Abstract
Drought stress is one of major factors resulting in maize yield loss. The roles of abscisic acid (ABA) have been widely studied in crops in response to drought stress. However, more attention is needed to identify key ABA-related proteins and also gain deeper molecular insights about drought stress in maize. Based on this need, the physiology and proteomics of the ABA-deficient maize mutant vp5 and its wild-type Vp5 under drought stress were examined and analyzed. Malondialdehyde content increased and quantum efficiency of photosystem II decreased under drought stress in both genotypes. However, the magnitude of the increase or decrease was significantly higher in vp5 than in Vp5. A total of 7051 proteins with overlapping expression patterns among three replicates in the two genotypes were identified by Multiplex run iTRAQ-based quantitative proteomic and liquid chromatography-tandem mass spectrometry methods, of which the expression of only 150 proteins (130 in Vp5, 27 in vp5) showed changes of at least 1.5-fold under drought stress. Among the 150 proteins, 67 and 60 proteins were up-regulated and down-regulated by drought stress in an ABA-dependent way, respectively. ABA was found to play active roles in regulating signaling pathways related to photosynthesis, oxidative phosphorylation (mainly related to ATP synthesis), and glutathione metabolism (involved in antioxidative reaction) in the maize response to drought stress. Our results provide an extensive dataset of ABA-dependent, drought-regulated proteins in maize plants, which may help to elucidate the underlying mechanisms of ABA-enhanced tolerance to drought stress in maize.
Keywords: Zea mays L., drought stress, abscisic acid (ABA), quantitative proteome, iTRAQ, LC-MS/MS, ABA signaling pathways
Introduction
Humans are heavily dependent on cereals as a main food crop. Among cereal crops, maize is widely cultivated all over the world, in addition to rice and wheat. Yet maize production is prominently affected by drought stress, which has been proved to be one of the major stress factors causing yield loss (Gong et al., 2014). It is predicted that the reduction of maize yield will reach 39.3% when water reduction is ~40% (Daryanto et al., 2016). Many drought-tolerant maize varieties have been cultivated by breeding efforts and technology improvement, which can increase the production of crops. Nevertheless, the yield increase may be counteracted by the prolonged drought stress caused by global climate change and uncertainties of precipitation patterns. Food security is therefore becoming more vulnerable in comparison with the past (FAO, 2008). For food security, it is necessary to determine more deeply the effects of drought stress on maize development and physiology, and the molecular mechanisms at different stages.
Both abscisic acid (ABA)-dependent and ABA-independent signaling are involved in plant responses to stress (Roychoudhury et al., 2013; Yoshida et al., 2014). For example, in Arabidopsis, high temperature inhibited the function of the ubiquitin proteasome system, but the inhibition was reduced in ABA biosynthetic mutants and in plants by the treatment of fluridone (ABA biosynthesis inhibitor), suggesting that it was regulated by high temperature in an ABA-dependent manner (Chiu et al., 2016). In Physcomitrella patens, the accumulation of low-molecular-weight soluble (LMS) sugars had no significant effect on the ABA-deficient mutant ppaba1 and its wild type after hyperosmotic and ABA treatments, suggesting that LMS sugars were regulated by hyperosmotic stress in an ABA-independent way (Takezawa et al., 2015).
When maize plants are subjected to adverse conditions, various mechanisms are evoked to deal with stress challenges, which include antioxidant capabilities, osmotic adjustment, photosynthetic rate reduction, and ABA accumulation (Gong et al., 2014; Sah et al., 2016). These processes involve the expression of stress-response genes, many of which are regulated by ABA (Fan et al., 2016). Therefore, ABA as an important messenger regulates the adaptive response of plants to abiotic stress (Sah et al., 2016). Currently, although the identification of ABA receptors has greatly increased our understanding of ABA perception in plants, this is not enough to identify the proteins regulated by ABA when maize plants are subjected to drought stress.
For this reason, this study aimed to better identify the key proteins regulated by ABA when maize seedlings were exposed to drought stress, and ultimately use this information to guide agricultural planning and minimize maize yield reduction caused by drought stress. Maize mutant viviparous-5 (vp5) is deficient in ABA biosynthesis and suffers photo-bleaching of leaves under normal light conditions (Robichaud et al., 1980; Hable et al., 1998), and has a much reduced ABA content in maize plants compared with wild-type Vp5 plants (Hu et al., 2015). Thus, the mutant vp5 and its wild-type Vp5 are ideal materials in which to identify key drought-response proteins regulated by ABA in maize plants. In this study, the differentially expressed proteins in maize plants exposed to drought stress were identified by multiplex run iTRAQ-based quantitative proteomic analysis and liquid chromatography-tandem mass spectrometry (LC-MS/MS) methods. As a result, 150 proteins with significant changes in expression level were identified as being significantly regulated by drought stress in an ABA-dependent or -independent way.
Methods
Plant material and treatments
Maize mutant vp5 and its wild-type Vp5 seedlings were used in the present study. As described previously (Hu et al., 2015), the vp5 mutant has reduced amounts of ABA because of a deficiency in ABA biosynthesis (Robichaud et al., 1980). Homozygous recessive kernels (vp5/vp5) lack carotenoids, causing white endosperm and embryos, which are easily distinguishable from the yellow wild-type kernels (Vp5/–). Because the recessive mutation is lethal in the homozygous state, it is maintained as a heterozygote. Seeds of vp5 and Vp5 plants were obtained by selfing plants grown from heterozygous seeds (Maize Genetics Stock Center, Urbana, IL, USA).
According to our previous description (Hu et al., 2015), Vp5 and vp5 seeds were germinated on moistened filter paper after being surface-sterilized for 10 min in 2% hypochlorite and then rinsed in distilled water. After germination for 2 days, both vp5 and Vp5 seedlings were cultured in Hoagland's nutrient solution in a light chamber (day 28°C/night 22°C, relative humidity 75%) under 400 μmol/m2/s photosynthetically active radiation with a 14/10 h (day/night) cycle. After 2 weeks, the seedlings were subjected to drought stress by placing them in a −0.7 MPa PEG6000 solution for 8 h at 28°C under relative humidity of 40%. Control seedlings were maintained at 28°C under relative humidity of 75%. Subsequently, leaves of treated and untreated seedlings were sampled, immediately frozen in liquid N2, and stored at −80°C until analysis. Three or five replicates were performed for each treatment.
The quantum efficiency of photosystem II (ΦPSII)
ΦPSII was measured by using an OS-30p Chlorophyll Fluorometer (Opti-Sciences, Tyngsboro, Massachusetts, USA) in the second full expand leaf.
Malondialdehyde (MDA)
MDA contents were measured according to a previous description (Hodges et al., 1999). Fifty milligrams fresh weight (FW) leaves in 1 ml 80% (v/v) ethanol were homogenized in a mortar. After centrifugation, the supernatant was reacted with thiobarbituric acid to produce the pinkish-red chromogen, thiobarbituric acid-malondialdehyde. Absorbance was measured at 440, 532, and 600 nm by using a UV-vis spectrophotometer. MDA content was calculated as nmol/g FW tissue.
Protein extraction
According to our previous description (Hu et al., 2015), total proteins from the second newly expanded leaf of the maize seedlings were extracted according to the following procedure. Approximately 0.5 g of fresh leaves from each biological replicate were ground into a fine power in liquid N2 in a mortar and further ground in 4 ml SDS buffer (30% sucrose, 2% SDS, 100 mM Tris-HCl, pH 8.0, 50 mM EDTA-Na2, 20 mM DTT) and 4 ml phenol (Tris-buffered, pH 8.0) in a 10 ml tube, followed by the addition of 1 mM phenylmethanesulfonyl fluoride and PhosSTOP Phosphatase Inhibitor Cocktail (one tablet/10 ml; Roche, Basel, Switzerland) to inhibit protease and phosphatase activity, respectively. The mixture was thoroughly vortexed for 30 s, and the phenol phase was separated by centrifugation at 14,000 × g and 4°C for 15 min. The upper phenol phase was pipetted into fresh 10 ml tubes, and 4-fold volumes of cold methanol plus 100 mM ammonium acetate were added. After centrifugation at 14,000 × g and 4 °C for 15 min, the supernatant was carefully discarded and the precipitated proteins were washed twice with cold acetone. Finally, the protein mixtures were harvested by centrifugation. Measurement of protein content was carried out using a 2-D Quant Kit (Amersham Bioscience, America) containing bovine serum albumin (2 mg/ml) as the standard. To enhance the quantitative accuracy, proteins extracted from every biological replicate were adjusted to the same concentration for the subsequent analysis (Wang et al., 2013; Hu et al., 2015; Zhang et al., 2015).
Protein digestion and iTRAQ labeling
As described previously (Hu et al., 2015), protein digestion was performed according to the FASP procedure (Umezawa et al., 2013), and the resulting peptide mixture was labeled using the 4-plex iTRAQ reagent according to the manufacturer's instructions (Applied Biosystems). Briefly, 200 μg proteins for each sample were incorporated into 30 μl STD buffer (4% SDS, 100 mM DTT, 150 mM Tris-HCl, pH 8.0). The detergent DTT and other low-molecular-weight components were removed using UA buffer (8 M urea, 150 mM Tris-HCl, pH 8.0) by repeated ultrafiltration (Microcon units, 30 kD). Then, 100 μl of 0.05 M iodoacetamide in UA buffer was added to block reduced cysteine residues, and the samples were incubated for 20 min in darkness. The filters were washed with 100 μl UA buffer three times and then washed twice with 100 μl DS buffer (50 mM trimethylammonium bicarbonate at pH 8.5). Finally, the protein suspensions were digested with 2 μg trypsin (Promega) in 40 μl DS buffer overnight at 37°C, and the resulting peptides were collected as a filtrate. The peptide content was estimated by UV light spectral density at 280 nm of a 0.1% solution using an extinction coefficient of 1.1 that was calculated on the basis of the frequency of tryptophan and tyrosine in vertebrate proteins.
For labeling, each iTRAQ reagent was dissolved in 70 μl ethanol and added to the respective peptide mixture. The samples, Vp5-control, Vp5-drought, vp5-control, and vp5-drought, were multiplexed and vacuum dried. Three independent biological experiments were performed.
Peptide fractionation with strong cation exchange chromatography
According to our previous description (Hu et al., 2015), iTRAQ-labeled peptides were fractionated by strong cation exchange chromatography using the AKTA Purifier system (GE Healthcare). The dried peptide mixture was reconstituted and acidified with 2 ml buffer A (10 mM KH2PO4 in 25% ACN, pH 2.7) and loaded onto a PolySULFOETHYL 4.6 × 100 mM column (5 μm, 200 Å, PolyLC Inc, Maryland, USA). The peptides were eluted at a flow rate of 1 ml/min with a gradient of 0–10% buffer B (500 mM KCl, 10 mM KH2PO4 in 25% of ACN, pH 2.7) for 2 min, 10–20% buffer B for 25 min, 20–45% buffer B for 5 min, and 50–100% buffer B for 5 min. The elution was monitored by absorbance at 214 nm, and fractions were collected every 1 min. The collected fractions (about 30 fractions) were finally combined into 10 pools and desalted on C18 Cartridges [Empore™ SPE Cartridges C18 (standard density), bed inner diameter 7 mm, volume 3 ml, Sigma]. Each fraction was concentrated by vacuum centrifugation and reconstituted in 40 μl 0.1% (v/v) trifluoroacetic acid. All samples were stored at −80°C until LC-MS/MS analysis.
Liquid chromatography (LC)-electrospray ionization (ESI) tandem MS (MS/MS) analysis by Q exactive
As described previously (Hu et al., 2015), experiments were performed on a Q Exactive mass spectrometer that was coupled to an Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific). Ten microliters of each fraction were injected for nanoLC-MS/MS analysis. The peptide mixture (5 μg) was loaded onto a C18 reversed-phase column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin) in buffer A (0.1% formic acid) and separated with a linear gradient of buffer B (80% acetonitrile and 0.1% formic acid) at a flow rate of 250 nl/min controlled by IntelliFlow technology over 140 min. MS data were acquired using a data-dependent top10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for higher-energy collisional dissociation fragmentation. Determination of the target value was based on predictive automatic gain control. Dynamic exclusion duration was 60 s. Survey scans were acquired at a resolution of 70,000 at m/z 200, and the resolution for higher-energy collisional dissociation spectra was set to 17,500 at m/z 200. Normalized collision energy was 30 eV, and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
Data analysis
According to our previous description (Hu et al., 2015), MS/MS spectra were searched using Mascot 2.2 (Matrix Science) embedded in Proteome Discoverer 1.4 against the uniprot_Zea_mays_87227_20150504.fasta (87227 sequences, download May 4th, 2015) and the decoy database. The parameters used in Mascot searches for normal peptides were as follows: Peptide mass tolerance: 20 ppm, MS/MS tolerance: 0.1 Da, Enzyme: Trypsin, max missed cleavage: 2, Fixed modification: Carbamidomethyl (C), iTRAQ4plex(K), iTRAQ4plex(N-term), Variable modification:Oxidation (M), FDR ≤ 0.01. The protein and peptide probabilities were set at 50 and 60%, respectively. Proteins with at least two unique peptides, a Mascot score of at least 25, and detection in at least two replicates were further used.
For each replicate of proteomics, iTRAQ ratios between drought stressed plants and controls for each run were converted to z-scores to normalize the data.
Bioinformatics
As described previously (Hu et al., 2015), the molecular functions of the identified proteins were classified according to their gene ontology annotations and their biological functions. The subcellular localization of the unique proteins identified in this study was predicted using the publicly available program WolfPsort (http://wolfpsort.org). Protein-protein interaction networks were analyzed using the publicly available program STRING (http://string-db.org/). STRING is a database of known and predicted protein-protein interactions. The interactions include direct (physical) and indirect (functional) associations, and they are derived from four sources: the genomic context, high-throughput experiments, coexpression, and previous knowledge. STRING quantitatively integrates the interaction data from these sources for a large number of organisms and, where applicable, transfers information between these organisms.
ABA assay
According to our previous description (Hu et al., 2012), maize leaves (0.5–1.0 g) were ground in liquid N2 with a mortar, extracted with 2 ml of ice-cold 80% methanol containing 1 mM butylated hydroxytoluene to prevent oxidation, and then stored overnight at 4°C. The extracts were centrifuged at 12,000 × g for 15 min at 4°C. The pellets were extracted once and stored at 4°C for 1 h. The two resulting supernatants were combined and passed through a C18 Sep-Pak cartridge (Waters, Milford, MA, USA). The efflux was collected and dried in N2. The residues were then dissolved in 10 mM phosphate buffer solution (pH 7.4), and concentrations of ABA were determined in enzyme-linked immunosorbent assay (ELISA). Statistical analyses of the physiological measurements were conducted using independent Student's t-tests with SPSS statistics software (version 17.0).
Gene expression analysis by reverse-transcription PCR
As described previously (Hu et al., 2010), total RNA was isolated from leaves by using an RNeasy mini kit according to the instructions supplied by the manufacturer (Qiagen, Valencia, CA). Approximately 3 μg of total RNA was reverse transcribed into cDNA using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). cDNA was amplified by PCR using the primers shown in Supplementary Table S1. To standardize the results, the relative abundance of β-actin was also determined and used as the internal standard. Aliquots of the PCR reactions were loaded on agarose gels and stained with ethidium bromide.
Statistical analysis
The proteins and ABA assays were the mean of three replicates. The means were compared by a one-way analysis of variance and Duncan's multiple range test at a 5% level of significance. False discovery rates attained by the Benjamini-Hochberg method were used to adjust p-values (correction for multiple comparisons). The significance of differences between Vp5 and vp5 were compared by t-test analysis at the 5% level.
Results
The influence of drought on ABA, MDA, and ΦPSII
After vp5 and Vp5 plants were subjected to drought treatment, the ABA content of leaves was measured by ELISA. Whether under normal or drought stress conditions, the ABA content in Vp5 leaves was more than 6-fold higher than that in vp5 leaves; compared with the corresponding control, the relative increase in ABA content was 120.45 and 21.98% in Vp5 and vp5 leaves under drought stress, respectively (Table 1). MDA is generated by lipid peroxidation, so the change in MDA content reflects the extent of membrane damage. Our results indicated that MDA content was prominently elevated by drought stress, and the relative increase was 193.1 and 232.6% in Vp5 and vp5 leaves, respectively, compared with the corresponding control (Table 1). ΦPSII is one of the chlorophyll fluorescence parameters and is classically used to monitor change in photosynthetic performance. The present results indicated that the relative decrease in ΦPSII caused by drought stress was 33.00 and 39.5% in Vp5 and vp5 leaves, respectively, compared with the respective controls. Regardless of drought stress level, the ΦPSII of Vp5 was significantly more than that of vp5. These results indicate that the ABA-deficient mutant vp5 is more susceptible to drought stress than Vp5 and is very useful material to study the key proteins regulated by ABA under drought stress.
Table 1.
ABA and MDA content, and ΦPSII in maize vp5 and Vp5 leaves under control and drought conditions.
| Gene type | Control | Drought | Relative increase/decrease (%) | |
|---|---|---|---|---|
| ABA | Vp5 | 76.50 (ng/gFW) b | 168.64 (ng/gFW) a | 120.45 ↑ |
| vp5 | 17.28 (ng/gFW) d | 21.08 (ng/gFW) c | 21.98 ↑ | |
| MDA | Vp5 | 5.12 (nmol/gFW) d | 15.01 (nmol/gFW) b | 193.1 ↑ |
| vp5 | 7.45 (nmol/gFW) c | 24.78 (nmol/gFW) a | 232.6 ↑ | |
| ΦPSII | Vp5 | 0.612 a | 0.410 b | 33.00 ↓ |
| vp5 | 0.210 c | 0.128 d | 39.05 ↓ |
Each value represents the average of three biological replicas. For Dun's Results, different characters are considered to be significant among different treatments. The symbols “↑and ↓” represent relative increase and decrease, respectively.
Identification of differentially expressed proteins under drought stress
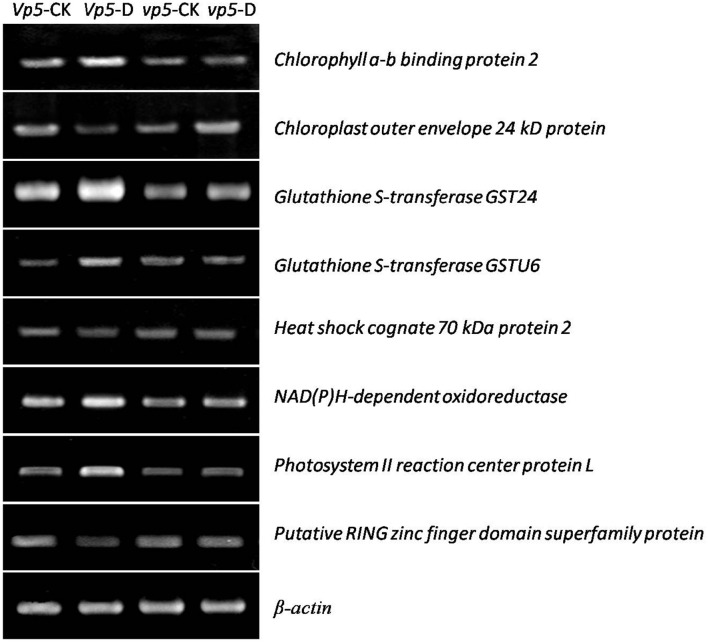
After vp5 and Vp5 seedlings were subjected to drought stress, total proteins from leaves were extracted and analyzed by multiplex run iTRAQ-based quantitative proteomic analysis and LC-MS/MS methods according to the workflow shown in Figure 1, resulting in the identification of 7051 proteins at a false discovery rate of 1%. Of the 7051 proteins (indentified peptides, see Supplementary Excels S1–S3), based on p < 0.01, there were 150 proteins with a ≥1.5-fold (increased) or ≤ 0.66-fold (decreased) expression change ratio under drought stress relative to the respective controls (Figure 2). In order to validate the above-mentioned results of protein abundance, the transcript expression levels of eight interesting proteins were analyzed by reverse-transcription PCR. The results indicated that the transcription patterns of the eight proteins were concomitant with protein expression levels, which supported the results attained by iTRAQ-based quantitative proteomic analysis and LC-MS/MS (Figure 3).
Figure 1.
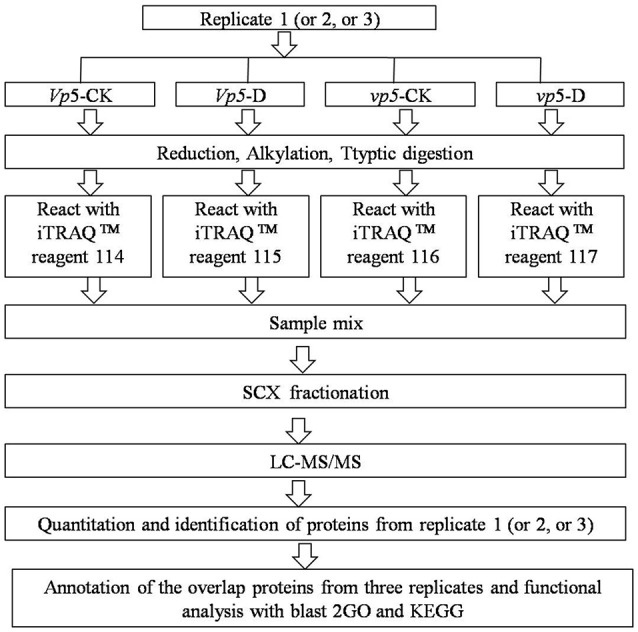
iTRAQ 4-plex labeling and LC MS/MS workflow for identifying proteins in the ABA-deficient mutant vp5 and wild-type Vp5 seedling leaves under drought conditions.
Figure 2.
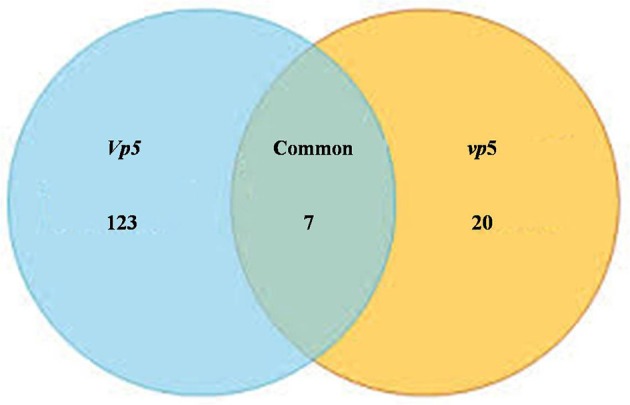
Venn diagram showing the number of significantly expressed proteins in maize leaves under drought stress. The diagram shows the overlap between the ABA-deficient mutant vp5 and the wild-type Vp5.
Figure 3.
Gene expression analysis of eight proteins in maize Vp5 and vp5 leaves under drought stress conditions. The eight proteins included chlorophyll a-b binding protein 2, chloroplast outer envelope 24 kD protein, glutathione S-transferase GST 24, glutathione S-transferase GSTU6, heat shock cognate 70 kDa protein 2, NAD(P)H-dependent oxidoreductase, photosystem II reaction center protein L, and putative RING zinc finger domain superfamily protein. Experiments were repeated at least three times.
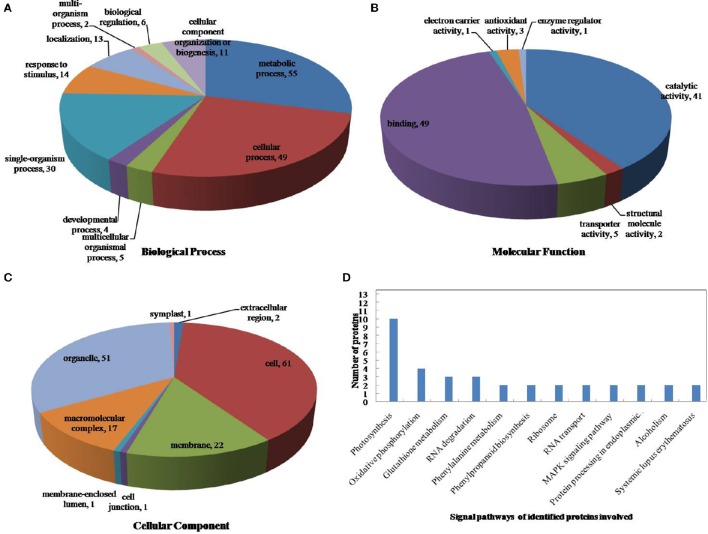
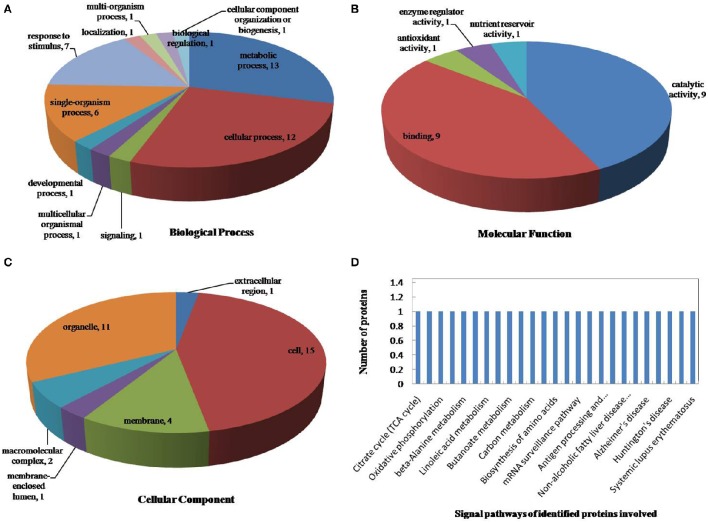
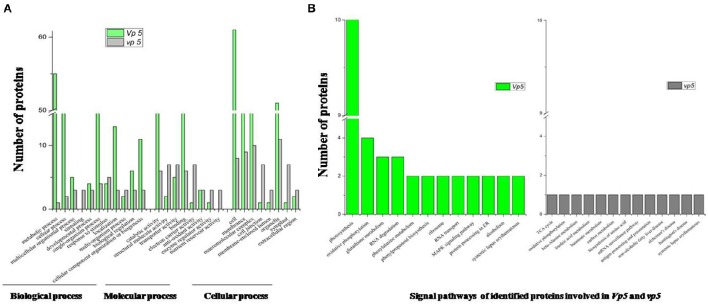
The drought-response proteins in vp5 and Vp5 were annotated using Blast2GO according to the biological process “cell component and molecular function” (Figures 4, 5A–C). The results showed that the differentially expressed proteins in Vp5 were mainly involved in such signal pathways as photosynthesis, oxidative phosphorylation, glutathione metabolism, and RNA degradation (Figure 4D), which was different from the signaling pathways identified in vp5 (Figure 5D). Figure 6 shows the comparisons of the biological process, molecular functions, cellular components, and signaling pathways of proteins identified in Vp5 and vp5 under drought stress, which clearly indicated a difference in significantly expressed proteins between Vp5 and vp5 under drought stress.
Figure 4.
Pie charts of the distribution of differentially expressed proteins based on their predicted molecular (A) biological process, (B) molecular functions, and (C) cellular components, and (D) the signaling pathways of the proteins identified in maize Vp5 leaves subjected to drought stress. In this study, 130 proteins were identified under drought stress and were classified by their known or predicted subcellular localization using Blast2Go (http://www.blast2go.com).
Figure 5.
Pie charts of the distribution of differentially expressed proteins by their predicted (A) biological process, (B) molecular functions, and (C) cellular components, and (D) the signaling pathways of the proteins identified in maize Vp5 leaves under drought stress. In this study, 27 proteins were identified under drought stress, and they were classified by their known or predicted cellular localization using Blast2Go (http://www.blast2go.com).
Figure 6.
Comparisons of the (A) biological process, molecular functions, cellular components, and (B) signaling pathways (from Figures 4, 5) of differentially expressed proteins identified in maize Vp5 and vp5 leaves under drought stress. The proteins were classified by their known or predicted cellular function using Blast2GO (http://www.blast2go.com).
ABA regulation for drought-response proteins
Plants respond to abiotic stress by ABA-dependent and ABA-independent mechanisms (Sah et al., 2016). In the present study, 150 proteins with a fold change ≥1.5 included 7 identified in both genotypes (Table 2), 123 only in Vp5 (Table 3), and 20 only in vp5 (Table 4). Specifically, among the 7 (Table 2), 123 (Table 3), and 20 (Table 4) proteins, there were 5, 64, and 8 uncharacterized proteins, respectively. Among the seven common proteins in the two genotypes (Table 2), the expression of four proteins (C0HF37, B7ZX39, B6T3J3, and K7TKJ3) was increased in Vp5 but decreased in vp5 under drought conditions, indicating that these proteins were up-regulated by drought stress in an ABA-dependent manner; drought stress decreased the expression of protein B4FMW4 by a similar extent in Vp5 and vp5, indicating that the protein was down-regulated by drought stress in an ABA-independent manner; drought stress significantly decreased the expression of two proteins (B6TM56, C0PBJ1) in Vp5, but significantly increased the expression of these two proteins in vp5, indicating that they were up-regulated by drought stress but down-regulated by ABA.
Table 2.
Proteins with significant expression level changes in both Vp5 and vp5 leaves under drought stress.
| Accession | Description | Vp5: D/control | vp5: D/control | Vp5: D/control | vp5: D/contro | T-test | Regulation of ABA and drought stress | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | Averagea | P-value | Averagea | P-value | ||||
| C0HF37 | Uncharacterized protein | 1.620 | 1.632 | 1.545 | 0.504 | 0.518 | 0.514 | 1.599 | 0.002 | 0.512 | 0.001 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FMW4 | Uncharacterized protein | 0.601 | 0.613 | 0.526 | 0.546 | 0.545 | 0.542 | 0.580 | 0.003 | 0.544 | 0.001 | 0.334 | Down-regulated by drought in an ABA-independent way |
| B7ZX39 | Uncharacterized protein | 1.615 | 1.627 | 1.540 | 0.541 | 0.552 | 0.546 | 1.594 | 0.001 | 0.546 | 0.001 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6T3J3 | Histone H2A | 1.672 | 1.691 | 1.590 | 0.561 | 0.575 | 0.571 | 1.651 | 0.001 | 0.569 | 0.002 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7TKJ3 | Uncharacterized protein | 5.551 | 5.063 | 5.076 | 0.621 | 0.635 | 0.632 | 5.230 | 0.000 | 0.629 | 0.003 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6TM56 | Chloroplast outer envelope 24 kD protein | 0.618 | 0.630 | 0.543 | 1.529 | 1.543 | 1.539 | 0.597 | 0.003 | 1.537 | 0.001 | 0.000 | Up-regulated by drought, but down-regulated by ABA |
| C0PBJ1 | Uncharacterized protein | 0.465 | 0.477 | 0.390 | 1.539 | 1.553 | 1.548 | 0.444 | 0.001 | 1.547 | 0.000 | 0.000 | Up-regulated by drought, but down-regulated by ABA |
Each value represents the average of three biological replicates. The average is significant at a p < 0.01 level. A t-test value <0.05 is considered to be significant between ABA-deficient mutant vp5 and wild-type Vp5. D, drought stress.
Table 3.
Proteins with significant expression level changes only in Vp5 leaves under drought stress.
| Accession | Description | Vp5: OS/control | vp5: OS/control | Vp5: OS/control | vp5: OS/contro | T-test | Regulation of ABA and drought stress | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | Averagea | P-value | Averagea | P-value | ||||
| B4F9L3 | Carbonic anhydrase | 0.627 | 0.592 | 0.602 | 0.970 | 0.962 | 0.984 | 0.607 | 0.003 | 0.972 | 0.655 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4F9Q3 | Uncharacterized protein | 1.825 | 1.890 | 1.850 | 0.916 | 0.898 | 0.910 | 1.855 | 0.000 | 0.908 | 0.188 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4F9U4 | Putative RING zinc finger domain superfamily protein | 0.534 | 0.499 | 0.509 | 1.084 | 1.076 | 1.098 | 0.514 | 0.001 | 1.086 | 0.213 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FA94 | Uncharacterized protein | 0.537 | 0.502 | 0.512 | 1.037 | 1.049 | 1.031 | 0.517 | 0.001 | 1.039 | 0.538 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FB53 | Uncharacterized protein | 1.527 | 1.492 | 1.502 | 1.060 | 1.052 | 1.074 | 1.507 | 0.001 | 1.062 | 0.346 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FB57 | Uncharacterized protein | 2.096 | 2.001 | 2.131 | 1.031 | 1.023 | 1.045 | 2.076 | 0.000 | 1.033 | 0.600 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FB96 | Uncharacterized protein | 0.661 | 0.626 | 0.636 | 0.979 | 0.971 | 0.993 | 0.641 | 0.004 | 0.981 | 0.760 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FBH1 | Uncharacterized protein | 0.677 | 0.642 | 0.652 | 1.015 | 1.007 | 1.029 | 0.657 | 0.004 | 1.017 | 0.784 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FBV4 | Chaperone protein dnaJ 10 isoform 1 | 1.705 | 1.793 | 1.740 | 0.934 | 0.926 | 0.948 | 1.746 | 0.000 | 0.936 | 0.332 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FBY1 | Chaperone protein dnaJ | 1.646 | 1.611 | 1.621 | 0.956 | 0.928 | 0.930 | 1.626 | 0.000 | 0.938 | 0.349 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FDK8 | Uncharacterized protein | 1.593 | 1.558 | 1.568 | 1.057 | 1.049 | 1.071 | 1.573 | 0.001 | 1.059 | 0.367 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FDY7 | Eukaryotic translation initiation factor 3 subunit E | 1.631 | 1.596 | 1.606 | 1.249 | 1.241 | 1.263 | 1.611 | 0.000 | 1.251 | 0.001 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FEE1 | Uncharacterized protein | 1.523 | 1.488 | 1.498 | 1.256 | 1.248 | 1.270 | 1.503 | 0.001 | 1.258 | 0.001 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FFW5 | Uncharacterized protein | 1.823 | 1.848 | 1.828 | 0.949 | 1.041 | 0.863 | 1.833 | 0.000 | 0.951 | 0.561 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FI24 | Uncharacterized protein | 0.681 | 0.646 | 0.656 | 1.275 | 1.167 | 1.089 | 0.661 | 0.004 | 1.177 | 0.009 | 0.001 | Down-regulated by drought in an ABA-dependent way |
| B4FJ71 | Uncharacterized protein | 0.577 | 0.542 | 0.552 | 1.020 | 1.012 | 1.034 | 0.557 | 0.002 | 1.022 | 0.724 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FJH1 | Uncharacterized protein | 0.500 | 0.465 | 0.475 | 1.024 | 1.016 | 1.038 | 0.480 | 0.001 | 1.026 | 0.678 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FK45 | Uncharacterized protein | 1.581 | 1.546 | 1.556 | 0.890 | 0.882 | 0.904 | 1.561 | 0.001 | 0.892 | 0.137 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FP86 | Uncharacterized protein | 2.291 | 2.456 | 2.366 | 0.999 | 0.991 | 1.013 | 2.371 | 0.000 | 1.001 | 0.987 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FPH3 | Uncharacterized protein | 1.610 | 1.575 | 1.585 | 1.028 | 1.020 | 1.042 | 1.590 | 0.001 | 1.030 | 0.633 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FPM5 | Uncharacterized protein | 1.814 | 1.789 | 1.809 | 1.017 | 1.009 | 1.031 | 1.804 | 0.000 | 1.019 | 0.760 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FPS3 | Uncharacterized protein | 1.637 | 1.602 | 1.612 | 0.900 | 0.862 | 0.854 | 1.617 | 0.000 | 0.872 | 0.018 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FQN6 | Sorbitol transporter | 0.682 | 0.647 | 0.657 | 1.332 | 1.324 | 1.346 | 0.662 | 0.005 | 1.334 | 0.005 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FRG9 | Uncharacterized protein | 0.620 | 0.585 | 0.595 | 0.952 | 0.944 | 0.966 | 0.600 | 0.002 | 0.954 | 0.473 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FWT5 | Soluble inorganic pyrophosphatase | 1.552 | 1.517 | 1.527 | 0.942 | 0.934 | 0.956 | 1.532 | 0.001 | 0.944 | 0.390 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FX06 | Uncharacterized protein | 0.568 | 0.533 | 0.543 | 0.909 | 0.901 | 0.923 | 0.548 | 0.002 | 0.911 | 0.200 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FX77 | Uncharacterized protein | 0.679 | 0.644 | 0.654 | 1.213 | 1.005 | 1.127 | 0.659 | 0.004 | 1.115 | 0.241 | 0.002 | Down-regulated by drought in an ABA-dependent way |
| B4FZ22 | Uncharacterized protein | 0.681 | 0.646 | 0.656 | 1.031 | 0.923 | 0.845 | 0.661 | 0.004 | 0.933 | 0.444 | 0.008 | Down-regulated by drought in an ABA-dependent way |
| B4G1A3 | Uncharacterized protein | 1.533 | 1.498 | 1.508 | 0.903 | 0.895 | 0.917 | 1.513 | 0.001 | 0.905 | 0.177 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4G1H1 | Uncharacterized protein | 1.599 | 1.564 | 1.574 | 1.295 | 1.287 | 1.309 | 1.579 | 0.001 | 1.297 | 0.007 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SHW9 | Ubiquitin fusion protein | 0.668 | 0.633 | 0.643 | 1.059 | 1.051 | 1.073 | 0.648 | 0.004 | 1.061 | 0.353 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6SJL2 | Thaumatin-like protein | 0.649 | 0.614 | 0.624 | 0.958 | 0.950 | 0.972 | 0.629 | 0.003 | 0.960 | 0.529 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6SK13 | Ribose-phosphate pyrophosphokinase 4 | 1.888 | 1.853 | 1.863 | 0.921 | 0.913 | 0.935 | 1.868 | 0.000 | 0.923 | 0.256 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SMB0 | Cytochrome b-c1 complex subunit 6 | 0.675 | 0.640 | 0.650 | 0.991 | 0.983 | 1.005 | 0.655 | 0.004 | 0.993 | 0.910 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6SP61 | Ferredoxin-1 | 0.655 | 0.620 | 0.630 | 0.762 | 0.754 | 0.776 | 0.635 | 0.003 | 0.764 | 0.015 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6SSB3 | Anthocyanidin 5,3-O-glucosyltransferase | 1.915 | 1.880 | 1.890 | 0.821 | 0.813 | 0.835 | 1.895 | 0.000 | 0.823 | 0.004 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6STN4 | Chlorophyll a-b binding protein 2 | 1.641 | 1.606 | 1.616 | 0.861 | 0.853 | 0.875 | 1.621 | 0.000 | 0.863 | 0.008 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SU20 | Tubby-like protein | 1.674 | 1.639 | 1.649 | 1.166 | 1.158 | 1.180 | 1.654 | 0.000 | 1.168 | 0.044 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SU31 | Glutathione peroxidase | 0.613 | 0.578 | 0.588 | 0.986 | 0.978 | 1.000 | 0.593 | 0.002 | 0.988 | 0.846 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6SV53 | Copper transporter 1 | 0.670 | 0.635 | 0.645 | 1.109 | 1.101 | 1.123 | 0.650 | 0.004 | 1.111 | 0.129 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6SXR2 | Photosystem I reaction center subunit XI | 1.604 | 1.569 | 1.579 | 0.837 | 0.829 | 0.851 | 1.584 | 0.001 | 0.839 | 0.005 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SZ69 | Heat shock cognate 70 kDa protein 2 | 0.674 | 0.639 | 0.649 | 0.868 | 0.860 | 0.882 | 0.654 | 0.004 | 0.870 | 0.019 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6SZK3 | NAD(P)H-dependent oxidoreductase | 1.857 | 1.822 | 1.832 | 0.933 | 0.925 | 0.947 | 1.837 | 0.000 | 0.935 | 0.326 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SZL9 | Leucine-rich repeat receptor protein kinase EXS | 1.559 | 1.524 | 1.534 | 1.066 | 1.058 | 1.080 | 1.539 | 0.001 | 1.068 | 0.307 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6T022 | Bowman-Birk type trypsin inhibitor | 1.694 | 1.659 | 1.669 | 0.833 | 0.825 | 0.847 | 1.674 | 0.000 | 0.835 | 0.047 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6T072 | Putative uncharacterized protein | 0.586 | 0.551 | 0.561 | 0.881 | 0.863 | 0.905 | 0.566 | 0.002 | 0.883 | 0.043 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6T1H8 | B12D protein | 0.584 | 0.549 | 0.559 | 1.011 | 1.003 | 1.025 | 0.564 | 0.001 | 1.013 | 0.683 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6T1Q2 | 4F5 protein family protein | 1.602 | 1.567 | 1.577 | 1.087 | 1.079 | 1.101 | 1.582 | 0.000 | 1.089 | 0.180 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6T531 | Ribonucleoprotein A | 0.618 | 0.583 | 0.593 | 0.748 | 0.740 | 0.762 | 0.598 | 0.001 | 0.750 | 0.004 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6T763 | Exosome complex exonuclease RRP41 | 0.376 | 0.341 | 0.351 | 1.079 | 1.071 | 1.093 | 0.356 | 0.000 | 1.081 | 0.222 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6T8S7 | Tonneau 1b | 0.622 | 0.587 | 0.597 | 0.916 | 1.108 | 1.030 | 0.602 | 0.001 | 1.018 | 0.986 | 0.002 | Down-regulated by drought in an ABA-dependent way |
| B6TGG7 | 3-oxoacyl-[acyl-carrier-protein] synthase | 0.624 | 0.589 | 0.599 | 1.355 | 1.347 | 1.369 | 0.604 | 0.001 | 1.357 | 0.002 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6TGS2 | Fb27 | 1.513 | 1.508 | 1.548 | 1.312 | 1.304 | 1.326 | 1.523 | 0.000 | 1.314 | 0.003 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6TLU5 | Lipid binding protein | 1.605 | 1.570 | 1.580 | 0.703 | 0.695 | 0.717 | 1.585 | 0.000 | 0.705 | 0.002 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6TP77 | Glutathione S-transferase GSTU6 | 1.531 | 1.496 | 1.506 | 0.987 | 0.979 | 1.001 | 1.511 | 0.000 | 0.989 | 0.568 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6TRQ5 | Tesmin/TSO1-like CXC domain containing protein | 0.513 | 0.478 | 0.488 | 0.970 | 0.962 | 0.984 | 0.493 | 0.000 | 0.972 | 0.373 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6TUT9 | Putative uncharacterized protein | 1.643 | 1.608 | 1.618 | 0.905 | 0.897 | 0.919 | 1.623 | 0.000 | 0.907 | 0.070 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6TVU2 | Prefoldin subunit 5 | 2.406 | 2.421 | 2.481 | 0.999 | 0.991 | 1.013 | 2.436 | 0.000 | 1.001 | 0.743 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6TWU1 | Putative uncharacterized protein | 0.662 | 0.627 | 0.637 | 1.064 | 1.056 | 1.078 | 0.642 | 0.001 | 1.066 | 0.330 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6TYB4 | Aldo-keto reductase yakc | 1.692 | 1.657 | 1.667 | 0.695 | 0.687 | 0.709 | 1.672 | 0.000 | 0.697 | 0.002 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6TYM4 | C2 domain containing protein | 0.650 | 0.615 | 0.625 | 1.121 | 1.113 | 1.135 | 0.630 | 0.001 | 1.123 | 0.075 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6U118 | T-complex protein 1 subunit zeta | 3.829 | 3.894 | 4.004 | 0.957 | 0.949 | 0.971 | 3.909 | 0.000 | 0.959 | 0.265 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6U4B9 | Dual specificity protein phosphatase 4 | 1.605 | 1.620 | 1.680 | 1.059 | 1.051 | 1.073 | 1.635 | 0.000 | 1.061 | 0.376 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6U5V4 | ABA-induced protein | 0.341 | 0.306 | 0.316 | 0.905 | 0.897 | 0.919 | 0.321 | 0.000 | 0.907 | 0.070 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6U7X1 | Maf-like protein CV_0124 | 1.567 | 1.572 | 1.562 | 0.935 | 0.927 | 0.949 | 1.567 | 0.000 | 0.937 | 0.148 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6UAH3 | Putative uncharacterized protein | 0.646 | 0.611 | 0.621 | 1.038 | 1.030 | 1.052 | 0.626 | 0.001 | 1.040 | 0.628 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6UEB0 | Lipid binding protein | 0.678 | 0.643 | 0.653 | 0.915 | 0.907 | 0.929 | 0.658 | 0.001 | 0.917 | 0.089 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6UG46 | Peroxin Pex14 | 0.649 | 0.614 | 0.624 | 1.328 | 1.020 | 1.042 | 0.629 | 0.001 | 1.130 | 0.036 | 0.007 | Down-regulated by drought in an ABA-dependent way |
| B7U627 | DHN2-like protein | 1.776 | 1.741 | 1.751 | 1.147 | 1.139 | 1.161 | 1.756 | 0.000 | 1.149 | 0.041 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B8A1T1 | Peroxidase | 0.666 | 0.631 | 0.641 | 1.055 | 1.047 | 1.069 | 0.646 | 0.001 | 1.057 | 0.417 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B8A310 | Uncharacterized protein | 1.898 | 2.063 | 1.973 | 1.096 | 1.088 | 1.110 | 1.978 | 0.000 | 1.098 | 0.142 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0HEI0 | Uncharacterized protein | 0.398 | 0.363 | 0.373 | 1.264 | 1.256 | 1.278 | 0.378 | 0.000 | 1.266 | 0.005 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0HFZ5 | Uncharacterized protein | 1.539 | 1.504 | 1.514 | 0.805 | 0.797 | 0.819 | 1.519 | 0.000 | 0.807 | 0.009 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0HG70 | Uncharacterized protein | 1.580 | 1.545 | 1.555 | 0.858 | 0.850 | 0.872 | 1.560 | 0.000 | 0.860 | 0.025 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0HGT5 | T-complex protein 1 subunit delta | 1.536 | 1.501 | 1.511 | 1.214 | 1.006 | 1.128 | 1.516 | 0.000 | 1.116 | 0.255 | 0.003 | Up-regulated by drought in an ABA-dependent way |
| C0HHB1 | Carboxypeptidase | 2.804 | 2.829 | 2.809 | 0.934 | 1.026 | 0.848 | 2.814 | 0.000 | 0.936 | 0.299 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0HJ24 | Uncharacterized protein | 1.608 | 1.573 | 1.583 | 1.046 | 1.038 | 1.060 | 1.588 | 0.000 | 1.048 | 0.521 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0P2V2 | Uncharacterized protein | 1.621 | 1.586 | 1.596 | 0.925 | 0.917 | 0.939 | 1.601 | 0.000 | 0.927 | 0.115 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0P2V7 | Uncharacterized protein | 1.534 | 1.499 | 1.509 | 1.046 | 1.038 | 1.060 | 1.514 | 0.000 | 1.048 | 0.521 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0P3Y3 | Uncharacterized protein | 0.682 | 0.647 | 0.657 | 1.037 | 1.029 | 1.051 | 0.662 | 0.001 | 1.039 | 0.643 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0P5M7 | Uncharacterized protein | 0.684 | 0.649 | 0.659 | 0.937 | 0.929 | 0.951 | 0.664 | 0.001 | 0.939 | 0.156 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0P727 | Uncharacterized protein | 0.674 | 0.639 | 0.649 | 1.076 | 1.068 | 1.090 | 0.654 | 0.001 | 1.078 | 0.241 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0PF36 | Uncharacterized protein | 0.648 | 0.613 | 0.623 | 0.990 | 0.982 | 1.004 | 0.628 | 0.001 | 0.992 | 0.609 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0PFW9 | Uncharacterized protein | 1.539 | 1.544 | 1.534 | 0.974 | 0.966 | 0.988 | 1.539 | 0.000 | 0.976 | 0.413 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0PIW4 | Uncharacterized protein | 0.656 | 0.621 | 0.631 | 1.001 | 0.993 | 1.015 | 0.636 | 0.001 | 1.003 | 0.774 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0PJ51 | Uncharacterized protein | 0.643 | 0.608 | 0.618 | 0.979 | 0.971 | 0.993 | 0.623 | 0.001 | 0.981 | 0.468 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0PKD1 | Uncharacterized protein | 1.839 | 1.804 | 1.814 | 0.949 | 0.941 | 0.963 | 1.819 | 0.000 | 0.951 | 0.215 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0PL82 | Uncharacterized protein | 1.594 | 1.559 | 1.569 | 1.018 | 1.010 | 1.032 | 1.574 | 0.000 | 1.020 | 0.944 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0PLS3 | Uncharacterized protein | 2.105 | 2.070 | 2.080 | 1.088 | 1.080 | 1.102 | 2.085 | 0.000 | 1.090 | 0.126 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0PMR0 | Uncharacterized protein | 0.580 | 0.545 | 0.555 | 0.968 | 0.960 | 0.982 | 0.560 | 0.001 | 0.970 | 0.555 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0PMS5 | Uncharacterized protein | 0.406 | 0.371 | 0.381 | 1.144 | 1.136 | 1.158 | 0.386 | 0.000 | 1.146 | 0.035 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0PMV2 | Uncharacterized protein | 1.814 | 1.779 | 1.789 | 1.004 | 0.996 | 1.018 | 1.794 | 0.000 | 1.006 | 0.904 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0PN61 | Uncharacterized protein | 0.393 | 0.358 | 0.368 | 0.900 | 0.892 | 0.914 | 0.373 | 0.000 | 0.902 | 0.103 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C4J1S1 | Uncharacterized protein | 0.675 | 0.640 | 0.650 | 0.916 | 0.908 | 0.930 | 0.655 | 0.002 | 0.918 | 0.154 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C4J3X0 | Uncharacterized protein | 0.448 | 0.413 | 0.423 | 0.809 | 0.801 | 0.823 | 0.428 | 0.000 | 0.811 | 0.015 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C4J8X4 | Uncharacterized protein | 0.679 | 0.644 | 0.654 | 1.002 | 0.994 | 1.016 | 0.659 | 0.002 | 1.004 | 0.936 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C4J9M6 | Uncharacterized protein | 1.990 | 1.955 | 1.965 | 0.993 | 0.985 | 1.007 | 1.970 | 0.000 | 0.995 | 0.920 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7TI96 | Uncharacterized protein | 4.551 | 4.516 | 4.526 | 1.001 | 0.993 | 1.015 | 4.531 | 0.000 | 1.003 | 0.952 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7U170 | Uncharacterized protein | 0.529 | 0.494 | 0.504 | 0.968 | 0.960 | 0.982 | 0.509 | 0.000 | 0.970 | 0.555 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| K7U195 | Uncharacterized protein | 0.639 | 0.604 | 0.614 | 0.874 | 0.866 | 0.888 | 0.619 | 0.001 | 0.876 | 0.014 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| K7U2V0 | Uncharacterized protein | 1.532 | 1.497 | 1.507 | 0.861 | 0.853 | 0.875 | 1.512 | 0.000 | 0.863 | 0.010 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7U658 | Uncharacterized protein | 2.081 | 2.246 | 2.156 | 1.195 | 1.187 | 1.209 | 2.161 | 0.000 | 1.197 | 0.003 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7UG66 | Oxygen evolving enhancer protein 3 | 1.713 | 1.678 | 1.688 | 0.860 | 0.852 | 0.874 | 1.693 | 0.000 | 0.862 | 0.010 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7UIV2 | FHA transcription factor | 1.879 | 1.844 | 1.854 | 0.904 | 0.896 | 0.918 | 1.859 | 0.001 | 0.906 | 0.114 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7UPC4 | Uncharacterized protein | 0.654 | 0.619 | 0.629 | 0.964 | 0.956 | 0.978 | 0.634 | 0.000 | 0.966 | 0.314 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| K7UTH3 | Putative DUF1296 domain containing family protein | 1.669 | 1.634 | 1.644 | 1.083 | 1.075 | 1.097 | 1.649 | 0.000 | 1.085 | 0.142 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7UUC8 | Uncharacterized protein (Fragment) | 0.671 | 0.636 | 0.646 | 1.129 | 1.121 | 1.143 | 0.651 | 0.002 | 1.131 | 0.048 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| K7UXB7 | Uncharacterized protein | 0.664 | 0.629 | 0.639 | 0.930 | 0.922 | 0.944 | 0.644 | 0.002 | 0.932 | 0.219 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| K7UZ21 | Peroxidase | 0.635 | 0.600 | 0.610 | 0.805 | 0.797 | 0.819 | 0.615 | 0.001 | 0.807 | 0.014 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| K7V2K4 | Uncharacterized protein | 2.104 | 2.109 | 2.099 | 1.150 | 1.142 | 1.164 | 2.104 | 0.000 | 1.152 | 0.031 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7V838 | Uncharacterized protein | 0.625 | 0.590 | 0.600 | 0.974 | 0.966 | 0.988 | 0.605 | 0.001 | 0.976 | 0.634 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| K7VXC0 | Uncharacterized protein | 1.989 | 1.954 | 1.964 | 0.973 | 0.965 | 0.987 | 1.969 | 0.000 | 0.975 | 0.620 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7W2T3 | Uncharacterized protein (Fragment) | 1.695 | 1.660 | 1.670 | 0.952 | 0.944 | 0.966 | 1.675 | 0.000 | 0.954 | 0.380 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| P00835 | ATP synthase epsilon chain, chloroplastic | 1.747 | 1.712 | 1.722 | 0.930 | 0.922 | 0.944 | 1.727 | 0.000 | 0.932 | 0.219 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| P09138 | Cytochrome c oxidase subunit 3 | 0.478 | 0.443 | 0.453 | 0.939 | 0.931 | 0.953 | 0.458 | 0.000 | 0.941 | 0.274 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| P11647 | NAD(P)H-quinone oxidoreductase chain 4, chloroplastic | 0.650 | 0.615 | 0.625 | 0.914 | 0.906 | 0.928 | 0.630 | 0.001 | 0.916 | 0.146 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| P26566 | 50S ribosomal protein L20, chloroplastic | 0.543 | 0.548 | 0.538 | 0.808 | 0.900 | 0.722 | 0.543 | 0.001 | 0.810 | 0.023 | 0.007 | Down-regulated by drought in an ABA-dependent way |
| P49120 | Histone H2B.4 | 0.685 | 0.650 | 0.660 | 0.842 | 0.834 | 0.856 | 0.665 | 0.000 | 0.844 | 0.006 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| P60138 | Photosystem II reaction center protein L | 2.448 | 2.413 | 2.423 | 0.685 | 0.677 | 0.699 | 2.428 | 0.000 | 0.687 | 0.000 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| Q2XX14 | Non-specific lipid-transfer protein | 2.569 | 2.634 | 2.744 | 1.041 | 1.033 | 1.055 | 2.649 | 0.000 | 1.043 | 0.220 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| Q49HD9 | 12-oxo-phytodienoic acid reductase | 0.634 | 0.599 | 0.609 | 1.344 | 1.336 | 1.358 | 0.614 | 0.000 | 1.346 | 0.000 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| Q8W4W3 | Putative gamma-glutamylcysteine synthetase | 0.604 | 0.569 | 0.579 | 0.913 | 1.105 | 1.027 | 0.584 | 0.000 | 1.015 | 0.823 | 0.002 | Down-regulated by drought in an ABA-dependent way |
| Q9FQB5 | Glutathione S-transferase GST 24 | 1.523 | 1.508 | 1.478 | 1.011 | 1.203 | 0.825 | 1.503 | 0.000 | 1.013 | 0.914 | 0.011 | Up-regulated by drought in an ABA-dependent way |
Each value represents the average of three biological replicates. The average is significant at a p < 0.01 level. A t-test value <0.05 is considered to be significant between ABA-deficient mutant vp5 and wild-type Vp5. D, drought stress.
Table 4.
Proteins with significant expression level changes only in vp5 leaves under drought stress.
| Accession | Description | Vp5: OS/control | vp5: OS/control | Vp5: OS/control | vp5: OS/contro | T-test | Regulation of ABA and drought stress | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | Averagea | P-value | Averagea | P-value | ||||
| B4FCS4 | Uncharacterized protein | 1.083 | 1.079 | 1.105 | 2.250 | 2.264 | 2.266 | 1.089 | 0.202 | 2.260 | 0.000 | 0.000 | Down-regulated by ABA |
| B4FUE3 | Glutamate decarboxylase | 1.111 | 1.107 | 1.133 | 1.554 | 1.547 | 1.561 | 1.117 | 0.115 | 1.554 | 0.001 | 0.000 | Down-regulated by ABA |
| B4G1E6 | Uncharacterized protein | 1.129 | 1.025 | 1.101 | 1.879 | 1.889 | 1.899 | 1.085 | 0.265 | 1.889 | 0.000 | 0.000 | Down-regulated by ABA |
| B4G227 | Uncharacterized protein | 1.104 | 1.100 | 1.126 | 1.634 | 1.637 | 1.650 | 1.110 | 0.132 | 1.640 | 0.000 | 0.000 | Down-regulated by ABA |
| B6SGT3 | Xylanase inhibitor protein 1 | 1.081 | 1.077 | 1.103 | 1.739 | 1.757 | 1.763 | 1.087 | 0.210 | 1.753 | 0.000 | 0.000 | Down-regulated by ABA |
| B6TDN0 | NADH-ubiquinone oxidoreductase 10.5 kDa subunit | 0.974 | 0.970 | 0.996 | 1.577 | 1.576 | 1.598 | 0.980 | 0.749 | 1.584 | 0.001 | 0.000 | Down-regulated by ABA |
| B6TNI6 | DREPP4 protein | 1.302 | 1.218 | 1.284 | 0.575 | 0.585 | 0.595 | 1.268 | 0.013 | 0.585 | 0.002 | 0.000 | Up-regulated by ABA |
| B6TSK6 | Oleoyl-acyl carrier protein thioesterase | 0.973 | 0.969 | 0.995 | 1.640 | 1.653 | 1.661 | 0.979 | 0.737 | 1.651 | 0.000 | 0.000 | Down-regulated by ABA |
| B6TTY1 | Germin-like protein subfamily 1 member 11 | 0.980 | 0.976 | 1.002 | 0.413 | 0.409 | 0.426 | 0.986 | 0.822 | 0.416 | 0.001 | 0.000 | Up-regulated by ABA |
| B6U297 | Lipoxygenase | 1.224 | 1.280 | 1.276 | 0.649 | 0.649 | 0.678 | 1.260 | 0.013 | 0.659 | 0.004 | 0.000 | Up-regulated by ABA |
| C0PF28 | Uncharacterized protein | 1.077 | 1.073 | 1.099 | 0.647 | 0.657 | 0.673 | 1.083 | 0.228 | 0.659 | 0.004 | 0.000 | Up-regulated by ABA |
| C4J6M5 | Uncharacterized protein | 0.871 | 0.867 | 0.893 | 1.901 | 1.892 | 1.912 | 0.877 | 0.004 | 1.902 | 0.000 | 0.000 | Down-regulated by ABA |
| C4JC43 | Putative VHS/GAT domain containing family protein | 0.725 | 0.721 | 0.747 | 0.623 | 0.634 | 0.644 | 0.731 | 0.010 | 0.634 | 0.003 | 0.001 | UP-regulated by ABA |
| E9NMH5 | Glutamic-acid and lysine rich protein | 0.979 | 0.975 | 1.001 | 1.555 | 1.569 | 1.577 | 0.985 | 0.810 | 1.567 | 0.001 | 0.000 | Down-regulated by ABA |
| K7U8Z5 | Uncharacterized protein | 0.948 | 0.944 | 0.970 | 1.485 | 1.475 | 1.515 | 0.954 | 0.474 | 1.492 | 0.001 | 0.000 | Down-regulated by ABA |
| K7V0W5 | Uncharacterized protein | 0.739 | 0.735 | 0.761 | 2.030 | 2.059 | 2.060 | 0.745 | 0.012 | 2.050 | 0.000 | 0.000 | Down-regulated by ABA |
| K7WEH3 | Uncharacterized protein | 1.057 | 1.053 | 1.079 | 0.619 | 0.638 | 0.621 | 1.063 | 0.341 | 0.626 | 0.003 | 0.000 | Up-regulated by ABA |
| Q9FER7 | Putative legumain | 0.820 | 0.816 | 0.842 | 1.569 | 1.579 | 1.580 | 0.826 | 0.041 | 1.576 | 0.001 | 0.000 | Down-regulated by ABA |
| Q9LL87 | Beta-glucosidase aggregating factor | 1.123 | 1.049 | 1.005 | 1.584 | 1.577 | 1.590 | 1.059 | 0.430 | 1.584 | 0.001 | 0.000 | Down-regulated by ABA |
| Q9ZTS9 | Peroxidase J (Fragment) | 0.715 | 0.719 | 0.723 | 0.414 | 0.424 | 0.434 | 0.719 | 0.008 | 0.424 | 0.001 | 0.000 | Up-regulated by ABA |
Each value represents the average of three biological replicates. The average is significant at a p < 0.01 level. A t-test value <0.05 is considered to be significant between ABA-deficient mutant vp5 and wild-type Vp5. D, drought stress.
Among the 123 proteins with 1.5-fold changes of expression level only in Vp5 (Table 3), the expression of 63 proteins was increased in Vp5 under drought conditions, indicating that these proteins were up-regulated by drought stress in an ABA-dependent manner; the expression of 60 proteins was decreased under drought conditions, indicating that these proteins were down-regulated by drought stress in an ABA-dependent manner.
Among the 20 proteins with 1.5-fold changes of expression level only in vp5 (Table 4), drought stress increased the expression of 13 proteins in vp5 but only slightly affected their expression in Vp5, indicating that these proteins were down-regulated by ABA. By contrast, drought stress significantly decreased the expression of seven proteins in vp5 but only slightly affected their expression in Vp5, indicating that these proteins were up-regulated by ABA.
Chloroplast proteins involved in ABA signaling under drought stress
In the present study, 27 chloroplast proteins were found to be related to ABA in maize leaves under drought conditions (Table 5). Among these, three proteins related to the transcriptional process included an uncharacterized protein (B4FJH1) with nucleotide binding function, ribonucleoprotein A (B6T531), and FHA transcription factor (K7UIV2), of which the uncharacterized protein and ribonucleoprotein A were down-regulated by drought in an ABA-dependent manner while the FHA transcription factor was up-regulated by drought in an ABA-dependent manner; two proteins related to protein synthesis included eukaryotic translation initiation factor 3 subunit E (B4FDY7) and 50S ribosomal protein L20 (P26566), which were up- and down-regulated by drought in an ABA-dependent manner, respectively; nine proteins related to the photosynthesis process included uncharacterized proteins (B4FB57 and C0PLS3) with chlorophyll binding function, chlorophyll a-b binding protein 2 (B6STN4), photosystem I reaction center subunit XI (B6SXR2), an uncharacterized protein (K7U2V0) with electron transport function, oxygen evolving enhancer protein 3 (K7UG66), NAD(P)H-quinone oxidoreductase (P11647), photosystem II reaction center protein L (P60138), and ATP synthase epsilon chain (P00835), of which NAD(P)H-quinone oxidoreductase chain 4 was down-regulated by drought stress in an ABA-dependent manner while the other eight proteins were up-regulated by drought stress in an ABA-dependent manner. In addition, there were five enzymes—an uncharacterized protein (B4F9Q3) with glutathione_S-Trfase_N, 3-oxoacyl-[acyl-carrier-protein] synthase (B6TGG7), aldo-keto reductase yakc (B6TYB4), an uncharacterized protein (B8A310) with hydrolase activity, and putative gamma-glutamylcysteine synthetase (Q8W4W3)—and seven proteins with unknown function (B4FBH1, B4FDK8, B4FMW4, B4FPH3, C0HFZ5, K7WEH3, and B4FMW4).
Table 5.
Chloroplast proteins with significant expression level changes in Vp5 or vp5 leaves under drought stress.
| Accession | Description | Vp5: OS/control | vp5: OS/control | Vp5: OS/control | vp5: OS/contro | T-test | Regulation of ABA and drought stress | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | Averagea | P-value | Averagea | P-value | ||||
| B4F9Q3 | Uncharacterized protein | 1.825 | 1.890 | 1.850 | 0.916 | 0.898 | 0.910 | 1.855 | 0.000 | 0.908 | 0.188 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FB57 | Uncharacterized protein | 2.096 | 2.001 | 2.131 | 1.031 | 1.023 | 1.045 | 2.076 | 0.000 | 1.033 | 0.600 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FBH1 | Uncharacterized protein | 0.677 | 0.642 | 0.652 | 1.015 | 1.007 | 1.029 | 0.657 | 0.004 | 1.017 | 0.784 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FDK8 | Uncharacterized protein | 1.593 | 1.558 | 1.568 | 1.057 | 1.049 | 1.071 | 1.573 | 0.001 | 1.059 | 0.367 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FDY7 | Eukaryotic translation initiation factor 3 subunit E | 1.631 | 1.596 | 1.606 | 1.249 | 1.241 | 1.263 | 1.611 | 0.000 | 1.251 | 0.001 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FJH1 | Uncharacterized protein | 0.500 | 0.465 | 0.475 | 1.024 | 1.016 | 1.038 | 0.480 | 0.001 | 1.026 | 0.678 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4FMW4 | Uncharacterized protein | 0.601 | 0.613 | 0.526 | 0.546 | 0.545 | 0.542 | 0.580 | 0.003 | 0.544 | 0.001 | 0.334 | Down-regulated by drought in an ABA-independent way |
| B4FPH3 | Uncharacterized protein | 1.610 | 1.575 | 1.585 | 1.028 | 1.020 | 1.042 | 1.590 | 0.001 | 1.030 | 0.633 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6STN4 | Chlorophyll a-b binding protein 2 | 1.641 | 1.606 | 1.616 | 0.861 | 0.853 | 0.875 | 1.621 | 0.000 | 0.863 | 0.008 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SXR2 | Photosystem I reaction center subunit XI | 1.604 | 1.569 | 1.579 | 0.837 | 0.829 | 0.851 | 1.584 | 0.001 | 0.839 | 0.005 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6T531 | Ribonucleoprotein A | 0.618 | 0.583 | 0.593 | 0.748 | 0.740 | 0.762 | 0.598 | 0.001 | 0.750 | 0.004 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6TGG7 | 3-oxoacyl-[acyl-carrier-protein] synthase | 0.624 | 0.589 | 0.599 | 1.355 | 1.347 | 1.369 | 0.604 | 0.001 | 1.357 | 0.002 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6TYB4 | Aldo-keto reductase yakc | 1.692 | 1.657 | 1.667 | 0.695 | 0.687 | 0.709 | 1.672 | 0.000 | 0.697 | 0.002 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B8A310 | Uncharacterized protein | 1.898 | 2.063 | 1.973 | 1.096 | 1.088 | 1.110 | 1.978 | 0.000 | 1.098 | 0.142 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0HFZ5 | Uncharacterized protein | 1.539 | 1.504 | 1.514 | 0.805 | 0.797 | 0.819 | 1.519 | 0.000 | 0.807 | 0.009 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0PKD1 | Uncharacterized protein | 1.839 | 1.804 | 1.814 | 0.949 | 0.941 | 0.963 | 1.819 | 0.000 | 0.951 | 0.215 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0PLS3 | Uncharacterized protein | 2.105 | 2.070 | 2.080 | 1.088 | 1.080 | 1.102 | 2.085 | 0.000 | 1.090 | 0.126 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7U2V0 | Uncharacterized protein | 1.532 | 1.497 | 1.507 | 0.861 | 0.853 | 0.875 | 1.512 | 0.000 | 0.863 | 0.010 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7UG66 | Oxygen evolving enhancer protein 3 | 1.713 | 1.678 | 1.688 | 0.860 | 0.852 | 0.874 | 1.693 | 0.000 | 0.862 | 0.010 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7UIV2 | FHA transcription factor | 1.879 | 1.844 | 1.854 | 0.904 | 0.896 | 0.918 | 1.859 | 0.001 | 0.906 | 0.114 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| P00835 | ATP synthase epsilon chain, chloroplastic | 1.747 | 1.712 | 1.722 | 0.930 | 0.922 | 0.944 | 1.727 | 0.000 | 0.932 | 0.219 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| P11647 | NAD(P)H-quinone oxidoreductase chain 4, chloroplastic | 0.650 | 0.615 | 0.625 | 0.914 | 0.906 | 0.928 | 0.630 | 0.001 | 0.916 | 0.146 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| P26566 | 50S ribosomal protein L20, chloroplastic | 0.543 | 0.548 | 0.538 | 0.808 | 0.900 | 0.722 | 0.543 | 0.001 | 0.810 | 0.023 | 0.007 | Down-regulated by drought in an ABA-dependent way |
| P60138 | Photosystem II reaction center protein L | 2.448 | 2.413 | 2.423 | 0.685 | 0.677 | 0.699 | 2.428 | 0.000 | 0.687 | 0.000 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| Q8W4W3 | Putative gamma-glutamylcysteine synthetase | 0.604 | 0.569 | 0.579 | 0.913 | 1.105 | 1.027 | 0.584 | 0.000 | 1.015 | 0.823 | 0.002 | Down-regulated by drought in an ABA-dependent way |
| K7WEH3 | Uncharacterized protein | 1.057 | 1.053 | 1.079 | 0.619 | 0.638 | 0.621 | 1.063 | 0.341 | 0.626 | 0.003 | 0.000 | Up-regulated by ABA |
| B4FMW4 | Uncharacterized protein | 0.601 | 0.613 | 0.526 | 0.546 | 0.545 | 0.542 | 0.580 | 0.003 | 0.544 | 0.001 | 0.334 | Down-regulated by drought in an ABA-independent way |
Each value represents the average of three biological replicates. The average is significant at a p < 0.01 level. A t-test value <0.05 is considered to be significant between ABA-deficient mutant vp5 and wild-type Vp5. D, drought stress.
ABA-regulated enzymes under drought stress
In this study, 28 enzymes were found to be implicated in drought and ABA signaling (Table 6). Six enzymes, including glutathione peroxidase (B6SU31), peroxidase (B8A1T1, K7UZ21), an uncharacterized protein (B4F9Q3) with glutathione_S-Trfase_N, glutathione S-transferase GSTU6 (B6TP77), putative gamma-glutamylcysteine synthetase (Q8W4W3), and glutathione S-transferase GST24 (Q9FQB5), were involved in glutathione synthesis and the removal of reactive oxygen species. Of the six enzymes, the expression levels of glutathione peroxidase, two peroxidases, and putative gamma-glutamylcysteine synthetase were down-regulated by drought stress in an ABA-dependent way while an uncharacterized protein, GSTU6, and GST24 were up-regulated by drought stress in an ABA-dependent way.
Table 6.
Enzymes with significant expression level changes in Vp5 or vp5 leaves under drought stress.
| Accession | Description | Vp5: OS/control | vp5: OS/control | Vp5: OS/control | vp5: OS/contro | T-test | Regulation of ABA and drought stress | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | Averagea | P-value | Averagea | P-value | ||||
| B4F9L3 | Carbonic anhydrase | 0.627 | 0.592 | 0.602 | 0.970 | 0.962 | 0.984 | 0.607 | 0.003 | 0.972 | 0.655 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B4F9Q3 | Uncharacterized protein | 1.825 | 1.890 | 1.850 | 0.916 | 0.898 | 0.910 | 1.855 | 0.000 | 0.908 | 0.188 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FB53 | Uncharacterized protein | 1.527 | 1.492 | 1.502 | 1.060 | 1.052 | 1.074 | 1.507 | 0.001 | 1.062 | 0.346 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FUE3 | Glutamate decarboxylase | 1.111 | 1.107 | 1.133 | 1.554 | 1.547 | 1.561 | 1.117 | 0.115 | 1.554 | 0.001 | 0.000 | Down-regulated by ABA |
| B4FWT5 | Soluble inorganic pyrophosphatase | 1.552 | 1.517 | 1.527 | 0.942 | 0.934 | 0.956 | 1.532 | 0.001 | 0.944 | 0.390 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SSB3 | Anthocyanidin 5,3-O-glucosyltransferase | 1.915 | 1.880 | 1.890 | 0.821 | 0.813 | 0.835 | 1.895 | 0.000 | 0.823 | 0.004 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SU31 | Glutathione peroxidase | 0.613 | 0.578 | 0.588 | 0.986 | 0.978 | 1.000 | 0.593 | 0.002 | 0.988 | 0.846 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6SZK3 | NAD(P)H-dependent oxidoreductase | 1.857 | 1.822 | 1.832 | 0.933 | 0.925 | 0.947 | 1.837 | 0.000 | 0.935 | 0.326 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SZL9 | Leucine-rich repeat receptor protein kinase EXS | 1.559 | 1.524 | 1.534 | 1.066 | 1.058 | 1.080 | 1.539 | 0.001 | 1.068 | 0.307 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6T763 | Exosome complex exonuclease RRP41 | 0.376 | 0.341 | 0.351 | 1.079 | 1.071 | 1.093 | 0.356 | 0.000 | 1.081 | 0.222 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6TDN0 | NADH-ubiquinone oxidoreductase 10.5 kDa subunit | 0.974 | 0.970 | 0.996 | 1.577 | 1.576 | 1.598 | 0.980 | 0.749 | 1.584 | 0.001 | 0.000 | Down-regulated by ABA |
| B6TGG7 | 3-oxoacyl-[acyl-carrier-protein] synthase | 0.624 | 0.589 | 0.599 | 1.355 | 1.347 | 1.369 | 0.604 | 0.001 | 1.357 | 0.002 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6TP77 | Glutathione S-transferase GSTU6 | 1.531 | 1.496 | 1.506 | 0.987 | 0.979 | 1.001 | 1.511 | 0.000 | 0.989 | 0.568 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6TSK6 | Oleoyl-acyl carrier protein thioesterase | 0.973 | 0.969 | 0.995 | 1.640 | 1.653 | 1.661 | 0.979 | 0.737 | 1.651 | 0.000 | 0.000 | Down-regulated by ABA |
| B6TYB4 | Aldo-keto reductase yakc | 1.692 | 1.657 | 1.667 | 0.695 | 0.687 | 0.709 | 1.672 | 0.000 | 0.697 | 0.002 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6U297 | Lipoxygenase | 1.224 | 1.280 | 1.276 | 0.649 | 0.649 | 0.678 | 1.260 | 0.013 | 0.659 | 0.004 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6U4B9 | Dual specificity protein phosphatase 4 | 1.605 | 1.620 | 1.680 | 1.059 | 1.051 | 1.073 | 1.635 | 0.000 | 1.061 | 0.376 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B8A1T1 | Peroxidase | 0.666 | 0.631 | 0.641 | 1.055 | 1.047 | 1.069 | 0.646 | 0.001 | 1.057 | 0.417 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0HHB1 | Carboxypeptidase | 2.804 | 2.829 | 2.809 | 0.934 | 1.026 | 0.848 | 2.814 | 0.000 | 0.936 | 0.299 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7UZ21 | Peroxidase | 0.635 | 0.600 | 0.610 | 0.805 | 0.797 | 0.819 | 0.615 | 0.001 | 0.807 | 0.014 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| P00835 | ATP synthase epsilon chain, chloroplastic | 1.747 | 1.712 | 1.722 | 0.930 | 0.922 | 0.944 | 1.727 | 0.000 | 0.932 | 0.219 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| P09138 | Cytochrome c oxidase subunit 3 | 0.478 | 0.443 | 0.453 | 0.939 | 0.931 | 0.953 | 0.458 | 0.000 | 0.941 | 0.274 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| P11647 | NAD(P)H-quinone oxidoreductase chain 4, chloroplastic | 0.650 | 0.615 | 0.625 | 0.914 | 0.906 | 0.928 | 0.630 | 0.001 | 0.916 | 0.146 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| Q49HD9 | 12-oxo-phytodienoic acid reductase | 0.634 | 0.599 | 0.609 | 1.344 | 1.336 | 1.358 | 0.614 | 0.000 | 1.346 | 0.000 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| Q9LL87 | Beta-glucosidase aggregating factor | 1.123 | 1.049 | 1.005 | 1.584 | 1.577 | 1.590 | 1.059 | 0.430 | 1.584 | 0.001 | 0.000 | Down-regulated by ABA |
| Q8W4W3 | Putative gamma-glutamylcysteine synthetase | 0.604 | 0.569 | 0.579 | 0.913 | 1.105 | 1.027 | 0.584 | 0.000 | 1.015 | 0.823 | 0.002 | Down-regulated by drought in an ABA-dependent way |
| Q9FQB5 | Glutathione S-transferase GST 24 | 1.523 | 1.508 | 1.478 | 1.011 | 1.203 | 0.825 | 1.503 | 0.000 | 1.013 | 0.914 | 0.011 | Up-regulated by drought in an ABA-dependent way |
| Q9ZTS9 | Peroxidase J (Fragment) | 0.715 | 0.719 | 0.723 | 0.414 | 0.424 | 0.434 | 0.719 | 0.008 | 0.424 | 0.001 | 0.000 | Down-regulated by drought, but up-regulated by ABA |
Each value represents the average of three biological replicas. The average is significant at a p < 0.01 level. A T-test value <0.05 is considered to be significant between ABA-deficient mutant vp5 and wild type Vp5. D, drought stress.
The expression levels of 12-oxo-phytodienoic acid reductase (Q49HD9, involved in jasmonic acid synthesis) and dual specificity protein phosphatase 4 (B6U4B9) were down- and up-regulated by drought stress in an ABA-dependent way, respectively; the expression levels of NAD(P)H-dependent oxidoreductase (B6SZK3) and aldo-keto reductase yakc (B6TYB4), belonging to the monomeric NADPH-dependent oxidoreductase family, were up-regulated by drought stress in an ABA-dependent manner; the expression levels of mitochondrion cytochrome c oxidase subunit 3 (P09138) and chloroplast NAD(P)H-quinone oxidoreductase chain 4 (P11647) were down-regulated by drought stress in an ABA-dependent manner, indicating that the electron transport of mitochondria and chloroplasts was inhibited under drought conditions; the expression levels of uncharacterized protein (B4FB53) belonging to the ubiquitin-conjugating enzyme family, soluble inorganic pyrophosphatase, leucine-rich repeat receptor protein kinase EXS (B6SZL9), and chloroplast ATP synthase epsilon chain (P00835) were up-regulated by drought stress in an ABA-dependent way.
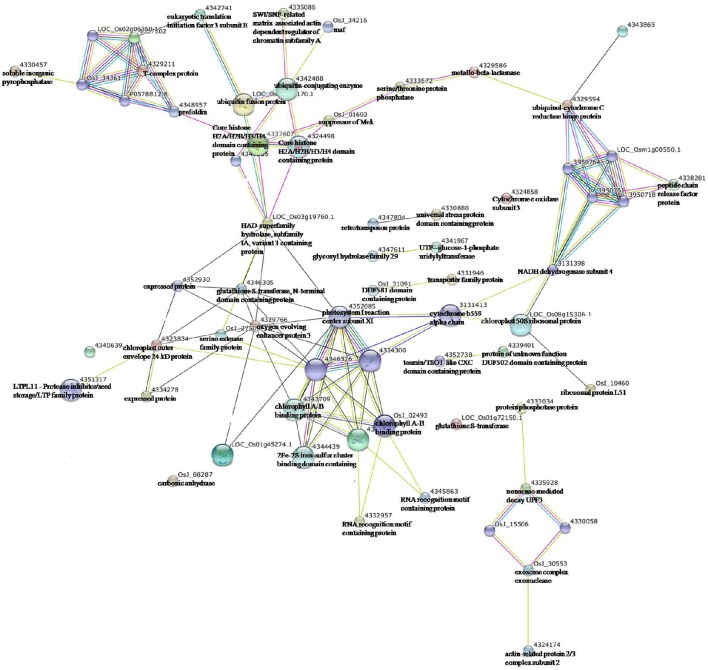
To uncover the interactions among these enzymes, especially protein kinases/phosphatases, with other proteins exhibiting significantly changed expression levels under drought stress, protein-protein interaction analysis was conducted using STRING software (Figure 7). In the network of interactions, five enzymes were found to play core roles in the response of maize leaves to drought stress, which included ubiquitin-conjugating enzyme (4342488), serine/threonine protein phosphatase (4333572), HAD-superfamily hydrolase (LOC_Os03g19760.1), NADH dehydrogenase subunit 4 (3131398), and glutathione S-transferase (4346305).
Figure 7.
Protein-protein interaction network analysis among the significantly expressed proteins in maize Vp5 and vp5 leaves under drought stress using String software.
Ubiquitin-conjugating enzyme (4342488) had interactions with core histone H2A/H2B/H3/H4 domain containing protein (4324498), SWI/SNF-related matrix-associated actin-dependent regulator (4335086) of chromatin subfamily A, and ubiquitin fusion protein (LOC_Os03g13170.1), which further interacted with core histone H2A/H2B/H3/H4 domain containing protein, eukaryotic translation initiation factor 3 subunit E (4342741), and maf (OsJ_34216). Serine/threonine protein phosphatase (4333572) had interactions with suppressor of Mek (OsJ_01603) and metallo-beta-lactamase (4329586), while they further interacted with core histone H2A/H2B/H3/H4 domain containing protein (4324498) and ubiquinol-cytochrome C reductase hinge protein (4329594), respectively; NADH dehydrogenase subunit 4 (3131398) had interactions with cytochrome b559 alpha chain (3131413) and chloroplast 50S ribosomal protein L20 (LOC_Os08g15306.1), while they further interacted with photosystem I reaction center subunit XI (4352085) and ribosomal protein L51 (OsI_10460), respectively.
HAD-superfamily hydrolase (LOC_Os03g19760.1) had interactions with core histone H2A/H2B/H3/H4 domain containing protein (4324498), expressed protein (4352930), glutathione S-transferase (4346305), photosystem I reaction center subunit XI (4352085), and oxygen evolving enhancer protein 3 (4329766), while glutathione S-transferase also had interactions with these proteins. In addition, exosome complex exonuclease (OsI_30553) had an interaction with actin-related protein 2/3 complex subunit 2 (4324174). The results indicated that the five enzymes might have an active role in protein defense/degradation and photosynthesis protection under drought stress (maize protein query sequences matched to rice protein query sequences; see Supplementary Table S2).
Proteins related to stimuli response under drought stress
Among 150 differentially expressed proteins, 21 proteins were characterized as “response to stimulus” (Figures 5, 6A, Table 7). Among the 21 proteins, the expression levels of three proteins (B4FBV4, B4FBY1, and C0HF37) belonging to the small heat shock protein (sHSP) family were up-regulated by drought stress in an ABA-dependent manner; two peroxidases (B8A1T1, K7UZ21) were down-regulated by drought in an ABA-dependent way, and peroxidase (Q9ZTS9) was down-regulated by ABA; DHN2-like protein (B7U627) and an uncharacterized protein (B4G1H1) of the dehydrin family, FHA transcription factor (K7UIV2) and DREPP4 protein (B6TNI6) were up-regulated by drought stress in an ABA-dependent manner. In particular, the expression of two uncharacterized proteins (C0PLS3 with ADP binding function, K7TI96 with nucleotide binding function) had 4.53- and 5.23-fold changes in Vp5 expression, while there was no obvious difference in expression level in vp5 under drought stress conditions.
Table 7.
Proteins related to stimuli response under drought stress.
| Accession | Description | Vp5: OS/control | vp5: OS/control | Vp5: OS/control | vp5: OS/contro | T-test | Regulation of ABA and drought stress | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | Averagea | P-value | Averagea | P-value | ||||
| B4FBV4 | Chaperone protein dnaJ 10 isoform 1 | 1.705 | 1.793 | 1.740 | 0.934 | 0.926 | 0.948 | 1.746 | 0.000 | 0.936 | 0.332 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FBY1 | Chaperone protein dnaJ | 1.646 | 1.611 | 1.621 | 0.956 | 0.928 | 0.930 | 1.626 | 0.000 | 0.938 | 0.349 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4FFW5 | Uncharacterized protein | 1.823 | 1.848 | 1.828 | 0.949 | 1.041 | 0.863 | 1.833 | 0.000 | 0.951 | 0.561 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B4G1H1 | Uncharacterized protein | 1.599 | 1.564 | 1.574 | 1.295 | 1.287 | 1.309 | 1.579 | 0.001 | 1.297 | 0.007 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6STN4 | Chlorophyll a-b binding protein 2 | 1.641 | 1.606 | 1.616 | 0.861 | 0.853 | 0.875 | 1.621 | 0.000 | 0.863 | 0.008 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6SU31 | Glutathione peroxidase | 0.613 | 0.578 | 0.588 | 0.986 | 0.978 | 1.000 | 0.593 | 0.002 | 0.988 | 0.846 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B6T531 | Ribonucleoprotein A | 0.618 | 0.583 | 0.593 | 0.748 | 0.740 | 0.762 | 0.598 | 0.001 | 0.750 | 0.004 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| B7U627 | DHN2-like protein | 1.776 | 1.741 | 1.751 | 1.147 | 1.139 | 1.161 | 1.756 | 0.000 | 1.149 | 0.041 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B8A1T1 | Peroxidase | 0.666 | 0.631 | 0.641 | 1.055 | 1.047 | 1.069 | 0.646 | 0.001 | 1.057 | 0.417 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| C0HF37 | Uncharacterized protein | 1.620 | 1.632 | 1.545 | 0.504 | 0.518 | 0.514 | 1.599 | 0.002 | 0.512 | 0.001 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| C0PLS3 | Uncharacterized protein | 2.105 | 2.070 | 2.080 | 1.088 | 1.080 | 1.102 | 2.085 | 0.000 | 1.090 | 0.126 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7TI96 | Uncharacterized protein | 4.551 | 4.516 | 4.526 | 1.001 | 0.993 | 1.015 | 4.531 | 0.000 | 1.003 | 0.952 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7TKJ3 | Uncharacterized protein | 5.551 | 5.063 | 5.076 | 0.621 | 0.635 | 0.632 | 5.230 | 0.000 | 0.629 | 0.003 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7UIV2 | FHA transcription factor | 1.879 | 1.844 | 1.854 | 0.904 | 0.896 | 0.918 | 1.859 | 0.001 | 0.906 | 0.114 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| K7UZ21 | Peroxidase | 0.635 | 0.600 | 0.610 | 0.805 | 0.797 | 0.819 | 0.615 | 0.001 | 0.807 | 0.014 | 0.000 | Down-regulated by drought in an ABA-dependent way |
| Q8W4W3 | Putative gamma-glutamylcysteine synthetase | 0.604 | 0.569 | 0.579 | 0.913 | 1.105 | 1.027 | 0.584 | 0.000 | 1.015 | 0.823 | 0.002 | Down-regulated by drought in an ABA-dependent way |
| B6TNI6 | DREPP4 protein | 1.302 | 1.218 | 1.284 | 0.575 | 0.585 | 0.595 | 1.268 | 0.013 | 0.585 | 0.002 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| B6U297 | Lipoxygenase | 1.224 | 1.280 | 1.276 | 0.649 | 0.649 | 0.678 | 1.260 | 0.013 | 0.659 | 0.004 | 0.000 | Up-regulated by drought in an ABA-dependent way |
| E9NMH5 | Glutamic-acid and lysine rich protein | 0.979 | 0.975 | 1.001 | 1.555 | 1.569 | 1.577 | 0.985 | 0.810 | 1.567 | 0.001 | 0.000 | Down-regulated by ABA |
| K7WEH3 | Uncharacterized protein | 1.057 | 1.053 | 1.079 | 0.619 | 0.638 | 0.621 | 1.063 | 0.341 | 0.626 | 0.003 | 0.000 | Up-regulated by ABA |
| Q9ZTS9 | Peroxidase J (Fragment) | 0.715 | 0.719 | 0.723 | 0.414 | 0.424 | 0.434 | 0.719 | 0.008 | 0.424 | 0.001 | 0.000 | Down-regulated by drought, but up-regulated by ABA |
Each value represents the average of three biological replicates. The average is significant at a p < 0.01 level. A t-test value <0.05 is considered to be significant between ABA-deficient mutant vp5 and wild-type Vp5. D, drought stress.
Main signaling pathways mediated by ABA under drought stress
Based on the KEGG analysis, signaling pathways in which the differentially expressed proteins were involved for Vp5 (Table 3) and vp5 (Table 4) under drought stress were classified into 48 and 26 categories, respectively (for proteins corresponding to each signaling pathway, see Supplementary Tables S3, S4). For Vp5 (Figure 4), the top three categories with the most proteins were photosynthesis (10), oxidative phosphorylation (4), glutathione metabolism (3), and RNA degradation (3). For vp5 (Figure 5), each of 26 categories had only one protein. Particularly, 10 signal pathways were in common in Vp5 and vp5; 38 were only found in Vp5; 16 were only found in vp5. Among the top 12 categories in Vp5 (Figure 5), signaling pathways such as photosynthesis, glutathione metabolism, RNA degradation, phenylalanine metabolism, phenylpropanoid biosynthesis, ribosome, MAPK signaling pathway, and protein processing in endoplasmic reticulum were found only in Vp5 while oxidative phosphorylation, RNA transport, alcoholism, and systemic lupus erythematosus were also found in vp5.
For Vp5, the top signaling pathway—photosynthesis—included 10 proteins (B6SP61, K7U2V0, B6SXR2, K7UG66, P00835, P60138, B6STN4, B4FB57, P11647, and C0PLS3), of which the expression of B6SP61 and P11647 was down-regulated by drought stress in an ABA-dependent manner while the expression of the other eight proteins was up-regulated by drought stress in an ABA-dependent way; the second signaling pathway—oxidative phosphorylation—included four proteins (P09138, P11647, B4FWT5, and P00835), of which P09138 and P11647 were down-regulated by drought stress in an ABA-dependent manner, and B4FWT5 and P00835 were up-regulated by drought stress in an ABA-dependent way; glutathione metabolism (including three proteins: Q8W4W3, B6SU31, and B6TP77) and RNA degradation (including three proteins: B6T763, B4FPS3, and C0PMV2) were the third top signaling pathways. Among the six proteins, the expression of Q8W4W3, B6SU31, and B6T763 was down-regulated by drought stress in an ABA-dependent manner while that of B6TP77, B4FPS3, and C0PMV2 was up-regulated by drought stress in an ABA-dependent way. These results indicated that drought stress caused major disturbances to crop photosynthesis and antioxidant defense.
Discussion
Due to increased food demand with increasing population and crop yield loss by abnormal environmental changes, increased or stable production of crops under normal or stress conditions is necessary. Therefore, it will be helpful for food security to explore the mechanisms of crop tolerance to stress. ABA is a vital hormone and confers tolerance to abiotic stress (Sah et al., 2016). Although there are some studies concerning proteomic analysis of maize tolerance to drought stress (Benešová et al., 2012; Yang et al., 2014), there are still few studies investigating the roles of ABA in proteomic changes during maize tolerance to drought stress. In this regard, the present study focused on the roles of ABA in drought-induced proteomic changes by comparing the ABA-deficient maize mutant vp5 and its wild-type Vp5.
ABA-mediated chloroplast proteins and photosynthesis under drought stress
It has been proved that photosynthesis of C4 plants is highly sensitive to drought stress. With sustained drought, stomatal conductance, and CO2 assimilation rate decrease rapidly (for a review, see Ghannoum, 2009). Our previous results showed that ABA regulated the phosphorylation of 21 chloroplast proteins under osmotic stress conditions (Hu et al., 2015). In this study, photosynthesis was the top signaling pathway affected by drought stress. Twenty-seven chloroplast proteins were regulated by ABA in maize plants exposed to drought stress, of which 10 proteins were involved in photosynthesis. Among these 10 proteins, drought down-regulated the expression of ferredoxin-1 and NAD(P)H-quinone oxidoreductase chain 4, but up-regulated the expression of photosystem I reaction center subunit XI, oxygen evolving enhancer protein 3, ATP synthase epsilon chain, photosystem II reaction center protein L, chlorophyll a-b binding protein 2, and two uncharacterized proteins in an ABA-dependent way.
In rice, ABA and salt treatment enhanced the expression of oxygen evolving enhancer protein 2 (Abbasi and Komatsu, 2014). In Arabidopsis, drought stress down- and up-regulated the expression of ferredoxin-1 and ferredoxin-2 genes, respectively (Lehtimäki et al., 2010). In maize, drought enhanced the expression of ferredoxin and PsaK (photosystem I reaction center subunit) in the drought-tolerant genotype CE704 but down-regulated their expression in the drought-sensitive 2023 genotype and the expression of PsbP (23 kDa extrinsic polypeptide of photosystem II) in two genotypes (Benešová et al., 2012). These results indicate that the expression patterns of chloroplast proteins related to photosynthesis are complex under drought conditions and highly dependent on the species and genotypes of plants, and the length and severity of stress.
Of the 27 chloroplast proteins, five proteins were involved in synthesis of chloroplast proteins, including eukaryotic translation initiation factor 3 subunit E and FHA transcription factor (up-regulated by drought in an ABA-dependent way), and uncharacterized protein, ribonucleoprotein A, and 50S ribosomal protein L20 (down-regulated by drought in an ABA-dependent way). In Arabidopsis, over-expressing wheat eukaryotic translation initiation factor 3 subunit G enhanced the survival rate and photosynthetic efficiency of plants under drought stress (Singh et al., 2013). In maize, drought down-regulated the expression of ribonucleoprotein A in the drought-tolerant CE704 and drought-sensitive 2023 genotypes, but down-regulated the expression of ribosomal protein S18 in CE704 and up-regulated its expression in the 2023 genotype (Benešová et al., 2012). These results indicate that drought stress disturbs chloroplast protein synthesis, and eukaryotic translation initiation factor 3 could play an active role in enhancing the tolerance of crops to drought stress.
ABA involved in drought-induced protein homeostasis and degradation
In order to gain functional activity, it is necessary for most proteins to fold into fine three-dimensional structures. However, under stress conditions, many proteins are at great risk of aberrant folding and aggregation. In order to avoid these hazards, cells need to depend on a complex network of molecular chaperones to prevent aggregation and promote efficient folding (Hartl et al., 2011; Kim et al., 2013). Recent advances showed that drought increased the expression levels of several sHSPs in maize leaves but decreased their expression in maize kernels (Benešová et al., 2012; Yang et al., 2014). Our previous study indicated that ABA regulated the changes in phosphorylation level of several sHSPs in maize leaves under drought stress (Hu et al., 2015). In the present study, drought stress enhanced the expression of three sHSPs (C0HF37, B4FBV4, and B4FBY1) but decreased the expression of HSP70 (B6SZ69) in an ABA-dependent manner. Taken together, these results show that HSPs as chaperons play extensive roles in maize endurance to drought stress.
Chaperone T-complex protein 1 has been extensively studied in humans and animals (Sinha et al., 2014; Wu et al., 2015), but the function of chaperone T-complex protein 1 remains poorly understood in plants. In this study, drought significantly elevated the expression of T-complex protein 1 subunits zeta (B6U118) and delta (C0HGT5) in an ABA-dependent way. This study therefore extends our understanding of T-complex protein 1 function in plants and is also the first research to our knowledge identifying the role of T-complex protein 1 in the maize response to drought stress.
The ubiquitin-dependent proteolytic pathway degrades most proteins and is the primary proteolysis mechanism in eukaryotic cells. Ubiquitination involves the successive action of at least three enzymes: ubiquitin-activating enzyme, ubiquitin-conjugating enzyme, and ubiquitin ligase (Zhang et al., 2015). In the current study, ubiquitin-conjugating enzyme (B4FB53) was obviously increased by drought stress in an ABA-dependent manner and had interactions with six proteins under drought conditions, including ubiquitin fusion protein (B6SHW9). In addition, drought increased the expression levels of carboxypeptidase (C0HHB1) and an uncharacterized protein (B4FEE1) with serine-type endopeptidase activity. In amaranth leaves, the content of ubiquitin conjugating enzyme was also enhanced under drought stress (Huerta-Ocampo et al., 2009). Our previous study indicated that the phosphorylation levels of eight ubiquitin proteins were significantly changed in maize leaves under drought stress (Hu et al., 2015). Taken together, these results suggest a higher rate of unnecessary or damaged protein degradation under drought conditions.
Antioxidative proteins regulated by ABA under drought stress
Oxidative stress refers to the imbalance of reactive oxygen species removal and the antioxidant defense system, which can result in the oxidative damage of many macromolecules and an excessive reduction of electron transport chain components in chloroplasts, mitochondria, and various detoxification processes (Cruz de Carvalho, 2008). Therefore, it is necessary for plants to start fine regulation mechanisms to remove reactive oxygen species under stress conditions. In the present study, nine proteins involved in processes related to redox homeostasis were differentially regulated: three isoforms of glutathione S-transferase, glutathione peroxidase, NADH-ubiquinone oxidoreductase 10.5 kDa subunit, NAD(P)H-quinone oxidoreductase chain 4, and three isoforms of peroxidase (Table 6).
Glutathione S-transferase can reduce H2O2 to its corresponding hydroxyl compounds, which may remediate oxidative damage of membranes. Glutathione peroxidase can active with and remove H2O2. In this study, drought stress increased the expression levels of three glutathione S-transferase isoforms but decreased the expression levels of glutathione peroxidase and three peroxidase isoforms in an ABA-dependent manner. The protein-protein interaction network analysis indicated that glutathione S-transferase had interactions with six proteins, suggesting its important role in antioxidative defense. Other studies indicated that the expression levels of glutathione S-transferase and glutathione peroxidase were scarcely affected in drought-tolerant maize genotypes but were significantly increased in drought-sensitive maize genotypes under drought conditions (Benešová et al., 2012; Yang et al., 2014). Taken together, these results show that the cooperative relations among the antioxidative proteins play an important role in redox homeostasis under drought conditions.
Dehydrins and xylanase inhibitor proteins act as cell rescue/defense-related proteins. Dehydrins play important roles in protecting the stability of membrane proteins, adjusting cell osmotic pressure, and stabilizing and preventing the denaturation of macromolecules (Close, 1996; Mohammadkhani and Heidari, 2008). Xylanase inhibitor proteins are believed to play a role in plant defensive reactions (Dornez et al., 2010; Vasconcelos et al., 2011). A previous study indicated that the expression levels of dehydrin and xylanase inhibitor protein 1 were barely affected in the drought-tolerant maize line Lo964 but were significantly increased in the drought-sensitive line B73 under drought conditions (Yang et al., 2014). In the maize drought-tolerant CE704 and drought-sensitive 2023 genotypes, the expression of dehydrin RAB-17 was increased under drought stress, but the increase was greater in CE704 than in the 2023 genotype. However, drought stress increased the expression of xylanase inhibitor (TAXI-IV) in the drought-tolerant CE704 genotype but decreased its expression in the drought-sensitive 2023 genotype (Benešová et al., 2012). In the current study, two isoforms of dehydrin were up-regulated by drought in an ABA-dependent manner; xylanase inhibitor protein 1 was up-regulated by drought but down-regulated by ABA. These results indicate that the regulation of drought stress by dehydrin and xylanase inhibitor protein depends on the plant species, genotype, and the severity/length of drought stress.
Overall, the application of the maize ABA-deficient mutant vp5 and its wild-type Vp5 was very useful to identify drought-response proteins involved in ABA signaling pathways. Among the 150 proteins identified, 67 proteins were up-regulated by drought in an ABA-dependent way; 60 proteins were down-regulated by drought in an ABA-dependent way. Under drought stress, the top three signaling pathways affected were photosynthesis, oxidative phosphorylation (mainly chloroplast-mediated ATP synthesis), and glutathione metabolism, indicating that ABA plays an important role in regulating photosynthesis, ATP synthesis, and antioxidative reactions in maize under drought stress. In addition, stimuli-response proteins (such as chaperone proteins and dehydrins), antioxidative proteins (such as glutathione S-transferase), and proteins for protein degradation (such as ubiquitin-conjugating enzyme, carboxypeptidase and endopeptidase) might have more effective roles in maize tolerance to drought stress. The present study provides a basis for further understanding the mechanisms and roles of ABA in maize responses to drought stress.
Author contributions
XH conceived the study and participated in its design. YZ and YW carried out the experiments. JW contributed samples. WW revised the manuscript. XH and HY analyzed the data and drafted the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Program for Scientific Innovation Talent for Henan Province (Grant 154100510005 to XH), Science and Technology Innovation Talents (13HASTIT001 to XH), and the National Natural Science Foundation of China (Grant 31171470 to XH).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01827/full#supplementary-material
Primers used in this study.
Maize proteins corresponding to rice proteins in the network of protein interaction in maize Vp5 seedlings under drought stress.
Map name of pathways induced by drought stress in maize Vp5.
Map name of pathways induced by drought stress in maize vp5.
Indentified peptides for the first independent biological experiment.
Indentified peptides for the second independent biological experiment.
Indentified peptides for the third independent biological experiment.
References
- Abbasi F. M., Komatsu S. (2014). A proteomic approach to analyze salt-responsive proteins in rice leaf sheath. Proteomics 4, 2072–2081. 10.1002/pmic.200300741 [DOI] [PubMed] [Google Scholar]
- Benešová M., Holá D., Fischer L., Jedelský P. L., Hnilička F., Wilhelmová N., et al. (2012). The physiology and proteomics of drought tolerance in maize: early stomatal closure as a cause of lower tolerance to short-term dehydration? PLoS ONE 7:e38017. 10.1371/journal.pone.0038017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R. S., Pan S., Zhao R., Gazzarrini S. (2016). ABA-dependent inhibition of the ubiquitin proteasome system during germination at high temperature in Arabidopsis. Plant J. 10.1111/tpj.13293. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Close T. J. (1996). Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plant. 97, 795–803. 10.1111/j.1399-3054.1996.tb00546.x [DOI] [Google Scholar]
- Cruz de Carvalho M. H. (2008). Drought stress and reactive oxygen species: production, scavenging and signaling. Plant Signal. Behav. 3, 156–165. 10.4161/psb.3.3.5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daryanto S., Wang L., Jacinthe P. A. (2016). Global synthesis of drought effects on maize and wheat production. PLoS ONE 11:e0156362. 10.1371/journal.pone.0156362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornez E., Croes E., Gebruers K., Carpentier S., Swennen R., Laukens K., et al. (2010). 2-D DIGE reveals changes in wheat xylanase inhibitor protein families due to Fusarium graminearum DeltaTri5 infection and grain development. Proteomics 10, 2303–2319. 10.1002/pmic.200900493 [DOI] [PubMed] [Google Scholar]
- Fan W. Q., Zhao M. Y., Li S. L., Bai X., Li J., Meng H., et al. (2016). Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biol. 16:99. 10.1186/s12870-016-0764-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2008). Climate Change and Food Security: A Framework Document. Rome: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Ghannoum O. (2009). C4 photosynthesis and water stress. Ann. Bot. 103, 635–644. 10.1093/aob/mcn093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F., Yang L., Tai F., Hu X., Wang W. (2014). “Omics” of maize stress response for sustainable food production: opportunities and challenges. OMICS 18, 714–732. 10.1089/omi.2014.0125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hable W. E., Oishi K. K., Schumaker K. S. (1998). Viviparous-5 encodes phytoene desaturase, an enzyme essential for abscisic acid (ABA) accumulation and seed development in maize. Mol. Gen. Genet. 257, 167–176. 10.1007/s004380050636 [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Bracher A., Hayer-Hartl M. (2011). Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Hodges D. M., DeLong J. M., Forney C. F., Prange R. K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. 10.1007/s004250050524 [DOI] [PubMed] [Google Scholar]
- Hu X. L., Li N. N., Wu L. J., Li C. Q., Li C. H., Zhang L., et al. (2015). Quantitative iTRAQ-based proteomic analysis of phosphoproteins and ABA-regulated phosphoproteins in maize leaves under osmotic stress. Sci. Rep. 5:15626. 10.1038/srep15626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X. L., Li Y. H., Li C. H., Yang H. R., Wang W., Lu M. H. (2010). Characterization of small heat shock proteins associated with maize tolerance to combined drought and heat stress. J. Plant Growth Regul. 29, 455 10.1007/s00344-010-9157-9 [DOI] [Google Scholar]
- Hu X., Wu X., Li C., Lu M., Liu T., Wang W., et al. (2012). Abscisic acid refines the synthesis of chloroplast proteins in maize (Zea mays) in response to drought and light. PLoS ONE 7:e49500 10.1371/journal.pone.0049500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Ocampo J. A., Briones-Cerecero E. P., Mendoza-Hernandez G., De Leon-Rodriguez A., de la Rosa A. P. B. (2009). Proteomic analysis of Amaranth (Amaranthus Hypochondriacus L.) leaves under drought stress. Int. J. Plant Sci. 170, 990–998. 10.1086/605119 [DOI] [Google Scholar]
- Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., Hartl F. U. (2013). Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355. 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- Lehtimäki N., Lintala M., Allahverdiyeva Y., Aro E. M., Mulo P. (2010). Drought stress-induced upregulation of components involved in ferredoxin-dependent cyclic electron transfer. J. Plant Physiol. 167, 1018–1022. 10.1016/j.jplph.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Mohammadkhani N., Heidari R. (2008). Effects of drought stress on soluble proteins in two maize varieties. Turk. J. Biol. 32, 23–30. [Google Scholar]
- Robichaud C. S., Wang J., Sussex I. M. (1980). Control of in vitro growth of viviparous embryo mutants of maize by abscisic acid. Dev. Genet. 1, 325–330. 10.1002/dvg.1020010405 [DOI] [Google Scholar]
- Roychoudhury A., Paul S., Basu S. (2013). Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 32, 985–1006. 10.1007/s00299-013-1414-5 [DOI] [PubMed] [Google Scholar]
- Sah S. K., Reddy K. R., Li J. X. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. 10.3389/fpls.2016.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Chauhan H., Khurana J. P., Khurana P., Singh P. (2013). Evidence for the role of wheat eukaryotic translation initiation factor 3 subunit g (TaeIF3g) in abiotic stress tolerance. Gene 532, 177–185. 10.1016/j.gene.2013.09.078 [DOI] [PubMed] [Google Scholar]
- Sinha S., Belcastro M., Datta P., Seo S., Sokolov M. (2014). Essential role of the chaperonin CCT in rod outer segment biogenesis. Invest. Ophthalmol. Vis. Sci. 55, 3775–3785. 10.1167/iovs.14-13889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa D., Watanabe N., Ghosh T. K., Saruhashi M., Suzuki A., Ishiyama K., et al. (2015). Epoxycarotenoid-mediated synthesis of abscisic acid in Physcomitrella patens implicating conserved mechanisms for acclimation to hyperosmosis in embryophytes. New Phytol. 206, 209–219. 10.1111/nph.13231 [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Takahashi F., Anderson J. C., Ishihama Y., Peck S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6:rs8. 10.1126/scisignal.2003509 [DOI] [PubMed] [Google Scholar]
- Vasconcelos E. A., Santana C. G., Godoy C. V., Seixas C. D., Silva M. S., Moreira L. R., et al. (2011). A new chitinase-like xylanase inhibitor protein (XIP) from coffee (Coffea arabica) affects Soybean Asian rust (Phakopsora pachyrhizi) spore germination. BMC Biotechnol. 11:14. 10.1186/1472-6750-11-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. C., Xue L., Batelli G., Lee S. Y., Hou Y. J., VanOosten M. J., et al. (2013). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U.S.A. 110, 11205–11210. 10.1073/pnas.1308974110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Z., Chang L. C., Lin Y. F., Hung Y. J., Pei D. (2015). Chaperonin-containing t-complex protein-1 subunit β as a possible biomarker for the phase of glomerular hyperfiltration of diabetic nephropathy. Dis. Markers 2015:548101. 10.1155/2015/548101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Jiang T., Fountain J. C., Scully B. T., Lee R. D., Kemerait R. C., et al. (2014). Protein profiles reveal diverse responsive signaling pathways in kernels of two maize inbred lines with contrasting drought sensitivity. Int. J. Mol. Sci. 5, 18892–18918. 10.3390/ijms151018892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Mogami J., Yamaguchi-Shinozaki K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. 10.1016/j.pbi.2014.07.009 [DOI] [PubMed] [Google Scholar]
- Zhang Z. Y., Li J. H., Liu H. H., Chong K., Xu Y. Y. (2015). Roles of ubiquitination-mediated protein degradation in plant responses to abiotic stresses. Environ. Exp. Bot. 114, 92–103. 10.1016/j.envexpbot.2014.07.005 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study.
Maize proteins corresponding to rice proteins in the network of protein interaction in maize Vp5 seedlings under drought stress.
Map name of pathways induced by drought stress in maize Vp5.
Map name of pathways induced by drought stress in maize vp5.
Indentified peptides for the first independent biological experiment.
Indentified peptides for the second independent biological experiment.
Indentified peptides for the third independent biological experiment.