Abstract
Tumors have exceptionally high demands for energy and anabolism because of their rapid growth. The de novo serine synthesis pathway initiated by phosphoglycerate dehydrogenase (PHGDH) has been recognized as a hallmark of metabolic adaption in carcinogenesis. The oncogenic role and prognostic value of PHGDH have been investigated in multiple cancer types, including breast cancer, melanoma, cervical cancer, and colon cancer. Due to the importance of PHGDH in cancer, we attempted to determine the clinical significance of PHGDH in 319 patients with non–small cell lung cancer (NSCLC). We evaluated the mRNA and protein expression levels of PHGDH gene, using quantitative reverse transcriptase polymerase chain reaction and tissue array–based immunohistochemistry, respectively. Significantly increased PHGDH expression in mRNA and protein levels was identified in tumor tissues versus matched adjacent nontumor tissues. More interestingly, immunohistochemical expression of PHGDH was significantly associated with lymph node metastasis (P = .021) and TNM stage (P = .016). Kaplan-Meier survival analysis indicated that NSCLC patients with low levels of PHGDH outperformed patients with high levels of PHGDH regarding 5-year overall survival. Significantly longer survival in the former suggested the prognostic implication of PHGDH in NSCLC. Multivariate survival analysis using Cox regression model demonstrated that high PHGDH levels and advanced TNM stage (III + IV) were independent predictors of prognosis in NSCLC. Moreover, bioinformatics analysis confirmed the increase in PHGDH transcripts (data from The Cancer Genome Atlas) and its prognostic value (Kaplan-Meier plotter) in NSCLC. In conclusion, this study suggested the clinical implication of PHGDH in NSCLC. PHGDH may be a promising therapeutic target in NSCLC.
Introduction
Some metabolic alterations occur during carcinogenesis, which facilitate transformed cells to continuously grow and proliferate [1], [2], [3]. Tumor cells metabolize glucoses differently from normal cells. Most tumor cells tend to generate energy through aerobic glycolysis to utilize glucose mainly through a fermentative metabolism, generating lactate products even when exposed to abundant oxygen (aerobic glycolysis) [4], [5], [6], [7]. The tendency toward increased aerobic glycolysis in tumor cells is known as the Warburg effect [5]. In the normal cells, glycolysis usually yields pyruvate, alanine, and lactase. However, in tumor cells, the final step in glycolysis catalyzed by pyruvate kinase is blocked during the aerobic glycolysis. Alternatively, glycolytic metabolites upstream of pyruvate kinase are rechanneled into other metabolic pathways in malignant cells [8]. The de novo serine synthesis pathway is one of most remarkable pathways branching from routine glycolysis. Resulting serines and glycines serve as intermediates for the production of amino acids other than alanine, lipids, and nucleic acids, which in turn support rapid expansion of biomass [3], [4], [8]. Phosphoglycerate dehydrogenase (PHGDH) plays a critical role in redirecting the glycolytic intermediate, 3-phosphoglycerate (3PG), to serine synthesis pathway. PHGDH oxidizes 3PG into phosphohydroxypyruvate, which is a rate-limiting step in the process of the generation of serine from 3PG. Phosphohydroxypyruvate can be subsequently converted into serine, which is further processed to form glycine by losing a carbon. Due to increased aerobic glycolysis in most tumors, PHGDH has recently emerged as a potential therapeutic target and putative metabolic oncogene.
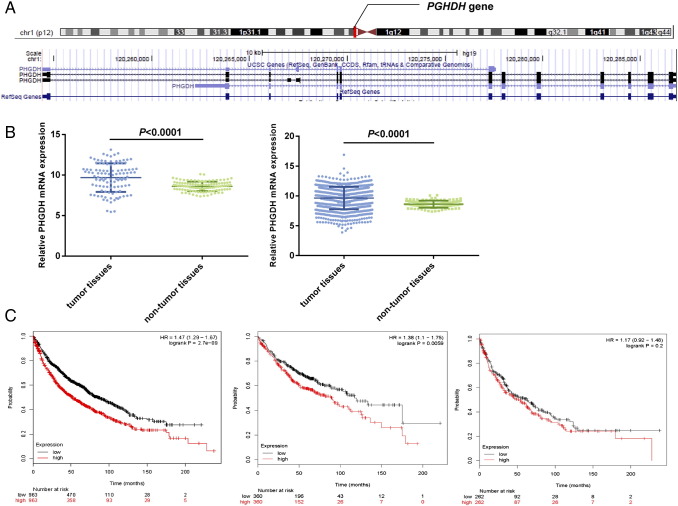
The PHGDH gene located on chromosome 1p12 contains 11 exons (Figure 1). Upregulated biosynthesis of serine and glycine was first noticed in lymphomas back in the 1950s [9]. Over the past few years, a similar phenomenon has also been found in breast cancer [10], [11] and melanoma [8] PHGDH is recurrently amplified in melanoma in which the amplification often leads to elevated protein expression. Overexpression of PHGDH was also observed in the certain subtypes of breast cancer. PHGDH overexpression predisposed mammary epithelial cell line (MCF-10a) to malignant transformation [8]. Inversely, knockdown of PHGDH by RNA interference inhibited the growth of PHGDH-amplified cell lines, as well as serine synthesis pathway [8].
Figure 1.
Bioinformatics analysis. (A) PHGDH gene. (B) TCGA data indicated a significantly higher PHGDH mRNA expression in tumor tissue than in normal tissues. (Left panel) 108 pairs of tumor and lung tissues (paired t test: P < .0001); (right panel) 108 tumor tissues versus 1014 normal tissues (Student's t test: P < .0001). (C) (left panel) Overall, the KM-plotter showed that patients with PHGDHhigh tumors had significantly shorter survival than those with PHGDHlow tumors (n = 1926, log-rank test: P < .001). (Middle panel) PHGDHhigh was significantly associated with reduced survival in adenocarcinoma (n = 720, HR = 1.38, 95% CI = 1.1-1.75; log-rank test: P = .006); (right panel) no significant difference in survival was observed in squamous cell carcinoma (n = 524, HR = 1.17, 95% CI = 0.92-1.48; log-rank test: P = .2).
Nonetheless, the role of PHGDH in lung cancer remains unclarified. Lung cancer is notorious for its high incidence and low 5-year survival rate. According to GLOBOCAN 2012 [12], lung cancer is the most commonly diagnosed cancer in men worldwide, with the highest lung cancer incidence rates in Europe, Eastern Asia, and Northern America. Eastern Asia also has the highest lung cancer incidence for women. Given the high prevalence of lung cancer in Eastern Asia and its poor prognosis, it is imperative to discover druggable molecular targets and reliable prognostic biomarkers to improve the management of non–small cell lung cancer (NSCLC) patients. In this study, we attempted to compare the expression of PHGDH in NSCLC and in adjacent normal tissues, and evaluate its clinicopathological association and prognostic implications.
Materials and Methods
Patients and Tissue Samples
We consecutively recruited a group of patients (n = 319) with pathologically confirmed NSCLC for this study. A total of 319 NSCLC samples and 143 matched pertumor specimens were obtained from patients immediately after surgical resection at the Affiliated Hospital of Nantong University, Jiangsu, China, from 2004 to 2009. All tissues were formalin fixed and paraffin embedded and archived in the Department of Pathology until further utilization. Median age at onset of NSCLC was 62.9 years, with ages varying from 35 to 83 years. None of patients underwent chemotherapy or radiotherapy prior to the surgical resection. Clinical information was retrieved from the archives at the hospital, including patient sex, age, smoking status, tumor size, tumor differentiation, histological type, tumor status (T), lymph node metastasis (N), distant metastasis (M), and TNM stage. Follow-up information was available on these cases. The last follow-up was performed on May 30, 2013. NSCLC was strictly staged according to the guidelines of the seventh edition of TNM staging in lung cancer [13]. All these patients signed informed consent before surgery, and this investigation was approved by the Research Ethics Committee of the Affiliated Hospital of Nantong University.
Tissue Microarray Construction and Immunohistochemistry (IHC)
A representative tissue core (2 mm in diameter) was picked out from each of the studied formalin-fixed, paraffin-embedded tissue blocks, and these selected tissue cores were assembled in array fashion using a tissue arraying instrument (Quick-Ray, UT06; UNITMA, Korea). We cut 4-μm histological sections from each of the tissue microarray specimens and have them adhered to the super frost-charged glass microscope slides. IHC analysis was carried out as described elsewhere [14]. The slides were deparaffinized and pretreated with citric acid (10 mmol/l, pH 6.0) for antigen retrieval using a microwave protocol. Primary PHGDH antibody was purchased from Abcam Inc. (Cambridge, MA). The slides subsequently underwent the standard procedure including the blockage of endogenous peroxidase, sequential incubation in primary and second antibodies, and diaminobenzidine staining. Hematoxylin was chosen as nuclear counterstain.
Evaluation of IHC Reaction Intensity
The immunostaining of PHGDH was graded on a semiquantitative basis as published previously [14] by integrating intensity and distribution. Intensity fell into categories of 0, 1+, 2+, and 3+, denoting no, weak, moderate, and strong staining, respectively. Distribution of staining was referred to as the percentage of positive tumor cells (0% to 100%). The final PHGDH expression score was obtained by multiplying two variables together, with 0 and 300 corresponding to no staining and 100% of cells with 3+ staining intensity, respectively. All samples were further divided into “low or negative” PHGDH expression (PHGDHlow) and “high or positive” protein expression (PHGDHhigh) groups by a cutoff point. The cutoff point for the PHGDH expression score was computed using the X-tile software program (http://www.tissuearray.org/rimmlab) [15]. In the program, every potential separation of PHGDH expression generated a χ2 value from log-rank χ2 statistics regarding overall survival (OS), with the highest χ2 value representing the optimal cutoff point.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Analysis for PHGDH Transcripts
Fresh NSCLC cancer tissue and matched normal, tumor-adjacent tissue samples (n = 20) were acquired for qRT-PCR analysis from the Department of Pathology at the Affiliated Hospital of Nantong University. It should be noted that samples examined by qRT-PCR were different from those used for IHC. Total RNA extraction and quality control were carried out according to published protocols [16]. Onestep qRT-PCR analysis was performed on an Applied Biosystems 7500 Real-Time PCR System using the LightCycler FastStart DNA Master SYBR Green I Kit (Roche Diagnostics, Tokyo, Japan). The primers for PHGDH (NM_006623) were as follows: forward primer 5′-CACATTCTTGGGCTGAAC-3′ and reverse primer 5′-TTATTAGACGGTTATTGCTGTA-3′. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as a internal control gene for the quantification of PHGDH mRNA levels in tissues (forward primer 5′-TGCACCACCAACTGCTTAGC-3′ and reverse primer 3′-GGCATGGACTGTGGTCATGAG-5′). Reverse transcription was achieved by an incubation for 30 minutes at 42°C. Taq DNA polymerase was activated by a subsequent incubation for 2 minutes at 94°C. Finally, 35 amplification cycles of 95°C for 20 seconds and 56°C for 20 seconds, and elongation at 72°C for 30 seconds were run to generate 82-bp PHGDH PCR products. All tests were performed in triplicate. Double delta Ct analysis was used to analyze qPCR data.
Bioinformatic Analysis
The PHGDH gene expression data were retrieved from The Cancer Genome Atlas (TCGA) database. The level 3 TCGA data (TCGA_LUNG_exp_HiSeqV2–2015–02-24) were obtained at the website of UCSC cancer browser (http://genome-cancer.ucsc.edu). Using an IlluminaHiSeq platform, the data set was collected from 1124 tissue samples consisting of both pathologically diagnosed NSCLC and matched normal tissue. Among them, 108 NSCLC tissues had matched normal tissue samples. The mRNA expression of PHGDH was obtained from the “genomic Matrix” file (using editplus software), and then the whole data set was separated into tumor tissue and normal tissue groups. Comparison of the relative mRNA expression levels between two groups of samples was performed.
Online survival analysis tool, Kaplan-Meier plotter (KM-plotter), was employed to evaluate the prognostic value of PHGDH in NSCLC with the utilization of the transcriptomic data [17]. The KM-plotter contains data of 54,675/22,277 genes from 4142 breast, 1648 ovarian, 2437 lung, and 1065 gastric cancer patients. The mean follow-up time for the 3 types of cancers was 69, 40, 49, and 33 months, respectively.
Statistical analysis
A χ2 test was adopted to analyze the correlation of GHPDH expression with demographic and clinicopathological variables. The Kaplan-Meier survival curves were plotted for subgroups defined by pathology or GHPDH expression, and the log-rank test was performed to check the significance of differences between groups. Univariate and multivariate Cox regression models were used to investigate the effects of tested variables on the survival. Differences were regarded as statistically significant at P < .05. All statistical analyses were performed using SPSS version 20.0 statistical software (SPSS Inc., Chicago, IL).
Results
Differential Expression of PHGDH between Tumor and Matched Normal Tissues
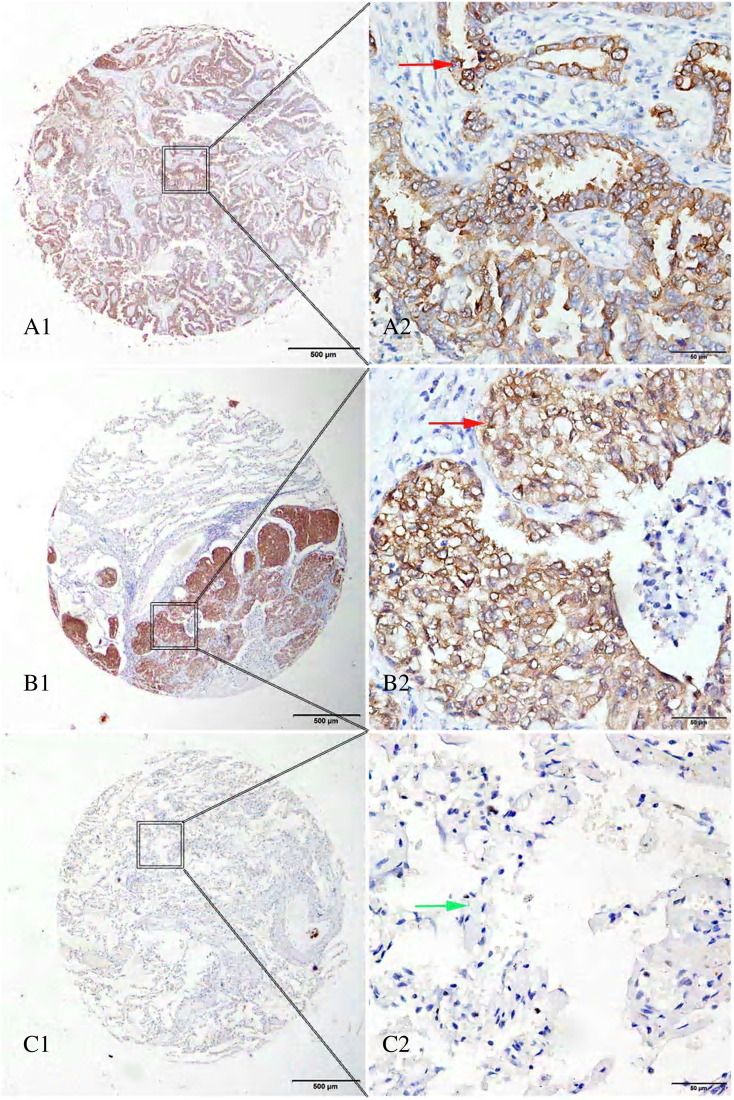
The representative images of PHGDH immunostaining were shown in Figure 2. We found that 55.2% of tumor tissues expressed high levels of PHGDH, whereas only 20% (21/132) of matched adjacent lung tissues had high levels of PHGDH. The PHGDH staining was significantly more prevalent in tumors than in matched adjacent lung tissues (P < .0001). Using optimal cutoff point of 160, patients with NSCLC were separated into PHGDHlow (scores < 160) and PHGDHhigh (scores ≥ 160) groups irrespective of other factors (e.g., age, sex, and TNM).
Figure 2.
Immunohistochemical expression of PHGDH in lung tissue microarray cores. (A1, A2) lung adenocarcinoma, strong cytoplasmic expression (red arrow); (B1, B2) squamous cell carcinoma of the lung, strong positive staining in cytoplasm (red arrow); (C1, C2) normal lung tissue, cytoplasm of alveolar epithelial cells, negative staining (green arrow). Original magnification ×40 (bar = 500 μm) in A1, B1, and C1; ×400 (bar = 50 μm) in A2, B2, and C2.
Association of PHGDH Expression with Clinicopathologic Characteristics in NSCLC
To further dissect the role of PHGDH in NSCLC carcinogenesis, we evaluated its potential correlation with demographic factors (age, sex) and clinicopathologic parameters in patients with NSCLC. High-level expression of PHGDH was positively associated with TNM stage (stage I, 48.00%; stage II, 62.50%; stage III + IV, 66.07%; P = .016, Table 1) but not with histological type, differentiation, and smoking habit. Similarly, the percentage of tumor with high PHGDH expression increased with the severity of lymph node invasion (pN0, 50.00%; pN1, 57.97%; pNII + III, 71.15%, P = .021, Table 1).
Table 1.
Histoclinical Characteristics of NSCLC Patients and PHGDH Expression
| Variables | No. | PHGDH Expression |
|||
|---|---|---|---|---|---|
| Low (%) |
High (%) |
??2 | P Value | ||
| 143 (44.83) | 176 (55.17) | ||||
| T stage | 0.4123 | .814 | |||
| T1 | 141 | 66 (46.81) | 75 (53.19) | ||
| T2 | 160 | 69 (43.13) | 91 (56.88) | ||
| T3+ 4 | 18 | 8 (44.44) | 10 (55.56) | ||
| N stage | 7.730 | .021⁎ | |||
| N0 | 198 | 99 (50.00) | 99 (50.00) | ||
| N1 | 69 | 29 (42.03) | 40 (57.97) | ||
| N2+ 3 | 52 | 15 (28.85) | 37 (71.15) | ||
| TNM stage | 8.2401 | .016⁎ | |||
| I | 175 | 91 (52.00) | 84 (48.00) | ||
| II | 88 | 33 (37.50) | 55 (62.50) | ||
| III + IV | 56 | 19 (33.93) | 37 (66.07) | ||
| Age | 0.0451 | .832 | |||
| Age ≤ 60 | 134 | 61 (45.52) | 73 (54.48) | ||
| Age> 60 | 185 | 82 (44.32) | 103 (55.68) | ||
| Sex | 0.3786 | .538 | |||
| Male | 211 | 92 (43.60) | 119 (56.40) | ||
| Female | 108 | 51 (47.22) | 57 (52.78) | ||
| Histological type | 4.0935 | .129 | |||
| Squamous | 109 | 42 (38.53) | 67 (61.47) | ||
| Adenocarcinoma | 163 | 82 (50.31) | 81 (49.69) | ||
| Other | 47 | 19 (40.43) | 28 (59.57) | ||
| Smoking | 0.6843 | .408 | |||
| Yes | 144 | 70 (48.61) | 74 (51.39) | ||
| No | 95 | 41 (43.16) | 54 (56.84) | ||
| Unknown | 80 | 32 | 48 | ||
| Differentiation | |||||
| Poorly | 68 | 27 (39.71) | 41 (60.29) | 5.3908 | .068 |
| Moderately | 202 | 87 (43.07) | 115 (56.93) | ||
| Well | 26 | 17 (65.38) | 9 (34.62) | ||
| Unknown | 23 | 12 | 11 | ||
P < .05.
Enhanced PHGDH mRNA Expression in NSCLC
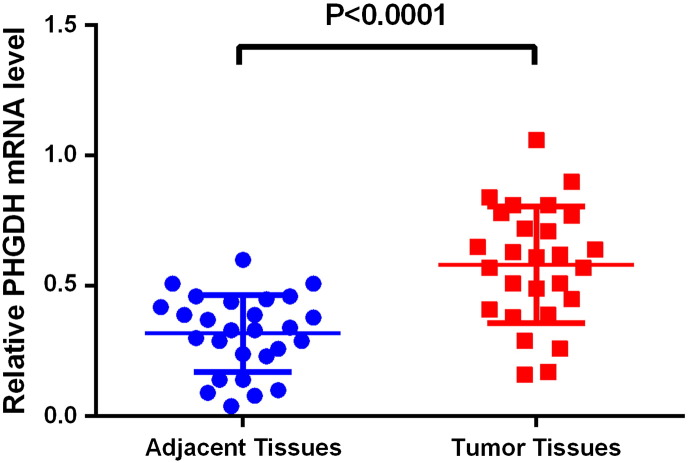
Quantitative RT-PCR analysis was conducted to identify differences in PHGDH mRNA expression between NSCLC and matched adjacent normal tissue samples (n = 20). PHGDH transcripts were significantly increased in NSCLC tissues when compared with adjacent control tissues (P < .0001, Figure 3).
Figure 3.
Significantly higher mRNA expression levels of PHGDH were observed in NSCLC than in tumor adjacent normal tissues (P < .0001). One-step qRT-PCR was used to examine PHGDH mRNA expression levels. Relative mRNA expression of PHGDH was normalized to GAPDH.
We next took advantage of TCGA genomic database to explore the differences in the PHGDH mRNA expression between NSCLCs and lung tissues. Analysis of 108 pairs of tumors and matched lung tissue revealed that the PHGDH mRNA expression level in tumors was significantly unregulated [9.68 ± 1.76 vs. 8.61 ± 0.59, P < .0001, Figure 1B (left panel)]. The increased PHGDH mRNA expression in tumors was further confirmed when we compared 1014 tumor tissues and 108 normal tissues [9.64 ± 1.87 vs. 8.61 ± 0.59, P < .0001, Figure 1B (right panel)].
Prognostic Value of PHGDH Protein Expression in NSCLC
In the univariate Cox regression analysis, several variables showed a major prognostic value with respect to 5-year OS, including PHGDH expression (P = .001), sex (P = .048), differentiation (P = .001), and TNM (Table 2). However, age, histology type, and smoking habit were not prognostic for clinical outcome (Table 2). Particularly, PHGDH adversely influenced OS in NSCLC, and patients with PHGDHhigh tumors showed an inferior 5-year OS [hazard ratio (HR) = 3.004, 95% confidence interval (CI) = 2.168-4.163, P = .001]. This observed survival disadvantage in NSCLCs with PHGDHhigh was maintained in the multivariate analysis (HR = 2.997, 95% CI = 2.105-4.267, P < .001, Table 2), suggesting that PHGDH is an independent predictor of OS in NSCLC.
Table 2.
Univariate and Multivariate Analysis of Prognostic Factors in NSCLC for 5-Year OS
| Variable | Univariate Analysis |
Multivariate Analysis |
||||
|---|---|---|---|---|---|---|
| HR | P Value | 95% CI | HR | P Value | 95% CI | |
| Expression of PHGDH | ||||||
| High vs low | 3.004 | .001⁎ | 2.168-4.163 | 2.997 | <.0001⁎ | 2.105-4.267 |
| Sex | ||||||
| Male vs female | 0.721 | .048⁎ | 0.522-0.996 | |||
| Age (years) | ||||||
| ≤ 60 vs >60 | 1.257 | .136 | 0.930-1.700 | |||
| Differentiation | ||||||
| Poorly vs moderately vs well | 0.631 | .001⁎ | 0.482-0.826 | 0.840 | .335 | 0.590-1.196 |
| Pathologic type | ||||||
| Squamous vs adenocarcinoma vs other | 1.016 | .081 | 0.808-1.278 | |||
| Smoking | ||||||
| Yes vs No | 1.153 | .429 | 0.809-1.645 | |||
| T | ||||||
| 1 vs 2 vs 3 + 4 | 1.554 | .001⁎ | 1.247-1.936 | 1.705 | .608 | 0.221-13.120 |
| N | ||||||
| 0 vs 1 vs 2 + 3 | 1.663 | .001⁎ | 1.398-1.977 | 2.167 | .003⁎ | 1.299-3.613 |
| TNM stage | ||||||
| I vs II vs III + IV | 1.649 | .001⁎ | 1.387-1.961 | 17.818 | .046⁎ | 1.050-301.890 |
P < .05.
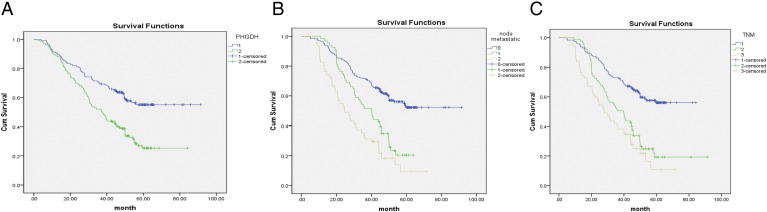
Survival Analysis
Kaplan-Meier survival analysis was performed and K-M curve was plotted for groups classified by IHC PHGDH expression (PHGDHhigh vs. PHGDHlow). Approximately, the 5-year cumulative survival rates were 23% and 57% for patients with PHGDHhigh and PHGDHlow tumors, respectively (Figure 4). Log-rank test was performed to compare the difference in the survival distributions between two groups. Analysis revealed a significant survival disadvantage for NSCLC patients with PHGDHhigh tumors. OS was significantly shorter in the PHGDHhigh group than PHGDHlow group, with a median survival of 38 months versus >60 months (log-rank test: P < .001, Figure 4). Because the PHGDH expression was significantly increased in NSCLC at both mRNA and protein levels, we further explored the effects of PHGDH mRNA expression levels on the survival by making use of online big data [17]. In line with our findings at the protein level, class distinctions based on the PHGDH gene expression were statistically significantly (n = 1926), leading to a clear separation between survival curves of PHGDHhigh and PHGDHhigh tumors. PHGDHhigh was significantly associated with unfavorable OS as compared with PHGDHlow (log-rank test: P < .001; Figure 1C, left panel). We next stratified data by histological type and found that PHGDHhigh was significantly associated with reduced survival in adenocarcinoma (n = 720, HR = 1.38, 95% CI = 1.1-1.75; log-rank test: P = .006; Figure 1C, middle panel) rather than survival in squamous cell carcinoma (n = 524, HR = 1.17, 95% CI = 0.92-1.48; log-rank test: P = .20, Figure 1C, right panel).
Figure 4.
Kaplan-Meier survival curves of NSCLC patients. (A) Patients with PHGDHhigh (green) had significantly shortened survival time when compared with those with PHGDHlow tumor (blue). (B) Patients with node metastasis (green) tended to survive shorter than patients without node metastasis (blue). (C) Advanced TNM stages (brown, II; green, III + IV) were significantly correlated with inferior OS.
Discussion
Metabolic characteristics of malignant cells are distinct from those of normal cells. One of the most remarkable distinctions is that cancer cells preferentially consume dramatically large amounts of glucoses, which are subsequently metabolized via glycolysis pathway. Accumulating works have shown mechanistic links between augmented glycolysis and well-established driver mutations, i.e., the excessive glycolysis may be a result of the abnormal signal transduction caused by transforming mutations [18] Alternatively, such metabolic reprogramming can also result from genomic modification of metabolic enzymes as exemplified by PHGDH [8], [10]. Amplification of PHGDH gene was identified in ~6% of breast cancers and 40% of melanomas, concurrent with corresponding increase in glycolysis [8], [10].
In this study, we found significantly increased PHGDH protein in NSCLC tissues as compared with controls. PHGDHhigh was correlated with TNM stage and lymph node invasion and also conferred a significant survival disadvantage over PHGDHlow. Consistenly, we also observed a significant increase in the PHGDH mRNA expression in tumor tissue, which was validated by TCGA data. Moreover, the KM-plotter substantiated that patients with PHGDHhigh tumors had significantly shorter survival than those with PHGDHlow tumors.
Our findings were in line with previous publications in various types of cancer. High PHGDH mRNA expression showed association with inferior clinical outcomes in breast cancer [11]. In a different study, Locasale et al. measured PHGDH expression in 106 breast cancers using IHC. It was intriguing that PHGDH expression appeared to be subtype specific, and high PHGDH expression was significanlty more prevalent in triple-negative and basal subtypes [8]. Moreover, in vitro experiments revealed that PGHDH protein expression was essential for growth in three breast cancer cell lines (BT-20, SK-BR-3, MCF-7) but not in nontumorigenic breast epithelial cells (MCF-10a) [8]. Interestingly, PHGDH overexpression disturbed acinar morphogenesis in the epithelial cells (MCF-10a), interrupted the formation of polarity, and supported anchorage-independent survival of cells. These alterations may make cells susceptible to transformation [8]. The role of PHGDH in cervical adenocarcinoma was assessed by Jing and coworkers [19]. They observed significantly elevated PHGDH expression in tumors versus normal cervical glandular epithelium by determining IHC expression of PHGDH in 54 human cervical adenocarcinoma samples. High PHGDH expression was predictive of poor prognosis in cervical adenocarcinoma. Furthermore, PHGDH cervical adenocarcinoma inhibited proliferation in cervical adenocarcinoma cells (HeLa cells). Knockdown of PHGDH in Hela cells by shRNA significantly impaired their capacity of forming tumor in nude mice when compared with control Hela-vector cells [19]. Yoon et al. evaluated the expression and prognostic value of several serine biosynthesis-associated enzymes in 486 patients with colon cancer, including PHGDH [20]. Consistent with results in other types of cancer, they confirmed significant increase in PHGDH expression level in the tumor tissue of colon but failed to provide evidence of its prognostic value in terms of relapse-free survival and OS [20].
In an esophageal squamous cell carcinoma cell line (T.T. cells. Haboring) with focal copy number gain in PHGDH, stable PHGDH knockdown repressed cell proliferation [8]. The same study showed that depletion of downstream enzymes phosphoserine aminotransferase (PSAT) and phosphoserine phosphatase in cell lines using shRNA led to comparable reduction in proliferation, suggesting that the reduced proliferation was related to the defects in the serine biosynthesis pathway [8]. A bioinformatics analysis involving 3131 human cancers identified amplification of PHGDH gene in 40% of melanomas [8]. In a validation study, 21% of human melanon tissues showed high IHC expression of PHGDH, whereas copy number gain of the gene was detected in 21 of a subset of 42 of the tissue collection [8]. Notebly, the study reveled the robust association of PHGDH copy number gain and high protein expression in human melanoma (P = .0045) [8]. In vitro experiemnts indicated that PHGDH knockdown via genetic manipulation hindered proliferation and redirecton of glycosis into serine biosythesis in PHGDH-amplified cell lines rather than in cell lines lacking a PHGDH copy number gain [8].
Consistently, there is some evidence indicating that serine/glycine biosynthesis might be implicated in carcinogenesis. Regarding breast cancer, expression of PHGDH and several other enzymes in the pathway was associated with metastasis in mice and unfavorable prognosis in humans [11]. Moreover, two isoforms of serine hydroxymethyltransferase are transcriptional targets of the oncogene c-Myc, and overexpressing this enzyme could arouse proliferation of c-Myc–deficient cells [21]. The role of PHGDH in breast cancer has been explored, and preliminary results were obtained [10], [11], [22], [23]. Functional genomics demonstrated the fundamental role of the serine synthesis pathway in breast cancer [10]. In contrast, a different group found that PHGDH overexpression stimulated the proliferation of breast cancer cell lines but was not indispensable for the maintenance and growth of breast tumor [22]. Most importantly, DeNicola et al. recently have reported that the transcription factor nuclear factor erythroid-2–related factor 2, commonly deregulated in NSCLC, closely regulates serine/glycine biosynethesis-related genes (e.g., PHGDH and PSAT1) in NSCLC cell lines [24]. These results highlighted the important role of PHGDH in NSCLC and advanced the understanding of PHGDH regulation.
The high expression of PHGDH in tumors and its biological function in carcinogenesis suggest that PHGDH may be a druggable metabolic enzyme. Indeed, the antimetabolite drugs have begun to be applied in cancer treatment over half a century ago. They have been successfully and widely used since then, including antifolate agents, inhibitors of thymidylate and purine biosynthesis (methotrexate, pralatrexate, 5-fluorouracil, and pemetrexed) [25]. The increasing understanding of the role of serine/glycine/one-carbon metabolism in carcinogenesis may define PHGDH and other relevant metabolic enzymes (e.g., phosphoserine phosphatase and PSAT1) as new potential therapeutic targets. Recently, several PHGDH inhibitors have been discovered through a quantitative high-throughput screen [26], [27]. It was demonstrated that CBR-5884, a small molecule, could noncompetitively inhibit PHGDH activity and suppress de novo synthesis of serine in malignant cells [26]. Consequentially, this inhibitor selectively killed tumor cells characterized by elevated biosynthetic activity [26]. Moreover, great achievements have been made recently in the treatment of NSCLC based on different pathways, including drugs targeting PI3K/Akt pathway and anti–PD-L1/anti–PD-1 agents. [32], [33], [34], [35] Combining these approaches and PDGDH inhibitors may hold great promise for NSCLC treatment.
Several facts may help to rationalize tumor cells' propensity for high aerobic glycolysis. First, PHGDH pathway provides plenty of de novo serines and glycines, which are precursors for several fundamental biosynthetic pathways. Second, whereas serine hydroxymethyltransferase simultaneously converts L-serine to glycine (retro-aldol cleavage) and tetrahydrofolate to 5,10-methylenetetrahydrofolate (hydrolysis), methyl groups were generated. This reaction provides the majority of methyl groups for the one-carbon pools, which are prerequisite for biosynthesis and DNA methylation [28]. Additionally, activation of PHGDH pathway also led to some consequences that were demonstrated to favor cell growth via multiple processes, including reduction in ATP production, direct changes in cellular redox status due to the oxidation of 3PG, and the production of αKG from glutamate. [29], [30], [31].
In conclusion, we provided evidence of clinical implication of PHGDH in NSCLC. Our results suggested that PHGDH might be an independent predictor of clinical outcomes in NSCLC.
Conflict of Interest
None.
Acknowledgement
This study was supported by funds from the Scientific Research Starting Foundation for Returned Overseas Chinese Scholars, Ministry of Education, China (to Jinhong Zhu), and the Six talent peaks project (WSW-049) in Jiangsu, China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
This study was supported by funds from the Scientific Research Starting Foundation for Returned Overseas Chinese Scholars, Ministry of Education, China (to Jinhong Zhu), and the Six talent peaks project (WSW-049) in Jiangsu, China.
Contributor Information
Xiaoyu Zhou, Email: z730303@163.com.
Jiahai Shi, Email: happysjh167@163.com.
References
- 1.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 6.Sotgia F, Martinez-Outschoorn UE, Lisanti MP. Genetic induction of the Warburg effect inhibits tumor growth. Oncotarget. 2012;3(11):1266–1267. doi: 10.18632/oncotarget.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galluzzi L, Kepp O, Kroemer G. Reverse Warburg: straight to cancer. Cell Cycle. 2012;11(6):1059. doi: 10.4161/cc.11.6.19746. [DOI] [PubMed] [Google Scholar]
- 8.Locasale JW, Grassian AR, Melman T. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43(9):869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kit S. The biosynthesis of free glycine and serine by tumors. Cancer Res. 1955;15(11):715–718. [PubMed] [Google Scholar]
- 10.Possemato R, Marks KM, Shaul YD. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollari S, Kakonen SM, Edgren H. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011;125(2):421–430. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- 12.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 13.Goldstraw P. The 7th edition of TNM in lung cancer: what now? J Thorac Oncol. 2009;4(6):671–673. doi: 10.1097/JTO.0b013e31819e7814. [DOI] [PubMed] [Google Scholar]
- 14.Lu C, Wang X, Zhu H, Feng J, Ni S, Huang J. Over-expression of ROR2 and Wnt5a cooperatively correlates with unfavorable prognosis in patients with non–small cell lung cancer. Oncotarget. 2015;6(28):24912–24921. doi: 10.18632/oncotarget.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun R, Wang X, Zhu H. Prognostic value of LAMP3 and TP53 overexpression in benign and malignant gastrointestinal tissues. Oncotarget. 2014;5(23):12398–12409. doi: 10.18632/oncotarget.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen R, Lu M, Wang J. Increased expression of Trop2 correlates with poor survival in extranodal NK/T cell lymphoma, nasal type. Virchows Arch. 2013;463(5):713–719. doi: 10.1007/s00428-013-1475-4. [DOI] [PubMed] [Google Scholar]
- 17.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non–small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Jing Z, Heng W, Xia L. Downregulation of phosphoglycerate dehydrogenase inhibits proliferation and enhances cisplatin sensitivity in cervical adenocarcinoma cells by regulating Bcl-2 and caspase-3. Cancer Biol Ther. 2015;16(4):541–548. doi: 10.1080/15384047.2015.1017690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon S, Kim JG, Seo AN. Clinical Implication of serine metabolism-associated enzymes in colon cancer. Oncology. 2015;89(6):351–359. doi: 10.1159/000439571. [DOI] [PubMed] [Google Scholar]
- 21.Nikiforov MA, Chandriani S, O'Connell B. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22(16):5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Chung F, Yang G. Phosphoglycerate dehydrogenase is dispensable for breast tumor maintenance and growth. Oncotarget. 2013;4(12):2502–2511. doi: 10.18632/oncotarget.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gromova I, Gromov P, Honma N. High level PHGDH expression in breast is predominantly associated with keratin 5–positive cell lineage independently of malignancy. Mol Oncol. 2015;9(8):1636–1654. doi: 10.1016/j.molonc.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeNicola GM, Chen PH, Mullarky E. NRF2 regulates serine biosynthesis in non–small cell lung cancer. Nat Genet. 2015;47(12):1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeBerardinis RJ. Serine metabolism: some tumors take the road less traveled. Cell Metab. 2011;14(3):285–286. doi: 10.1016/j.cmet.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullarky E, Lucki NC, Beheshti Zavareh R. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc Natl Acad Sci U S A. 2016;113(7):1778–1783. doi: 10.1073/pnas.1521548113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacold ME, Brimacombe KR, Chan SH. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12(6):452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vander Heiden MG, Locasale JW, Swanson KD. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329(5998):1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Locasale JW, Cantley LC. Altered metabolism in cancer. BMC Biol. 2010;8:88. doi: 10.1186/1741-7007-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010;3(119):ra31. doi: 10.1126/scisignal.2000911. [DOI] [PubMed] [Google Scholar]
- 32.Fumarola C, Bonelli MA, Petronini PG, Alfieri RR. Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochem Pharmacol. 2014;90(3):197–207. doi: 10.1016/j.bcp.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Zhang XD, Proud C. Dissecting the signaling pathways that mediate cancer in PTEN and LKB1 double-knockout mice. Sci Signal. 2015;8(392):pe1. doi: 10.1126/scisignal.aac8321. [DOI] [PubMed] [Google Scholar]
- 34.Heavey S, O'Byrne KJ, Gately K. Strategies for co-targeting the PI3K/AKT/mTOR pathway in NSCLC. Cancer Treat Rev. 2014;40(3):445–456. doi: 10.1016/j.ctrv.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Soria JC, Marabelle A, Brahmer JR, Gettinger S. Immune checkpoint modulation for non-small cell lung cancer. Clin Cancer Res. 2015;21(10):2256–2262. doi: 10.1158/1078-0432.CCR-14-2959. [DOI] [PubMed] [Google Scholar]