Abstract
Metastasis is a multistep process starting with the dissemination of tumor cells from a primary site and ending with secondary tumor development in an anatomically distant location. The epithelial–mesenchymal transition (EMT), a process that endows epithelial tumor cells with mesenchymal properties including reduced adhesion and increased motility, is considered a critical step driving the early phase of cancer metastasis. Although significant progress has been made in understanding the molecular characteristics of EMT, the intracellular mechanisms driving transition through the various stages of EMT remain unclear. In recent years, an increasing number of studies have demonstrated the involvement of long non-coding RNAs (lncRNAs) in tumor metastasis through modulating EMT. LncRNAs and their associated signaling networks have now emerged as new players in the induction and regulation of EMT during metastasis. Here we summarize the recent findings and characterizations of several known lncRNAs involved in the regulation of EMT. We will also discuss the potential use of these lncRNAs as diagnostic and prognostic biomarkers as well as therapeutic targets to slow down or prevent metastatic spread of malignant tumors.
Facts
EMT facilitates cancerous epithelial cells to enter into a mesenchymal-like state by endowing them migratory and invasive properties, which enables primary tumor cells to move and colonize distant organs and form secondary tumor metastases.
EMT is closely linked to carcinogenesis, invasion, metastasis, recurrence, and resistance. Understanding the molecular mechanisms that control EMT will shed lights to the metastatic processes of tumor cells, and provide new therapeutic targets and treatment options for effective cancer therapy.
EMT is regulated by a complex signaling network involving both transcriptional and post-transcriptional regulatory pathways related to cancer metastasis.
LncRNAs are generally defined as non-protein-coding RNA transcripts as indicated by the lack of a discernable open reading frame. Many identified lncRNAs are polyadenylated, and locate within nuclear or cytosolic fractions.
An increasing number of reports during the past few years support the involvement of lncRNAs in regulating tumor metastasis and progression by controlling EMT through specific ligands, receptors, or multiple EMT-related signaling pathways.
The distribution and levels of lncRNAs in various locations, such as distal metastases, have been exploited as potential diagnostic and prognostic biomarkers for cancer.
Open Questions
Although there are growing interests in lncRNAs as potential biomarkers and therapeutic targets of EMT and cancer metastasis, it remains largely unknown how they are regulated in cancer cells and how they affect EMT and metastasis.
It remains to be determined the different and precise molecular mechanisms by which functional lncRNAs switch EMT on and off during tumor development.
Technologies should be advanced to achieve more sensitive and reliable detection and effective targeting of lncRNAs for cancer treatment.
The effective delivery of lncRNA-targeted therapeutics to respective tumor sites, the challenges of stability, immunogenicity, and bioavailability are some of the main obstacles to be overcome in the clinical translation of lncRNA-targeted therapy.
Metastasis is responsible for as much as 90% of cancer-induced mortality, yet this process remains one of the most elusive pathological process in cancer progression.1 The development of new therapeutic strategies targeting key factors driving metastasis remains a challenging goal for both clinicians and scientists. First described in embryogenesis, embryonic cells undergo a process known as epithelial-to-mesenchymal transition (EMT) that allows epithelial cells to migrate and travel long distances to form tissues and organs. Once migratory embryonic cells reach their destination, they undergo the reverse process, mesenchymal-to-epithelial transition (MET) to settle, proliferate, and differentiate into different organs.2 Likewise, cancer cells follow the similar process to establish metastases. EMT facilitates cancerous epithelial cells to enter into a mesenchymal-like state by endowing them migratory and invasive properties, which enables a primary tumor to move and colonize distant organs and form secondary tumors metastases.2, 3 Moreover, these post-EMT cancer cells are often resistant to novel tumor-targeted radiotherapeutic/chemotherapeutic drugs and survive standard cancer therapies, and associate with tumor relapse and metastasis. Deeper understanding the molecular mechanisms that control EMT will not only shed lights to the metastatic processes of tumor cells, but also provide new therapeutic targets and treatment options for effective cancer therapy.
LncRNAs are commonly referred to as non-protein-coding RNA transcripts longer than 200 nt. Emerging evidence have shown that lncRNAs are dysregulated in multiple cancer types and have an important role in tumorigenesis and cancer progression.4 Recent studies have also demonstrated an essential role of lncRNAs in regulating EMT and cancer metastasis. In this article, we will provide a comprehensive review of the known lncRNAs relevant to EMT in cancer metastasis and discuss the molecular mechanisms underlying their regulation of EMT and their therapeutic implications as biomarkers and potential drug targets.
Key Regulators of EMT
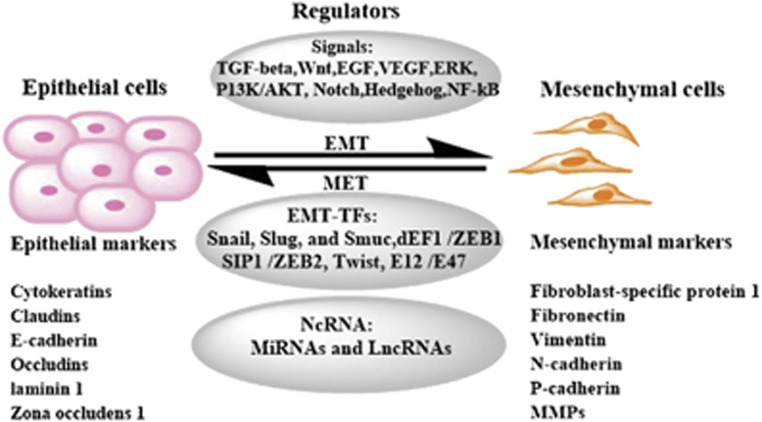
The EMT process is accompanied by loss of epithelial marker proteins and dissolution of adherent junction proteins, such as cytokeratin, E-cadherin, β-catenin, γ-catenin, which have key roles in cell–cell adhesion.3 Concomitantly, mesenchymal marker proteins, such as N-cadherin, P-cadherin, fibronectin, and intermediate filament protein vimentin, are frequently overexpressed and contribute to cell migration as well as invasion-associated gene expression in different types of cancer.5 The most important regulators of cell adhesion are the cadherin family of proteins including E-, N-, and P-cadherin. E-cadherin as the best characterized cadherins in particular has a key role in epithelial cell–cell adhesion. It acts by binding to E-cadherins from neighboring cells and providing a physical link between their cytoskeletons.6 The E-cadherin is replaced by abnormal expression of N- or P-cadherin is a hallmark for EMT. The downregulation of E-cadherin leads to the release of β-catenin, and the latter translocates to the nucleus and functions as an activator for transcription factors, such as ZEB, Twist, Snail, and Slug, which are known to act as the repressor of E-cadherin expression. The induction of these transcription factors promotes cell migration, tissue morphogenesis, and cancer development.7 Other proteins that mediate EMT include vimentin and fibronectin. Vimentin and fibronectin are upregulated in cells undergoing EMT, resulting in epithelial cells to acquire a mesenchymal shape and increased motility. Fibronectin mediates cellular interactions with the extracellular matrix and is important for migration, differentiation, growth, and cell adhesion (Figure 1).8
Figure 1.
Regulatory network in EMT. EMT can be regulated by many signaling pathways, transcription factors, and transcriptional/post-transcriptional regulators.
Regulations of EMT Signaling Networks in Tumor Cells
EMT is regulated by a complex signaling network at both transcriptional and post-transcriptional levels. Many growth factors, such as transforming growth factor-β (TGF-β), fibroblast growth factor (FGF), epidermal growth factor (EGF), vascular endothelial growth factor (VEGF), and their associated signaling proteins, such as nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), ERK, PI3K/AKT, Hedgehog (Hh), Notch, and Wnt, are engaged to trigger and complete an EMT process.9 EMT-inducing signals are cell- or tissue-type-specific and require the cooperation between multiple signaling pathways and regulators (Figure 1). These signals usually activate one of the EMT-inducing transcription factors (EMT-TFs), known as key EMT regulators, which include the Snail family of zinc-finger transcription factors (Snail, Slug, and Smuc), the dEF1 family of two-handed zinc-finger factors (dEF1/ZEB1 and SIP1/ZEB2), and the basic helix–loop–helix factors Twist and E12/E47.3 With the exception of Twist, these EMT-TFs repress the expression of E-cadherin by direct binding to the E-box sites in the promoter of E-cadherin, trigger gene re-programing and alter protein expression, and dynamically modulate EMT.5 In addition to transcription factors, EMT can be regulated by non-coding RNAs such as lncRNA and miRNA at both transcriptional and post-transcriptional levels. Previous work has revealed that EMT induction in cancer depends on an intricate network of multiple signaling pathways (Figure 1). Here we discuss several major signaling pathways that participate in the initiation of cancer EMT.
TGF-β signaling pathway in EMT
TGF-β signaling has a predominant role in suppressing growth of normal epithelial cells, while promotes metastasis in many tumor cells, in part through tightly controlling the process of EMT.10 TGF-β activates Smads by binding to type II and type I serine-threonine kinase receptors (TβRII and TβRI), respectively. TβRI is phosphorylated by TβRII, and then activates Smads. Activated Smads translocate into the nucleus to interact with various EMT-TFs and transcriptional co-activators, and regulate the transcription of target genes. For example, TGF-β can activate the expression of Snail via Smads pathways as downstream effectors to repress the expression of E-cadherin and claudin.11 In addition to the canonical TGF-β /Smad pathway, TGF-β has been shown to regulate the expression of EMT markers through non-canonical (Smad-independent) TGF-β signaling pathways. It is reported that MEK/ERK, PI3K/AKT, p38-MAPK, as well as induction of miRNAs have important roles in TGF-β-induced EMT.12, 13, 14, 15
Wnt/β-catenin signaling pathway in EMT
The Wnt signaling pathway is an important regulator of EMT-TFs expression and the EMT process. Wnt couples with its cell surface receptors, the low-density lipoprotein receptor and membrane protein Frizzled, to activate and stabilize the protein β-catenin, which is the central component of the Wnt/β-catenin signaling moving from the cytoplasm to the nucleus to regulate the transcription of Wnt target genes. In the nucleus, β-catenin acts as a coactivator of T-cell factor/lymphoid-enhancing factor-1 (TCF/LEF-1) to promote the transcription of Snail, Slug, and Twist, which in turn represses E-cadherin.16, 17
Hh signaling in EMT
Studies have shown that the main role of Hh signaling pathway in tumor is to promote EMT and maintain cancer stem cells (CSCs).18 In humans, Hh signaling is orchestrated by two transmembrane receptors, Patched (Ptch), and Smoothened (Smo). The Hh family include three homologous Hh ligands: Sonic hedgehog (Shh), Indian hedgehog (Ihh), and Desert hedgehog (Dhh). Hh ligands can activate Ptch, release Smo, and then initiate an intracellular cascade that activates the Gli family of transcription factors. As marker of the Hh pathway activity, Gli promotes EMT by inducing the transcription of target genes such as Ptch, Wnt, and Snail.19 In addition, Hh signaling can cooperate with other signaling pathways such as Wnt, Notch, FGF, and TGF-β to modulate EMT-induced CSCs signaling network.19, 20
Hypoxic/hypoxia-inducible transcription factor 1 in EMT
Recent studies indicated that each step of the metastasis process, from the initial EMT to the ultimate organotropic colonization, can potentially be regulated by hypoxia, suggesting a master regulator role of hypoxia and hypoxia-inducible transcription factor 1 (HIF-1). HIF-1 consists of an unstable α-subunit and a stable β-subunit. Under hypoxic conditions, HIF-1α stabilizes and translocates to the nucleus, promotes EMT by upregulating EMT-associated transcription activators or repressors, modulating EMT-associated signaling pathways, EMT-associated inflammatory cytokines, and epigenetic regulators.21 It has been shown that the activation of the HIF-1α-mediated canonical hypoxia signaling leads to the upregulation of Twist, Snail, ZEB1, and E12/E47 and enhanced EMT in breast cancer.22, 23, 24
Regulation of EMT by lncRNAs
Non-coding RNAs (ncRNAs), as newcomers in genome biology, are initially regarded as transcriptional ‘noise', but accumulating evidence has demonstrated that ncRNAs such as microRNAs (miRNA), small inhibitory RNAs (siRNAs), piwi-interacting RNAs (piRNAs), circular RNAs, and lncRNAs have critical regulatory roles in gene expression.25 Recent studies have also elucidated the association between EMT and ncRNAs in tumor metastasis.15, 26
LncRNAs as a newer class of ncRNAs are divided into five broad categories: (1) sense, (2) antisense, (3) bidirectional, (4) intronic, or (5) intergenic with respect to the nearest protein-coding transcripts.27 Studies have demonstrated that lncRNAs are aberrantly expressed in a variety of human cancers, such as lung cancer, gastric cancer, pancreatic cancer, and breast cancer, and have important roles in various cancer-associated biological processes and signaling pathways.28, 29 They can act in cis or trans to modulate gene expression, for example, by binding miRNAs to protect the mRNAs.30 Through regulating gene expression by multiple distinct molecular mechanisms, including transcription, post-transcriptional processing, genomic imprinting, chromatin modification, and the regulation of protein function. There are growing interests in using lncRNAs for cancer diagnosis and prognosis, and lncRNAs are considered promising therapeutic targets for cancer treatment.29 Increasing evidence has demonstrated a potential role of lncRNAs in tumor metastasis by influence the EMT process.29 In the following section, we will provide an overview of the main mechanisms through which lncRNAs regulate EMT in tumor cells. A summary of EMT-associated lncRNAs and their pathophysiological functions and signaling mechanisms related to EMT is provided in Table 1.
Table 1. LncRNAs that regulate EMT and their targets in different cancer types.
| Cancer type | lncRNA | Function | Dysregulation of lncRNA | Potential mechanism | Ref. |
|---|---|---|---|---|---|
| Bladder cancer | H19 | Oncogenic | Up | Promotes EMT by interacting with EZH2 and repressing E-cadherin expression. | 49 |
| lncRNA-ZEB2NAT | Oncogenic | Up | Induces EMT and invasion through the TGFβ1-ZEB2NAT-ZEB2 axis in CAFs. | 39 | |
| MALAT1 | Oncogenic | Up | Mediates TGF-β induced EMT via suz12 or promotes EMT by activating Wnt/β-catenin signal pathway. | 38, 47 | |
| lncRNA-HIT | Oncogenic | Up | Activated by TGF-β and induces EMT. | 40 | |
| KIAA0125 | Oncogenic | Up | Promotes migration and invasion partly via induction of vimentin and suppression of β-catenin. | 102 | |
| TUG1 | Oncogenic | Up | Decreases miR-145 and induces EMT. | 73 | |
| UBC1 | Oncogenic | Up | Binds to PRC2 complex induces EMT. | 60 | |
| UCA1 | Oncogenic | Up | Promotes migration and invasion via hsa-miR-145/ZEB1/2 /FSCN1 pathway. | 75 | |
| Breast cancer | HOTAIR | Oncogenic | Up | Promotes EMT by suppressing miR-568 to maintain NFAT5 expression | 71 |
| LncRNA-ATB | Oncogenic | Up | Activated by TGF-β, binds to miR-200c, upregulates ZEB1 and ZNF-217, and induces EMT. | 36 | |
| LincRNA-ROR | Oncogenic | Up | Regulates EMT by acting as a sponge for mir-205. | 76 | |
| Linc00617 | Oncogenic | Up | Induces EMT via activating the transcription of Sox2. | 100 | |
| LncRNA-Hh | Oncogenic | Up | Activates the Hedgehog signaling pathway. | 53 | |
| Cervical cancer | lncRNA-EBIC | Oncogenic | Up | Promotes invasion by binding to EZH2 and represses E-cadherin expression. | 61 |
| Colon cancer | BANCR | Oncogenic | Up | Induces EMT through the MEK/ERK pathway. | 51 |
| H19 | Oncogenic | Up | Promotes EMT as a ceRNA for miR-138 and miR-200a. | 66 | |
| HOTAIR | Oncogenic | Up | Not determined | 45 | |
| lncRNA-ATB | Oncogenic | Up | Not determined | 34 | |
| Esophageal squamous cell carcinoma | HOTAIR | Oncogenic | Up | Inhibits WIF-1 expression and activates Wnt pathway to induce EMT. | 46 |
| Epithelial ovarian cancer | MANCR | Oncogenic | Up | Induces EMT through a MEK/ERK-dependent mechanism. | 52 |
| Gastric cancer | HOTAIR | Oncogenic | Up | Promotes EMT through regulating Snail via HER2/AKT/HSF-1/Slug pathway by inhibiting miR-331-3p or by silencing miR34a by binding to PRC2. | 69, 70, 79 |
| H19 | Oncogenic | Up | Induces EMT, promotes invasion and metastasis by binding to miR-141. | 67 | |
| HULC | Oncogenic | Up | Not determined | 93 | |
| LncRNA-ATB | Oncogenic | Up | Induces EMT, promotes invasion and metastasis through the TGF-β/miR-200s/ZEB axis. | 33 | |
| LEIGC | Tumor suppressor | Down | Not determined | 78 | |
| Linc00152 | Oncogenic | Up | Unknown mechanism | 84 | |
| SPRY4-IT1 | Oncogenic | Down | Contributes to metastasis via affecting EMT process. | 88 | |
| Lung cancer | BANCR | Oncogenic | Down | Promotes EMT and metastasis by regulating of EMT marker expression. | 85 |
| SPRY4-IT1 | Oncogenic | Down | Promotes proliferation and metastasis by affecting the EMT. | 62 | |
| ZEB1-AS1 | Oncogenic | Up | Induces EMT by upregulating ZEB1 expression. | 80 | |
| Hepatocellular carcinoma | AOC4P | Tumor suppressor | Down | Enhances vimentin degradation and suppresses EMT. | 83 |
| HOTAIR | Oncogenic | Up | Downregulates E-cadherin and induces EMT. | 59 | |
| H19 | Oncogenic | Up | Increases HMGA2-mediated EMT through antagonizing let-7. | 65 | |
| lncRNA-ATB | Oncogenic | Up | Activated by TGF-β, binds miR-200s, upregulates ZEB1/2ZEB2 to induce EMT and invasion. | 32 | |
| LncRNA-Dreh | Tumor suppressor | Down | Inhibits metastasis by repressing vimentin expression and changing the normal cytoskeleton structure. | 82 | |
| linc-RoR | Oncogenic | Up | Involves in miR-145/HIF-1α signaling module. | 43 | |
| lncTCF7 | Oncogenic | Up | Acts through IL-6/STAT3/lncTCF7 signaling axis leading to HCC aggressiveness through EMT induction. | 101 |
Role of lncRNAs in the regulation of EMT signaling networks in tumor cells
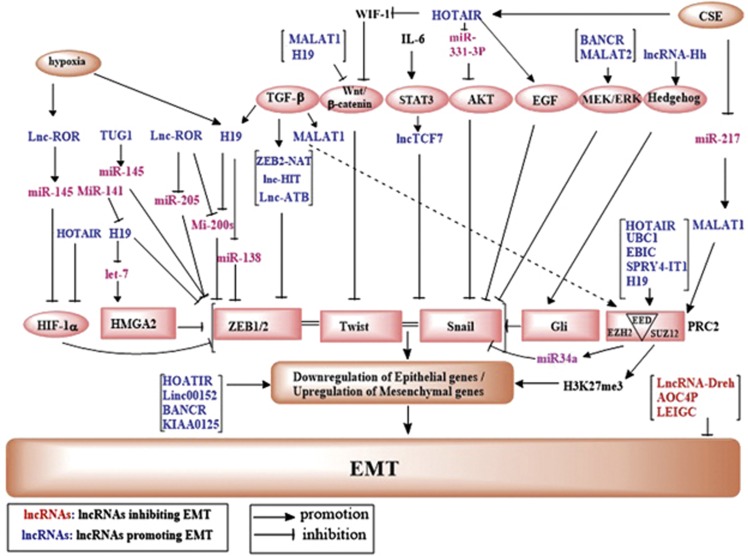
LncRNAs regulate EMT through a complex network of signaling events. A diagram of the signaling network of known lncRNAs relevant to EMT is shown in Figure 2 and several main pathways are discussed below:
Figure 2.
LncRNAs that are known to regulate EMT processes and their validated targets. A diagram depicts the major signaling pathways through which lncRNAs regulated EMT. Pink oval, names of the signaling pathways; pink square oval, EMT-TFs; blue text, lncRNAs that inhibit EMT; red text, lncRNAs that promote EMT; purple text, miRNAs.
TGF-β pathway
TGF-β is a well-known EMT initiator.31 Lnc-ATB (lncRNA activated by TGF-β) is a TGF-β-induced lncRNA that could mediate TGF-β-induced EMT and has been shown to promote metastasis in hepatocellular carcinoma, colorectal cancer, gastric cancer, and breast cancer.32, 33, 34, 35, 36, 37 In addition, Fan and colleagues showed that TGF-β induced a specific lncRNA called metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which leads to EMT in bladder cancer cells. Interestingly, MALAT1 is associated with the suppression of SUZ12, which prevents the ability of Snail1 from downregulating E-cadherin.38 Thus, the induction of MALAT1 by TGF-β results in decreased E-cadherin and increased N-cadherin/fibronectin, leading to enhanced EMT.38 Cancer-associated fibroblasts (CAFs) as one of the principal constituents of tumor stroma have an important role in tumor development. TGF-β1 secreted by CAFs induces EMT of urothelial bladder cancer through lncRNA-ZEB2NAT.39 In addition, LncRNA-HIT (HOXA-associated transcript induced by TGF-β) is also involved in TGFβ-induced EMT. The effects of lncRNA-HIT on EMT, migration, and invasion in breast cancer were rescued through introduction of ectopic E-cadherin.40 These findings suggest that lncRNAs can be induced by TGF-β and play a key role in TGF-β-induced EMT.
Hypoxia/HIF-1α pathway
As described earlier, HIF-1α regulates EMT at multiple fronts, including the expression of EMT-TFs, EMT-associated pathways, and cytokine. The oncofetal H19 lncRNA is concomitantly induced by both TGF-β and hypoxia in a mouse breast cancer model, which regulates E-cadherin expression and stimulates tumor metastasis through a positive feedback loop between Slug and H19/miR-675.41 The positive regulation of H19 by HIF-1α may partially explain its high expression in metastases.42 Linc-RoR (regulator of reprogramming) is a hypoxia-responsive lncRNA that modulates expression of miR-145 and HIF-1α and acts through a hypoxia/miR-145/HIF-1α signaling axis to modulate EMT in vitro and in vivo.43
Wnt signaling pathway
Activation of Wnt/β-catenin signaling pathway has been shown to induce EMT. Wnt inhibitory factor 1 (WIF-1) plays an important role in the Wnt/β-catenin signal pathway. Ge et al. demonstrated that HOX transcript antisense intergenic RNA (HOTAIR), an EMT-associated lncRNA and a powerful predictor of metastasis, inhibits WIF-1 expression and activates the Wnt pathway in esophageal squamous cell carcinoma cells.44, 45 HOTAIR can also directly decrease WIF-1 expression by promoting its histone H3K27 methylation in the promoter region and then activates the Wnt/β-catenin signaling pathway.46 MALAT1 can promote EMT by activating Wnt signaling in vitro, and knockdown of MALAT1 results in a decrease of the ZEB1, ZEB2, and Slug levels, and an increase of E-cadherin levels in bladder cancer cell or urothelial carcinoma.47, 48 H19 is associated with EZH2, and this association results in the activation of Wnt/β-catenin and subsequent inhibition of E-cadherin.49 Thus, Wnt signaling is an important target of lncRNAs to regulate EMT.
MEK/ERK pathway
The MEK/ERK pathway is another major pathway through which lncRNAs regulate EMT.50 Guo et al. demonstrated that overexpression of BRAF-activated non-coding RNA (BANCR) induces colorectal carcinoma migration by inducing EMT via the MEK/ERK signaling pathway since treatment with the MEK inhibitor affects the expression of epithelial and mesenchymal markers in colorectal cancer.51 Similarly, Chen et al. indicated that MANCR (MALAT2-activated lncRNA) contributes to gastric cancer migration by inducing EMT via a MEK/ERK-dependent mechanism as the MEK/ERK pathway inhibitor inhibits cancer metastasis.52
Hedgehog signaling pathway
CSCs are a subpopulation of neoplastic cells with self-renewal capacity and limitless proliferative potential as well as high invasive and migratory capacity. These cells are commonly associated with EMT and subsequent tumor metastasis. LncRNA-Hh contributes to Twist-induced EMT and enhances CSC-like stemness.53 Specifically, lncRNA-Hh transcriptionally regulated by Twist directly targets GAS1 to stimulate the activation of Hh. The activated Hh increases Gli expression, and enhances the expression of SOX2 and OCT4 to maintain CSCs. These results indicate that lncRNA-Hh impinges upon EMT and CSC stemness by modulating the hedgehog signaling.
Coordination with PRC2
A major mechanism through which lncRNAs regulate gene expression involves the interaction with the epigenetic silencing complex polycomb repressive complex 2 (PRC2), one of the two major classes of polycomb group protein complexes. It is estimated that about 20% of all lncRNA transcripts bind PRC2.54, 55, 56 PRC2 that comprises EZH2, embryonic ectoderm development (EED), and suppressor of zeste 12 (SUZ12), is a histone methyl-transferase that catalyzes the trimethylation of histone H3 lysine 27 (H3K27me3) to repress transcription of specific genes such as E-cadherin.55 LncRNA HOTAIR has been shown to interact with PRC2 to promote cancer progression in breast cancer,57 gastrointestinal cancer,58 and hepatocellular carcinoma.59 Similarly, lncRNA-UBC1 (upregulated in bladder cancer 1) physically associates with PRC2 subunit EZH2 and SUZ12 and contributes to increased cancer invasion and metastasis.60 LncRNA H19 can also regulate bladder cancer metastasis by interacting with EZH2 and subsequently repressing E-cadherin expression and tumor metastasis.49 LncRNA-EBIC (EZH2-binding lncRNA in cervical cancer) promotes tumor cell invasion by binding to EZH2 and inhibiting E-cadherin expression in cervical cancer.61 In addition, SPRY4-IT1 (SPRY4 intronic transcript 1) is a key regulatory factor underlying the EZH2 pathway. Knockdown of SPRY4-IT1 reverses the inhibition of the EZH2 expression-mediated impairment of non-small cell lung cancer cell migration, invasion, and EMT process.62 Collectively, lncRNA and PRC2 interaction plays a critical role in lncRNA-regulated EMT and tumor metastasis.
Cooperation with miRNAs
In recent years, a new regulatory mechanism has emerged that coding- and non-coding RNAs can regulate each other by competing for shared miRNA, which has been demonstrated in a variety of cancers. Abundant evidence indicates that miRNAs are capable of directly modulating EMT-TFs or EMT-activating signaling pathways.15 Competing endogenous RNAs (ceRNAs), also called natural microRNA sponges, are endogenous coding or non-coding transcripts including lncRNAs, circular RNAs and pseudogenes that share sequences with common microRNAs. These ceRNAs can bind and sequester miRNAs to protect their target mRNAs from being degraded.63 LncRNAs have been shown to regulate EMT and tumor metastasis through their ability to act as endogenous ceRNAs for EMT-regulatory miRNAs. For example, H19 promotes pancreatic cancer cell invasion and migration by increasing its target HMGA2-mediated EMT through antagonizing let-7, a microRNA and a well-known tumor suppressor in pancreatic ductal adenocarcinoma, thus H19 may repress let-7 function through competitive ceRNA network.64, 65 H19 also functions as a ceRNA for miR-138 and miR-200a, antagonized their functions, leading to the de-repression of their endogenous targets vimentin, ZEB1, and ZEB2 in colorectal cancer.66 On the other hand, miR-141 binds to H19 in a sequence-specific manner, and suppresses H19 expression and function including proliferation and invasion in gastric cancer.67 HOTAIR was reported to promote EMT via HER2/AKT/HSF-1/Slug pathway by inhibiting miR-331-3p in gastric cancer patients.68, 69 HOTAIR can also epigenetically downregulate miR34a by binding to PRC2 to activate miR34a target gene C-Met (HGF/C-Met/Snail pathway) and Snail, thereby promoting EMT in advanced stages of gastric cancer.70 HOTAIR also suppresses miR-568 to maintain NFAT5 expression which promotes invasion via EMT.71 The lncRNA TUG1 (taurine upregulated gene) can decrease the expression of miR-145 to regulate the activity of ZEB2 and EMT.72, 73 The family of miR-200s including miR-200a, miR-200b, miR-200c, miR-141, and miR-429 plays a key role in EMT by inhibiting EMT-TF ZEB1/2 and upregulating E-cadherin.15, 74 Yuan et al. found that lncRNA-ATB promotes metastasis of hepatoma cells through upregulating ZEB1 and ZEB2 via competitively binding to the miR-200 family members.32 Similarly, Lnc-ATB has also been shown to upregulate ZEB1 and ZNF-217 by competitively binding to miR-200c, leading to EMT in breast cancer cells.36 LncRNA UCA1 promotes bladder cancer cell migration and invasion via hsa-miR-145/ZEB1/2/FSCN1 pathway.75 Another lncRNA linc-ROR may function through regulating multiple miRNAs to affect EMT-associated signaling pathways. Linc-ROR modulates hypoxia signaling through a miR-145/HIF-1α signaling pathway in HCC cells.43 Similarly, linc-ROR regulates EMT by acting as a sponge for miR-205, and linc-ROR overexpression prevents the degradation of miR-205 target genes in breast cancer cells, including the EMT inducer ZEB2.76 In summary, miRNAs and lncRNAs can cooperate with each other in an lncRNA-miRNA functional network to regulate EMT.77
Regulation of the expression of EMT-TFs and EMT markers
An increasing number of reports in the past few years support the regulation of EMT-TFs by lncRNAs, although it remains to be determined if the effects were direct. For instance, lncRNA LEIGC is a critical regulator in preventing EMT in gastric cancer, as LEIGC knockdown results in highly elevated expression of Snail, Slug, Twist and Zeb (ZEB) genes.78 Recent studies have also highlighted the importance of HOATIR in the regulation of EMT through regulating Snail, Slug, and Twist expression.79 Similarly, ZEB1-AS1 (ZEB1 antisense1) induces EMT by upregulating ZEB1 expression in hepatocellular carcinoma.80
EMT is marked by the loss of epithelial markers and concomitant increased expression of mesenchymal markers (Figure 1) (see Sanchez-Tillo et al.9 for a complete list of EMT markers).8 Many lncRNAs have been linked to the regulation of EMT through modulating EMT markers either directly or indirectly and the mechanisms remain elusive. It has been shown that HOTAIR promotes malignant transformation of lymph node stromal cells through downregulating E-cadherin and inducing EMT in normal liver stem cells.81 Depletion of HOTAIR increased expression of E-cadherin while concomitantly decreasing expression of vimentin and MMP9.45 LncRNA-Dreh regulates tumor metastasis by modifying the expression and reorganization of vimentin.82 Ming Sun et al. has also shown that lncRNA AOC4P exerts a tumor-suppressive effect on hepatocellular carcinoma tumor progression by binding to vimentin and enhancing vimentin degradation and suppressing EMT.83 In another study, linc00152 knockdown suppresses EMT program by decreasing N-cadherin, vimentin and oncogenic AEG-1 protein levels, and increasing E-cadherin expression.84 Sun et al. has also demonstrated that overexpression of lncRNA BANCR modulates EMT through the regulation of E-cadherin, N-cadherin, and vimentin expression.85
In summary, EMT is regulated by an intricate network of signaling pathways associated with cancer metastasis. Owing to the complex interactions between these signaling pathways, many of these lncRNA regulated pathways converge on a few master regulatory molecules or parallel pathways to induce changes of EMT at various levels. Thus, understanding the crosstalks between EMT-inducing signaling pathways and their regulation by lncRNAs will provide fundamental knowledge to the molecular processes of EMT.
Therapeutic Implications
LncRNA as potential diagnostic and prognostic biomarkers of EMT and metastasis
Metastatic spread of malignant tumors accounts for majority of cancer-related deaths. Growing efforts are devoted to the discovery of biomarkers that can be used for predicting and measuring metastatic potential of tumors. Since EMT is a potential enabling early event during metastasis, EMT regulatory lncRNAs are not only functionally important but also valuable for predicting metastasis. An increasing number of lncRNAs discovered in the past few years have been found to be dysregulated in different types of cancers and their metastases including those of breast, colon, liver, bladder, and lung, which may serve as potential biomarkers for cancer diagnosis and prognosis (see a complete list of these lncRNAs in Table 2). For example, it has been shown that the abnormal expression of HOTAIR represses several tumors and metastasis suppressor genes and has a unique association with patient prognosis.44 The expression levels of HOTAIR can predict tumor recurrence in HCC patients who have undergone liver transplantation therapy.81 MALAT1 is another prominent lncRNA overexpressed in a wide range of cancers like osteosarcoma, colorectal cancer, lung cancer, and specifically linked to high metastasis rate and poor prognosis in non-small cell lung cancer patients.86 Furthermore, Ying et al. demonstrated that MALAT1 is increased in highly invasive subline of brain metastases from lung cancer cells. The increased level of MALAT1 promotes lung cancer brain metastasis by inducing EMT, suggesting that MALATI may be a promising prognostic factor and therapeutic target for lung cancer brain metastasese.87 The expression of MALAT2 (metastasis-associated lung adenocarcinoma transcript 2) is upregulated in gastric cancer tissues, and a higher expression level of MALAT2 might serve as a negative prognostic marker in stage II/III gastric cancer patients.52 SPRY4-IT1 expression is decreased in gastric cancer tissues and associates with larger tumor size, advanced pathological stage, deeper depth of invasion, and lymphatic metastasis. Patients with lower SPRY4-IT1 expression have a relatively poor prognosis.88 Highly upregulated in liver cancer (HULC) serves as a specific non-invasive biomarker for HCC due to its overexpression in both tumors and plasma of HCC patients.89 In colorectal cancer, it is not expressed in primary tumors but is detected in colorectal cancers metastasized to liver showing its specificity for metastases.90, 91 HULC overexpression in gastric cancer was found to be correlated with EMT, lymph node metastasis, distant metastasis, and advanced tumor node metastasis stage and silencing of HULC effectively reversed the EMT phenotype, indicating its potential value as a prognostic factor.92 Similarly, high lncRNA-ATB expression is significantly associated with greater tumor size, depth of tumor invasion, lymphatic invasion, vascular invasion, and lymph node metastasis.34 High level of lncRNA-ATB could also predispose breast cancer patients to EMT and trastuzumab resistance.36 Meanwhile, lncRNA H19 expresses at high levels in human cancer tissues, but is nearly undetectable in the surrounding normal tissue, indicating the potential diagnostic value of this lncRNA. Taken together, with growing numbers of lncRNAs being discovered and characterized, their value as diagnostic and prognostic markers of EMT and metastasis will be increasingly recognized.
Table 2. Overview of clinical lncRNA biomarkers relative to EMT in cancer metastasis.
| Cancer type | lncRNA | Biomarker usability potential | Ref. |
|---|---|---|---|
| Bladder cancer | UBC1 | High expression of UBC1 confers a worse prognosis, lymph node metastasis, and survival. | 60 |
| Breast cancer | lncRNA-ATB | High expression of LncRNA-ATB in breast cancer patients confers EMT and trastuzumab resistance. | 36 |
| Cervical cancer | lncRNA-EBIC | High expression of lncRNA-EBIC is associated with a recurrence and worse prognosis. | 61 |
| Colon cancer | BANCR | Overexpression of BANCR is associated with high lymph node metastasis and high tumor stage. | 51 |
| lncRNA-ATB | Overexpression of lncRNA-ATB confers bigger tumor size, and associates with high lymph node and hematogenous metastasis. | 34, 37 | |
| HOTAIR | High expression of HOTAIR is associated with high metastasis and worse prognosis. | 45 | |
| Epithelial ovarian cancer | HOTAIR | High expression of HOTAIR is associated with a worse prognosis. | 44 |
| Esophageal cancer | HOTAIR | High expression of HOTAIR is associated with a worse prognosis. | 46 |
| Gastric cancer | MANCR | High expression of MALAT2 is associated with a worse prognosis in stage II/III. | 52 |
| lncRNA-ATB | High expression of LncRNA-ATB is associated with a worse prognosis. | 33 | |
| HULC | Overexpression of HULC is associated with high lymph node metastasis. | 92 | |
| Linc00152 | Overexpression of Linc00152 is a diagnostic indicator of gastric cancer. | 84 | |
| SPRY4-IT1 | Low expression of SPRY4-IT1 confers a worse prognosis. | 88 | |
| HOTAIR | High expression of HOTAIR is a predictor of recurrence liver transplantation. | 79 | |
| Hepatocellular Carcinoma | HULC | Overexpression of HULC is associated with high lymph node metastasis. | 89, 90 |
| linc-RoR | Overexpression of linc-RoR is a diagnostic indicator of HCC and chemoresistance. | 43 | |
| ZEB1-AS1 | High expression of ZEB1-AS1 confers a worse prognosis. | 80 | |
| Lung cancer | BANCR | Low expression of BANCR confers a worse prognosis. | 85 |
| MALAT1 | Overexpression of MALAT1 is associated with high lung cancer brain metastasis. | 86, 87 | |
| SPRY4-IT1 | Low expression of SPRY4-IT1 confers a worse prognosis. | 62 |
Perspectives on therapeutic strategies targeting lncRNAs
With the emergence of lncRNAs as important regulators of EMT, there will be increasing demand for lncRNA-based cancer therapy. Currently, there are more than 100 clinical trials on cancer are ongoing using EMT as a keyword. These trials will help us to determine the clinical context where we could use EMT to optimize treatments for cancer patients.93 LncRNAs can be targeted therapeutically by a variety of approaches including (i) RNA interference (RNAi)-mediated downregulation of specific lncRNAs; (ii) antisense oligonucleotides (ASO)-based therapy; (iii) plasmid-based therapy; (iv) lncRNA mimics or small-molecule inhibitors; (v) gene therapy, and so on.94 Although RNAi technology offers immense therapeutic promise due to its high potency, the main obstacle of siRNA/shRNA-based therapeutics remains its poor delivery in vivo. In contrast to RNAi, ASOs are synthetic, short, single-stranded DNAs or RNAs (between 8 and 50 nt), another type of nucleic acid drugs, which are designed with sequence specificity to target lncRNAs. ASOs are widely used as gene knockdown reagents in tissue culture and in Xenopus, Zebrafish, and mouse model systems.95, 96 It has been shown that subcutaneous injection of MALAT1-targeting ASOs into a mouse xenograft model blocks the lung cancer metastasis effectively.97 In addition, small molecules can be synthesized to specifically bind to RNA-binding pockets of lncRNAs. They compete with protein factors or intracellular small ligands for binding of lncRNAs. Binding of small molecules may also induce conformational changes within lncRNA molecules and disrupt formation of important lncRNA structures.98 For instance, the interaction of HOTAIR with PRC2 or LSD1 can be inhibited with the help of HOTAIR-targeting small molecular inhibitors to reduce the metastasis in breast cancer.99 Overall, the development of new technologies to more efficient delivery of lncRNA-targeted therapeutics will help to bring lncRNA-based therapies closer to the clinic.
Conclusions and Future Perspectives
EMT is a complex, multifunctional, and tightly regulated process that plays a critical role in metastatic spread of cancer cells. EMT-activating signaling pathways and downstream transcription factors are responsible for driving EMT and conferring aggressive mesenchymal phenotypes to epithelial cells. Over the past few years, lncRNAs are emerging as promising biomarkers and therapeutic targets for EMT and metastasis. Accumulating evidence has indicated that lncRNAs as a new class of ncRNAs are dysregulated to impact epithelial plasticity by targeting different signaling pathways, EMT-TFs, and EMT-related targets in a variety of cancers.78, 100, 101, 102 The distribution and levels of lncRNAs in various locations including distal metastases, have been exploited as potential diagnostic and prognostic biomarkers for cancer. Technologies have been advanced to achieve more sensitive and reliable detection and effective targeting of lncRNAs for cancer treatment. Despite these advances, there remain many challenges, such as limited knowledge of lncRNA functional mechanisms, targets, and binding partners, the challenges of effective delivery, stability, immunogenicity, and bioavailability of lncRNA-targeted therapeutics, which all will be tremendous tasks to undertake for the future studies. Overall, lncRNAs have shed new lights on our understanding of cancer pathways and brought our understanding of oncogenesis to a new horizon. Understanding the different and precise molecular mechanisms by which functional lncRNAs switch EMT on and off is important for opening up new avenues in lncRNA-directed diagnosis, prognosis, and therapeutic intervention against cancer.
Acknowledgments
This work was supported in part by the National Institutes of Health grant R01CA142580 and R21NS096946 (QJW), National Nature Science Foundation of China grant NSFC81370449 (AJ), and Oversea Hong Kong & Macao Scholars Collaborative Research Fund of NSFC in China (grant no. 81328020, FD and QJW).
Glossary
- ASOs
antisense oligonucleotides
- BANCR
BRAF-activated non-coding RNA
- CAF
cancer-associated fibroblasts
- ceRNAs
competing endogenous RNAs
- CSCs
cancer stem cells
- EGF
epidermal growth factor
- EMT
epithelial-mesenchymal transition
- EMT-TFs
EMT-inducing transcription factors
- EZH2
enhancer of zeste homolog 2
- FGF
fibroblast growth factor
- HGF
hepatocyte growth factor
- HGF/SF
hepatocyte growth factor/scatter factor
- Hh
hedgehog signaling
- HIF-1α
hypoxia-inducible factor-1α
- HULC
highly upregulated in liver cancer
- H3K27me3
histone H3 lysine 27 trimethylation
- lncRNAs
long non-coding RNAs
- lncRNA TUG1
taurine upregulated gene
- lncRNA-UBC1
upregulated in bladder cancer 1
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- MALAT2
metastasis-associated lung adenocarcinoma transcript 2
- MALAT2 MANCR
MALAT2-activated lncRNA
- MET
mesenchymal-to-epithelial transition
- miRNA
microRNA
- ncRNAs
non-coding RNAs
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- PRC2
polycomb-repressive complex
- RNAi
RNA interference
- SPRY4-IT1
SPRY4 intronic transcript 1
- TF
transcription factors
- TGF-β
transforming growth factor-β
- TβRII and TβRI
type II and type I serine–threonine kinase receptors
- UBC
urothelial bladder cancer
- VEGF
vascular endothelial growth factor
- WIF-1
Wnt inhibitory factor 1
- ZEB1-AS1
ZEB1 antisense 1
The authors declare no conflict of interest.
Footnotes
Edited by R Johnstone
References
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011; 331: 1559–1564. [DOI] [PubMed] [Google Scholar]
- Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science 2013; 342: 1234850. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139: 871–890. [DOI] [PubMed] [Google Scholar]
- Serviss JT, Johnsson P, Grander D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet 2014; 5: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13: 97–110. [DOI] [PubMed] [Google Scholar]
- van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci 2008; 65: 3756–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SH, Wang LH. Regulation of cancer metastasis by microRNAs. J Biomed Sci 2015; 22: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 2009; 119: 1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Tillo E, Liu Y, de Barrios O, Siles L, Fanlo L, Cuatrecasas M et al. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci 2012; 69: 3429–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunen D, Willems SM, Kellner U, Midgley R, Simon I, Bernards R. TGF-beta: an emerging player in drug resistance. Cell Cycle 2013; 12: 2960–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res 2009; 19: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Hebert MC, Zhang YE. TGF-beta receptor-activated p38 MAP kinase mediates Smad-independent TGF-beta responses. EMBO J 2002; 21: 3749–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande M, Franzen A, Karlsson JO, Ericson LE, Heldin NE, Nilsson M. Transforming growth factor-beta and epidermal growth factor synergistically stimulate epithelial to mesenchymal transition (EMT) through a MEK-dependent mechanism in primary cultured pig thyrocytes. J Cell Sci 2002; 115(Pt 22): 4227–4236. [DOI] [PubMed] [Google Scholar]
- Garg M. Targeting microRNAs in epithelial-to-mesenchymal transition-induced cancer stem cells: therapeutic approaches in cancer. Expert Opin Ther Targets 2015; 19: 285–297. [DOI] [PubMed] [Google Scholar]
- Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol 2006; 8: 1398–1406. [DOI] [PubMed] [Google Scholar]
- Howe LR, Watanabe O, Leonard J, Brown AM. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res 2003; 63: 1906–1913. [PubMed] [Google Scholar]
- Bailey JM, Mohr AM, Hollingsworth MA. Sonic hedgehog paracrine signaling regulates metastasis and lymphangiogenesis in pancreatic cancer. Oncogene 2009; 28: 3513–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Ma L, Zhang Z, Liu X, Gao H, Zhuang Y et al. Hedgehog signaling regulates epithelial-mesenchymal transition in pancreatic cancer stem-like cells. J Cancer 2016; 7: 408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev 2007; 3: 30–38. [DOI] [PubMed] [Google Scholar]
- Bao B, Azmi AS, Ali S, Ahmad A, Li Y, Banerjee S et al. The biological kinship of hypoxia with CSC and EMT and their relationship with deregulated expression of miRNAs and tumor aggressiveness. Biochim Biophys Acta 2012; 1826: 272–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren K, Nordenskjold B, Landberg G. Hypoxia, Snail and incomplete epithelial-mesenchymal transition in breast cancer. Br J Cancer 2009; 101: 1769–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamachary B, Zagzag D, Nagasawa H, Rainey K, Okuyama H, Baek JH et al. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res 2006; 66: 2725–2731. [DOI] [PubMed] [Google Scholar]
- Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 2008; 27: 6958–6969. [DOI] [PubMed] [Google Scholar]
- Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell 2011; 145: 178–181. [DOI] [PubMed] [Google Scholar]
- Guo F, Parker Kerrigan BC, Yang D, Hu L, Shmulevich I, Sood AK et al. Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions. J Hematol Oncol 2014; 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meseure D, Drak Alsibai K, Nicolas A, Bieche I, Morillon A. Long noncoding RNAs as new architects in cancer epigenetics, prognostic biomarkers, and potential therapeutic targets. Biomed Res Int 2015; 2015: 320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget 2014; 5: 8027–8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014; 505: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu L, Wang Y, Zhao G, Xie R, Liu C et al. KLF8 involves in TGF-beta-induced EMT and promotes invasion and migration in gastric cancer cells. J Cancer Res Clin Oncol 2013; 139: 1033–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014; 25: 666–681. [DOI] [PubMed] [Google Scholar]
- Saito T, Kurashige J, Nambara S, Komatsu H, Hirata H, Ueda M et al. A long non-coding RNA activated by transforming growth factor-beta is an independent prognostic marker of gastric cancer. Ann Surg Oncol 2015; 22(Suppl 3): S915–S922. [DOI] [PubMed] [Google Scholar]
- Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res 2015; 35: 1385–1388. [PubMed] [Google Scholar]
- Li W, Kang Y. A new Lnc in metastasis: long noncoding RNA mediates the prometastatic functions of TGF-beta. Cancer Cell 2014; 25: 557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget 2015; 6: 11652–11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F et al. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol 2015; 31: 595–603. [DOI] [PubMed] [Google Scholar]
- Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res 2014; 20: 1531–1541. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Lu Q, Shen B, Huang X, Shen L, Zheng X et al. TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci Rep 2015; 5: 11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Zhang G, Li ZP, Permuth-Wey J, Challa S, Li Y et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) beta: LncRNA-hit-mediated TGFbeta-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem 2015; 290: 6857–6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouk IJ, Raveh E, Abu-lail R, Mezan S, Gilon M, Gershtain E et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta 2014; 1843: 1414–1426. [DOI] [PubMed] [Google Scholar]
- Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta 2010; 1803: 443–451. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci 2014; 127(Pt 7): 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Chen J, Tang W. The molecular mechanism of HOTAIR in tumorigenesis, metastasis, and drug resistance. Acta Biochim Biophys Sin (Shanghai) 2014; 46: 1011–1015. [DOI] [PubMed] [Google Scholar]
- Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep 2014; 32: 395–402. [DOI] [PubMed] [Google Scholar]
- Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci 2013; 104: 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Liu Y, Nie L, Gui Y, Cai Z. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology 2013; 81: 209.e201–207. [DOI] [PubMed] [Google Scholar]
- Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst 2012; 8: 2289–2294. [DOI] [PubMed] [Google Scholar]
- Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett 2013; 333: 213–221. [DOI] [PubMed] [Google Scholar]
- Leicht DT, Balan V, Kaplun A, Singh-Gupta V, Kaplun L, Dobson M et al. Raf kinases: function, regulation and role in human cancer. Biochim Biophys Acta 2007; 1773: 1196–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D et al. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett 2014; 8: 869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tian Y, Pang EJ, Wang Y, Li L. MALAT2-activated long noncoding RNA indicates a biomarker of poor prognosis in gastric cancer. Cancer Gene Ther 2015. (advance online publication; doi:10.1002/mc.22338). [DOI] [PubMed]
- Zhou M, Hou Y, Yang G, Zhang H, Tu G, Du YE et al. LncRNA-Hh strengthen cancer stem cells generation in twist-positive breast cancer via activation of hedgehog signaling pathway. Stem Cells 2016; 34: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008; 27: 7274–7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 2009; 106: 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 2013; 20: 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res 2012; 72: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res 2011; 39: 2119–2128. [DOI] [PubMed] [Google Scholar]
- He W, Cai Q, Sun F, Zhong G, Wang P, Liu H et al. linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim Biophys Acta 2013; 1832: 1528–1537. [DOI] [PubMed] [Google Scholar]
- Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang SB et al. Long noncoding RNA-EBIC promotes tumor cell invasion by binding to EZH2 and repressing E-cadherin in cervical cancer. PLoS One 2014; 9: e100340. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis 2014; 5: e1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495: 384–388. [DOI] [PubMed] [Google Scholar]
- Saito Y, Suzuki H, Matsuura M, Sato A, Kasai Y, Yamada K et al. MicroRNAs in Hepatobiliary and Pancreatic Cancers. Front Genet 2011; 2: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z et al. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol 2014; 35: 9163–9169. [DOI] [PubMed] [Google Scholar]
- Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015; 6: 22513–22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Ye F, Yin C, Zhuang Y, Yue G, Zhang G. The interaction between MiR-141 and lncRNA-H19 in regulating cell proliferation and migration in gastric cancer. Cell Physiol Biochem 2015; 36: 1440–1452. [DOI] [PubMed] [Google Scholar]
- Carpenter RL, Paw I, Dewhirst MW, Lo HW. Akt phosphorylates and activates HSF-1 independent of heat shock, leading to Slug overexpression and epithelial-mesenchymal transition (EMT) of HER2-overexpressing breast cancer cells. Oncogene 2015; 34: 546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014; 13: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH et al. Linc HOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis 2015; 6: e1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JT, Wang LF, Zhao YL, Yang T, Li W, Zhao J et al. Nuclear factor of activated T cells 5 maintained by HOTAIR suppression of miR-568 upregulates S100 calcium binding protein A4 to promote breast cancer metastasis. Breast Cancer Res 2014; 16: 454. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Inamoto T, Taniguchi K, Takahara K, Iwatsuki A, Takai T, Komura K et al. Intravesical administration of exogenous microRNA-145 as a therapy for mouse orthotopic human bladder cancer xenograft. Oncotarget 2015; 6: 21628–21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Qiu K, Li M, Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett 2015; 589(20 Pt B): 3175–3181. [DOI] [PubMed] [Google Scholar]
- Cong N, Du P, Zhang A, Shen F, Su J, Pu P et al. Downregulated microRNA-200a promotes EMT and tumor growth through the wnt/beta-catenin pathway by targeting the E-cadherin repressors ZEB1/ZEB2 in gastric adenocarcinoma. Oncol Rep 2013; 29: 1579–1587. [DOI] [PubMed] [Google Scholar]
- Xue M, Pang H, Li X, Li H, Pan J, Chen W. Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci 2016; 107: 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 2014; 5: e1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta 2015; 1859: 169–176. [DOI] [PubMed] [Google Scholar]
- Han Y, Ye J, Wu D, Wu P, Chen Z, Chen J et al. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. BMC Cancer 2014; 14: 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L et al. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci 2013; 9: 587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene 2015; 35: 1575–1584. [DOI] [PubMed] [Google Scholar]
- Ye P, Wang T, Liu WH, Li XC, Tang LJ, Tian FZ. Enhancing HOTAIR/MiR-10b drives normal liver stem cells toward a tendency to malignant transformation through inducing epithelial- to-mesenchymal transition. Rejuvenation Res 2015; 18: 332–340. [DOI] [PubMed] [Google Scholar]
- Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN et al. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology 2013; 57: 1882–1892. [DOI] [PubMed] [Google Scholar]
- Wang TH, Lin YS, Chen Y, Yeh CT, Huang YL, Hsieh TH et al. Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transitionl. Oncotarget 2015; 6: 23342–23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle 2015; 14: 3112–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer 2014; 13: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Huang C, Meng X, Li J. Long noncoding RNA MALAT1insights into its biogenesis and implications in human disease. Curr Pharm Des 2015; 21: 5017–5028. [DOI] [PubMed] [Google Scholar]
- Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol 2015; 121: 101–108. [DOI] [PubMed] [Google Scholar]
- Xie M, Nie FQ, Sun M, Xia R, Liu YW, Zhou P et al. Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J Transl Med 2015; 13: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007; 132: 330–342. [DOI] [PubMed] [Google Scholar]
- Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int 2013; 2013: 136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol 2009; 21: 688–692. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep 2014; 31: 358–364. [DOI] [PubMed] [Google Scholar]
- Pasquier J, Abu-Kaoud N, Al Thani H, Rafii A. Epithelial to mesenchymal transition in a clinical perspective. J Oncol 2015; 2015: 792182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S, Xiao YF, Tang B, Hu CJ, Xie R, Yang SM et al. Long noncoding RNA in digestive tract cancers: function, mechanism, and potential biomarker. Oncologist 2015; 20: 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Montague TG, Lennox KA, Behlke MA, Schier AF. Antisense oligonucleotide-mediated transcript knockdown in zebrafish. PLoS One 2015; 10: e0139504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 2012; 488: 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013; 73: 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Chua MS, Andrisani O, So S. Epigenetics in hepatocellular carcinoma: an update and future therapy perspectives. World J Gastroenterol 2014; 20: 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res 2011; 71: 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhu L, Xu L, Qin K, Liu C, Yu Y et al. Long noncoding RNA linc00617 exhibits oncogenic activity in breast cancer. Mol Carcinog 2015. [DOI] [PubMed]
- Wu J, Zhang J, Shen B, Yin K, Xu J, Gao W et al. Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J Exp Clin Cancer Res 2015; 34: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W, Wang L, Lu J, Mu J, Liu Y, Dong P. Long noncoding RNA KIAA0125 potentiates cell migration and invasion in gallbladder cancer. Biomed Res Int 2015; 2015: 108458. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]