, Hu Z, Jimenez-Jacinto V, Joshi-Tope G, Kandasamy K, Lopez-Fuentes AC, Mi H, Pichler E, Rodchenkov I, Splendiani A, Tkachev S, Zucker J, Gopinath G, Rajasimha H, Ramakrishnan R, Shah I, Syed M, Anwar N, Babur O, Blinov M, Brauner E, Corwin D, Donaldson S, Gibbons F, Goldberg R, Hornbeck P, Luna A, Murray-Rust P, Neumann E, Ruebenacker O, Samwald M, van Iersel M, Wimalaratne S, Allen K, Braun B, Whirl-Carrillo M, Cheung KH, Dahlquist K, Finney A, Gillespie M, Glass E, Gong L, Haw R, Honig M, Hubaut O, Kane D, Krupa S, Kutmon M, Leonard J, Marks D, Merberg D, Petri V, Pico A, Ravenscroft D, Ren L, Shah N, Sunshine M, Tang R, Whaley R, Letovksy S, Buetow KH, Rzhetsky A, Schachter V, Sobral BS, Dogrusoz U, McWeeney S, Aladjem M, Birney E, Collado-Vides J, Goto S, Hucka M, Le Novere N, Maltsev N, Pandey A, Thomas P, Wingender E, Karp PD, Sander C, Bader GD. The BioPAX community standard for pathway data sharing. Nat Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elabd S, Sabry I (2015) Two birds with one stone: possible dual-role of oxytocin in the treatment of diabetes and osteoporosis. Front Endocrinol (Lausanne) 6 [DOI] [PMC free article] [PubMed]
- Gimpl G, Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol Rev. 2001;81:629–683. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- Goel R, Raju R, Maharudraiah J, Sameer Kumar GS, Ghosh K, Kumar A, Lakshmi TP, Sharma J, Sharma R, Balakrishnan L, Pan A, Kandasamy K, Christopher R, Krishna V, Mohan SS, Harsha HC, Mathur PP, Pandey A, Keshava Prasad TS. A signaling network of thyroid-stimulating hormone. J Proteomics Bioinform. 2011;4:238–241. doi: 10.4172/jpb.1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Gao F, Li J, Li J, Yu X, Ma X, Zheng W, Cui S, Liu K, Zhang M, Kunze W, Liu CY. Oxytocin-induced membrane hyperpolarization in pain-sensitive dorsal root ganglia neurons mediated by Ca(2+)/nNOS/NO/KATP pathway. Neuroscience. 2015;289:417–428. doi: 10.1016/j.neuroscience.2014.12.058. [DOI] [PubMed] [Google Scholar]
- Grillon C, Krimsky M, Charney DR, Vytal K, Ernst M, Cornwell B. Oxytocin increases anxiety to unpredictable threat. Mol Psychiatry. 2013;18:958–960. doi: 10.1038/mp.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou MD, Camier M, Clamagirand C. Evidence for the presence of pro-oxytocin/neurophysin-converting enzyme in the human ovary. J Endocrinol. 1994;142:345–352. doi: 10.1677/joe.0.1420345. [DOI] [PubMed] [Google Scholar]
- Gutkowska J, Jankowski M. Oxytocin revisited: its role in cardiovascular regulation. J Neuroendocrinol. 2012;24:599–608. doi: 10.1111/j.1365-2826.2011.02235.x. [DOI] [PubMed] [Google Scholar]
- Jeng YJ, Liebenthal D, Strakova Z, Ives KL, Hellmich MR, Soloff MS. Complementary mechanisms of enhanced oxytocin-stimulated prostaglandin E2 synthesis in rabbit amnion at the end of gestation. Endocrinology. 2000;141:4136–4145. doi: 10.1210/endo.141.11.7761. [DOI] [PubMed] [Google Scholar]
- Jirikowski GF, Sanna PP, Bloom FE. mRNA coding for oxytocin is present in axons of the hypothalamo-neurohypophysial tract. Proc Natl Acad Sci U S A. 1990;87:7400–7404. doi: 10.1073/pnas.87.19.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy K, Keerthikumar S, Raju R, Keshava Prasad TS, Ramachandra YL, Mohan S, Pandey A. Path builder--open source software for annotating and developing pathway resources. Bioinformatics. 2009;25:2860–2862. doi: 10.1093/bioinformatics/btp453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD, Mathivanan S, Pecquet C, Gollapudi SK, Tattikota SG, Mohan S, Padhukasahasram H, Subbannayya Y, Goel R, Jacob HK, Zhong J, Sekhar R, Nanjappa V, Balakrishnan L, Subbaiah R, Ramachandra YL, Rahiman BA, Prasad TS, Lin JX, Houtman JC, Desiderio S, Renauld JC, Constantinescu SN, Ohara O, Hirano T, Kubo M, Singh S, Khatri P, Draghici S, Bader GD, Sander C, Leonard WJ, Pandey A. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, MacIntyre DA, Firmino Da Silva M, Blanks AM, Lee YS, Thornton S, Bennett PR, Terzidou V. Oxytocin activates NF-kappaB-mediated inflammatory pathways in human gestational tissues. Mol Cell Endocrinol. 2015;403:64–77. doi: 10.1016/j.mce.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Majumder J, Lodh R, Agarwala BK. Butterfly species richness and diversity in the Trishna wildlife sanctuary in South Asia. J Insect Sci. 2013;13:79. doi: 10.1673/031.013.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjappa V, Raju R, Muthusamy B, Sharma J, Thomas JK, Nidhina PAH, Gowda HC, Pandey A, Anilkumar G, Prasad TSK. A comprehensive curated reaction map of leptin signaling pathway. J Proteomics Bioinform. 2011;4:184–189. doi: 10.4172/jpb.1000188. [DOI] [Google Scholar]
- Neumann ID. Involvement of the brain oxytocin system in stress coping: interactions with the hypothalamo-pituitary-adrenal axis. Prog Brain Res. 2002;139:147–162. doi: 10.1016/S0079-6123(02)39014-9. [DOI] [PubMed] [Google Scholar]
- Okimoto N, Bosch OJ, Slattery DA, Pflaum K, Matsushita H, Wei FY, Ohmori M, Nishiki T, Ohmori I, Hiramatsu Y, Matsui H, Neumann ID, Tomizawa K. RGS2 mediates the anxiolytic effect of oxytocin. Brain Res. 2012;1453:26–33. doi: 10.1016/j.brainres.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Ondrejcakova M, Barancik M, Bartekova M, Ravingerova T, Jezova D. Prolonged oxytocin treatment in rats affects intracellular signaling and induces myocardial protection against infarction. Gen Physiol Biophys. 2012;31:261–270. doi: 10.4149/gpb_2012_030. [DOI] [PubMed] [Google Scholar]
- Prasad TS, Kandasamy K, Pandey A. Human protein reference database and human Proteinpedia as discovery tools for systems biology. Methods Mol Biol. 2009;577:67–79. doi: 10.1007/978-1-60761-232-2_6. [DOI] [PubMed] [Google Scholar]
- Rozek LS, Dolinoy DC, Sartor MA, Omenn GS. Epigenetics: relevance and implications for public health. Annu Rev Public Health. 2014;35:105–122. doi: 10.1146/annurev-publhealth-032013-182513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhya VK, Raju R, Verma R, Advani J, Sharma R, Radhakrishnan A, Nanjappa V, Narayana J, Somani BL, Mukherjee KK, Pandey A, Christopher R, Prasad TS. A network map of BDNF/TRKB and BDNF/p75NTR signaling system. J Cell Commun Signal. 2013;7:301–307. doi: 10.1007/s12079-013-0200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serradeil-Le Gal C, Valette G, Foulon L, Germain G, Advenier C, Naline E, Bardou M, Martinolle JP, Pouzet B, Raufaste D, Garcia C, Double-Cazanave E, Pauly M, Pascal M, Barbier A, Scatton B, Maffrand JP, Le Fur G. SSR126768A (4-chloro-3-[(3R)-(+)-5-chloro-1-(2,4-dimethoxybenzyl)-3-methyl-2-oxo-2,3-dihydro-1H-indol-3-yl]-N-ethyl-N-(3-yridylmethyl)-benzamide, hydrochloride): a new selective and orally active oxytocin receptor antagonist for the prevention of preterm labor. J Pharmacol Exp Ther. 2004;309:414–424. doi: 10.1124/jpet.103.061200. [DOI] [PubMed] [Google Scholar]
- Sharma J, Balakrishnan L, Datta KK, Sahasrabuddhe NA, Khan AA, Sahu A, Singhal A, Getnet D, Raju R, Chatterjee A, Gowda H, Keshava Prasad TS, Shankar S, Pandey A. A knowledgebase resource for interleukin-17 family mediated signaling. J Cell Commun Signal. 2015;9:291–296. doi: 10.1007/s12079-015-0297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MP, Ayad VJ, Mundell SJ, McArdle CA, Kelly E, Lopez Bernal A. Internalization and desensitization of the oxytocin receptor is inhibited by dynamin and clathrin mutants in human embryonic kidney 293 cells. Mol Endocrinol. 2006;20:379–388. doi: 10.1210/me.2005-0031. [DOI] [PubMed] [Google Scholar]
- Soloff MS, Alexandrova M, Fernstrom MJ. Oxytocin receptors: triggers for parturition and lactation? Science. 1979;204:1313–1315. doi: 10.1126/science.221972. [DOI] [PubMed] [Google Scholar]
- Subbannayya T, Balakrishnan L, Sudarshan G, Advani J, Kumar S, Mahmood R, Nair B, Sirdeshmukh R, Mukherjee KK, Umathe SN, Raju R, Prasad TS. An integrated map of corticotropin-releasing hormone signaling pathway. J Cell Commun Signal. 2013;7:295–300. doi: 10.1007/s12079-013-0197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summar ML, Phillips JA 3rd, Battey J, Castiglione CM, Kidd KK, Maness KJ, Weiffenbach B, Gravius TC (1990) Linkage relationships of human arginine vasopressin-neurophysin-II and oxytocin-neurophysin-I to prodynorphin and other loci on chromosome 20. Mol Endocrinol 4:947–950 [DOI] [PubMed]
- Tamma R, Colaianni G, Zhu LL, DiBenedetto A, Greco G, Montemurro G, Patano N, Strippoli M, Vergari R, Mancini L, Colucci S, Grano M, Faccio R, Liu X, Li J, Usmani S, Bachar M, Bab I, Nishimori K, Young LJ, Buettner C, Iqbal J, Sun L, Zaidi M, Zallone A. Oxytocin is an anabolic bone hormone. Proc Natl Acad Sci U S A. 2009;106:7149–7154. doi: 10.1073/pnas.0901890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzidou V, Blanks AM, Kim SH, Thornton S, Bennett PR. Labor and inflammation increase the expression of oxytocin receptor in human amnion. Biol Reprod. 2011;84:546–552. doi: 10.1095/biolreprod.110.086785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Balakrishnan L, Sharma K, Khan AA, Advani J, Gowda H, Tripathy SP, MAP S, Gandotra S, TS P, Shankar S. A network map of Interleukin-10 signaling pathway. J Cell Commun Signal. 2015;10:61–67. doi: 10.1007/s12079-015-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrachnis N, Malamas FM, Sifakis S, Deligeoroglou E, Iliodromiti Z. The oxytocin-oxytocin receptor system and its antagonists as tocolytic agents. Int J Endocrinol. 2011;2011:350546. doi: 10.1155/2011/350546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YF, Hatton GI. Mechanisms underlying oxytocin-induced excitation of supraoptic neurons: prostaglandin mediation of actin polymerization. J Neurophysiol. 2006;95:3933–3947. doi: 10.1152/jn.01267.2005. [DOI] [PubMed] [Google Scholar]
- Wang W, Fan YQ, Lv Z, Yao XJ, Wang W, Huang KW, Meng Q, Fang CL, Lee TH, Corrigan CJ, An YQ, Ying S. Interleukin-25 promotes basic fibroblast growth factor expression by human endothelial cells through interaction with IL-17RB, but not IL-17RA. Clin Exp Allergy. 2012;42:1604–1614. doi: 10.1111/j.1365-2222.2012.04062.x. [DOI] [PubMed] [Google Scholar]
- Weeks A. The prevention and treatment of postpartum haemorrhage: what do we know, and where do we go to next? BJOG. 2015;122:202–210. doi: 10.1111/1471-0528.13098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 21 kb)


