Abstract
A DNA gap repair assay was used to determine the effect of mutations in the DNA damage checkpoint system on the efficiency and outcome (crossover/non-crossover) of recombinational DNA repair. In Saccharomyces cerevisiae gap repair is largely achieved by homologous recombination. As a result the plasmid either integrates into the chromosome (indicative of a crossover outcome) or remains extrachromosomal (indicative of a non-crossover outcome). Deletion mutants of the MEC1 and RAD53 checkpoint kinase genes exhibited a 5-fold decrease in gap repair efficiency, showing that 80% of the gap repair events depended on functional DNA damage checkpoints. Epistasis analysis suggests that the DNA damage checkpoints affect gap repair by modulating Rad51 protein-mediated homologous recombination. While in wild-type cells only ∼25% of the gap repair events were associated with a crossover outcome, Mec1-deficient cells exhibited a >80% crossover association. Also mutations in the effector kinases Rad53, Chk1 and Dun1 were found to affect crossover association of DNA gap repair to various degrees. The data suggest that the DNA damage checkpoints are important for the optimal functioning of recombinational DNA repair with multiple terminal targets to modulate the efficiency and outcome of homologous recombination.
INTRODUCTION
Maintaining the integrity of the genome is critical for the survival of any living organism. DNA double-stranded breaks (DSBs) are a potentially lethal form of DNA damage. Ionizing radiation and a host of drugs can induce DSBs directly or indirectly, which determines their therapeutic value in cancer treatment (1). DSBs may also result from intrinsically generated DNA damage (reactive oxygen species) and from the processing of stalled replication forks during DNA replication (2,3). Programmed DSBs are known to initiate regulated recombination events such as mating-type switching in Saccharomyces cerevisiae and meiotic recombination in presumably all eukaryotes as well as the assembly of functional B- and T-cell receptor genes (4,5). Eukaryotes maintain two major pathways for the repair of DSBs, homologous recombination and non-homologous endjoining (NHEJ) (4,6). Homologous recombination typically uses the sister chromatid or homolog as a template to repair DSBs in a largely error-free way, whereas NHEJ directly joins DNA ends in a template-independent manner that, depending on the exact DNA end structure, leads to error-prone DSB repair.
In the yeast S.cerevisiae, DSBs are primarily repaired by the RAD52-dependent homologous recombination pathway when a homologous DNA sequence is available. In the absence of available homology or in mutants affecting homologous recombination, NHEJ rejoins DSBs with varying efficiency depending on the exact end structure (4,6). Homologous recombination involves several sub-pathways with distinct substrate and genetic requirements. DSBs are processed to generate 3′-OH-ending single-stranded DNA tails, to which the single-stranded DNA binding protein RPA appears to bind first (4,7). In the absence of a homolog or sister chromatid, single-strand annealing (SSA) is an efficient pathway that utilizes repeated DNA sequences flanking a DSB leading to a deletion of the intervening sequence. SSA requires the Rad52 and Rad59 proteins but not the other members of the RAD52 epistasis group (4,6,8,9). In the presence of a homolog or sister chromatid, RPA is replaced on the single-stranded DNA by Rad51 protein in a reaction involving the Rad52 protein and the Rad55–Rad57 heterodimer (10–13). The Rad51 filament, presumably in conjunction with the dsDNA-specific ATPase Rad54, performs homology search and DNA strand invasion, forming a D-loop that connects the broken duplex to the intact template (14–18). After a poorly understood transition that probably involves the remodeling of the Rad51-heteroduplex DNA complex by Rad54 protein (19–21), DNA polymerase gains access to the invading 3′ end and initiates DNA synthesis, stabilizing the D-loop and copying the DNA missing on the broken chromosome. Capture of the second end by the displaced strand in the D-loop may lead to the establishment of a double Holliday junction in a pathway termed double-strand break repair (DSBR) (6). The double Holliday junction requires resolution by a structure-specific endonuclease, which may lead to a crossover or non-crossover outcome, or dissolution by the combined action of a DNA helicase with a type I topoisomerase, which will lead to a non-crossover outcome exclusively (22). Biochemical and genetic evidence suggests a role for RecQ-like DNA helicases in conjunction with topoisomerase III in the dissolution of double Holliday junctions to non-crossover products (23–26). Endonucleolytic resolution of double Holliday junctions in eukaryotes may involve multiple activities depending on the organism and the context (DNA repair in vegetative/somatic cells, meiotic recombination). Among these are the Mus81–Mms4/Eme1 structure-specific endonuclease, the presumable endonuclease encoded by the Drosophila MEI-9 and MUS312 genes, and an unidentified mammalian catalytic activity that depends on the Rad51 paralogs Xrcc3 and Rad51C (27–31). In the absence of second end capture, the invading strand may reverse the D-loop after DNA synthesis to anneal directly with the resected second end. This pathway has been termed synthesis-dependent strand annealing (SDSA) (4). SDSA also depends on the Rad51, Rad52, Rad55, Rad57 and Rad54 proteins but lacks a requirement to process double Holliday junctions, although some structure-specific nucleases may be required to process flap structures that can form during the annealing step (32).
Homologous recombination is not a constitutive DNA repair pathway but highly regulated at multiple levels. Single-stranded DNA gaps, nicks and DSBs (processed into single-stranded DNA tails) are known to be highly recombinogenic (33–35). The constant presence of single-stranded DNA at the replication fork during ongoing DNA replication necessitates negative regulation of recombination to avoid interference with ongoing DNA replication in the absence of replication fork stalling or DNA damage. The Srs2 DNA helicase is a negative regulator of homologous recombination, shunting potential substrates to other postreplication repair pathways including translesion synthesis (36–38). Srs2 functions by dissociating the Rad51 nucleoprotein filament, preventing the formation of D-loops (39,40). Positive regulation of recombinational repair at the level of nucleation of the Rad51 filament has been proposed to occur under genotoxic stress through the phosphorylation of the Rad55–Rad57 heterodimer (41). It is presently unclear, what regulatory processes are involved to decide usage of different DSB repair pathways (NHEJ versus homologous recombination) and different sub-pathways of homologous recombination (SSA, DSBR, SDSA).
The DNA damage and replication block checkpoints in eukaryotes monitor the structural integrity of the genome and coordinate the cellular responses to DNA damage and stalled replication forks (42–44). Several pathways activate DNA damage checkpoints during the S- and the G1/G2 phases of the cell cycle to control a complex web of protein kinases that govern the genotoxic stress response including transient cell cycle arrests in G1 and G2, the suppression of late-firing origins, transcriptional induction of target genes, the relocalization of telomeric proteins and in complex organisms DNA-damage-induced apoptosis (42–44). The PI3 kinase-like-kinase Mec1 (ATR in mammals; Rad3 in Schizosaccharomyces pombe) is the central checkpoint protein in S.cerevisiae. The known checkpoint responses are virtually eliminated in mec1 null mutant cells. Activated Mec1 kinase transduces the checkpoint signal by direct phosphorylation of the FHA-domain kinase Rad53 (Chk2 in mammals; Cds1 in S.pombe) and Chk1 kinase (Chk1 in both mammals and S.pombe) (42,44). Dun1, another FHA-domain checkpoint kinase in budding yeast, can be directly activated by Rad53 kinase through direct phosphorylation but also exhibits Rad53-independent functions (45,46). Tel1 kinase (a related PI3 kinase-like-kinase, ATM in mammals; Tel1 in S.pombe) appears to play only a minor checkpoint role in wild-type S.cerevisiae, unlike its mammalian counterpart ATM. All checkpoint kinases are inferred to have specific terminal target proteins in the effector pathways controlled by the checkpoints. However, it is unclear, how much the individual pathways (e.g. cell cycle arrest, transcriptional induction) contribute to survival. Artificially imposed cell cycle arrest cannot rescue the DNA damage sensitive phenotype in budding yeast mec1 or mammalian ATM-defective cells, a phenomenon termed cell cycle arrest-independent radiosensitivity in mammals (41,47). This suggests that Mec1/ATM-dependent pathways other than cell cycle arrest make major contributions to survival after genotoxic stress.
The functioning of DNA repair pathways is paramount for cells to survive genotoxic stress. It has been proposed that the DNA damage checkpoints directly interface with DNA repair pathways in analogy to the SOS system of bacteria (43). A number of proteins involved in DNA repair have been identified as phosphorylation targets of checkpoint kinases, including the single-stranded DNA binding protein RPA (48), the Mre11–Rad50–Xrs2(Nbs1) complex (49) and the Srs2 helicase (50). The biological significance and the mechanistic consequences of these phosphorylation events remain unclear. These proteins also function in the DNA damage checkpoint and it is unclear if phosphorylation affects their checkpoint or their DNA repair function or both. A direct control of DNA repair pathways, specifically of homologous recombination, by the DNA damage checkpoints is suggested by the original discovery of Mec1 in budding yeast as a meiotic recombination mutant, esr1-1 (51). In vegetative cells, esr1-1 (mec1) cells were shown to be almost entirely defective for DNA damage-induced recombination (41). The Rad55–Rad57 heterodimer, which probably functions during nucleation of the central Rad51 filament during recombination (10), is a terminal target of the DNA damage checkpoints (41). Rad55–Rad57 is unlikely to have a function in the checkpoint itself, as the DNA damage-induced G2 cell cycle arrest and transcriptional response were determined to be normal in rad55- or rad57-mutant cells (41). The mechanistic impact of checkpoint-mediated control of homologous recombination through Rad55–Rad57 phosphorylation or additional DNA repair pathway through other target proteins remains to be determined.
Plasmid-based transformation assays have been pivotal in understanding the mechanism of homologous recombination and have led directly to the development of the DSBR model of recombination (33,52). Repair of DSBs and double-stranded DNA gaps in a plasmid-borne DNA sequence with homology to a chromosomal template recapitulates the genetic requirements and characteristics of the repair of chromosomal DSBs to a significant extent. As the repair of chromosomal DSBs, plasmid gap repair in wild-type S.cerevisiae cells is primarily executed by the RAD52 recombinational repair pathway involving the Rad51, Rad52, Rad54, Rad55 and Rad57 proteins (33,53–55). Like chromosomal recombination, plasmid gap repair can be associated with a crossover event (56) and the low frequency of crossover association resembles that found for chromosomal recombination in vegetative cells (4,6,55,57,58). Little is known about, how the crossover/non-crossover outcome of homologous recombination is regulated.
We are interested in the regulation of homologous recombination by the DNA damage checkpoints, and we followed up an earlier report of Symington and colleagues (55), who reported a small but significant 2-fold decrease in the efficiency of DNA gap repair in a hypomorphic mutation of the RAD53 checkpoint kinase gene using an elegant plasmid-based gap repair assay. This assay not only allowed a direct quantitation of the gap repair efficiency but also afforded the opportunity to easily distinguish between crossover and non-crossover gap repair events. Using null mutants in the major checkpoint kinase genes MEC1, RAD53, CHK1 and DUN1 we show that 80% of gap repair depends on functioning DNA damage checkpoints. Moreover, we demonstrate that the checkpoints control the crossover/non-crossover outcome during gap repair. The data provide evidence for multiple checkpoint targets in the recombinational repair pathway that influence repair efficiency and outcome.
MATERIALS AND METHODS
Strains and media
All strains used in this study are derivatives of W303 with the corrected RAD5 allele and are listed in Table 1. Strains containing the ADE2 wild type and met17-s alleles [wild type as LSY697, rad51::LEU2 as LSY826, rad52::LEU2 as LSY718 (55)] were kindly supplied by Dr L. Symington (Columbia University). Strains containing the TRP1::GAP-RNR1 cassette [wild type as DES460, mec1::HIS3 as DES459, rad53::HIS3 as DES453, dun1::HIS3 as MHY26 (59)] were kindly provided by Dr S. Elledge (Harvard University). PCR-product-mediated transformation was used to generate the chk1::KAN allele using pFA-kanMX6 and primers CHK1-F 5′-TATCATAAGTTGCTGTATATGGGCAGCACGTATTACTCGTACGCTGCAGGTCGACGG-3′ and CHK1-R 5′-GCATCTTAACCCTTCTTTTGTCTCCATTTTTTTCAGTCATCGATGAATTCGAGCTCG-3′ (60). The disruption was verified by Southern analysis. PCR was used to monitor the rad5-535/RAD5 alleles in genetic crosses (61). The final WDHY strains listed in Table 1 were generated by tetrad analysis and only strains from tetrads with four viable spores were chosen.
Table 1. S.cerevisiae strains used in this study.
| Strain | Relevant genotypea | Reference |
|---|---|---|
| LSY697 | (55) | |
| WDHY1800 | TRP1::GAP-RNR1 | This study |
| WDHY1814 | TRP1::GAP-RNR1 mec1::HIS3 | This study |
| WDHY1817 | TRP1::GAP-RNR1 rad53::HIS3 | This study |
| WDHY1819 | TRP1::GAP-RNR1 chk1::KAN | This study |
| WDHY1820 | TRP1::GAP-RNR1 rad53::HIS3 chk1::KAN | This study |
| WDHY1827 | TRP1::GAP-RNR1 dun1::HIS3 | This study |
| WDHY1832 | TRP1::GAP-RNR1 rad51::LEU2 | This study |
| WDHY1834 | TRP1::GAP-RNR1 rad52::LEU2 | This study |
| WDHY1928 | TRP1::GAP-RNR1 mec1::HIS3 rad51::LEU2 | This study |
aAll strains are W303 derivatives with the common genotype: MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 met17-s ADE2 RAD5. The WDHY strains contain TRP1::GAP-RNR1 to suppress the mec1 and rad53 lethality. LSY697 is a kind gift of Dr Symington (Columbia University).
Standard genetic techniques and media were used to grow and manipulate yeast strains (62). YPD medium contained 2% glucose, 2% peptone and 1% yeast extract. For solid media, 2% agar was added. Synthetic complete (SC) medium lacked cysteine, since this amino acid can be converted into methionine via MET17-independent pathways (63). SC-met or SC-ura indicates SC media from which uracil or methionine, respectively, was selectively omitted to allow selection for the corresponding wild-type genes. Lead plates contained 4% glucose, 0.3% peptone, 0.5% yeast extract, 0.02% ammonium sulfate, 2% agar (55). After autoclaving the agar solution was cooled to 65°C and 0.1% lead nitrate (64), Pb(NO3)2, was added from a 10% (w/v) stock solution. 5-FOA plates were prepared by dissolving 1 g of 5-FOA powder in 1 l of SC held at 60°C. All experiments were performed at 30°C.
Yeast transformation
Yeast cells were transformed using the lithium acetate method (65). Specifically, 10 ml log-phase cells (OD 0.6–0.9) per transformation were sedimented at 4000 r.p.m. (Beckman Allegra 6), washed once in 100 mM lithium acetate (LiOAc) solution and resuspended in 100 μl of 100 mM LiOAc. The cell suspension was incubated at 30°C with gentle agitation for 1 h. Circular- or linear plasmid DNA (100 ng) along with 5 μl of carrier DNA (10 μg/μl) were added to the 100 μl cell suspension. The cell suspension was further incubated with gentle agitation at 30°C for 30 min. A freshly prepared 40% polyethylene glycol (PEG) solution (700 μl) was then added to the transformation tube and it was incubated with gentle agitation at 30°C for 1 h. Cells were then heat shocked in a 42°C water bath for 15 min. Afterwards, cells were sedimented at 3000 r.p.m., twice, and the PEG was removed by aspiration. Cells were then resuspended in 200 μl of liquid YPD and incubated at 30°C for 20 min with gentle agitation. Subsequently, cells were diluted and plated onto the appropriate media.
Gap repair assay
The gap repair assay developed by Dr Symington (Columbia University) was used essentially as described (55). Briefly, plasmid pSB110, an ars plasmid containing the URA3 and MET17 markers, was digested with EcoNI and BspEI to create a 238 bp gap within MET17 open reading frame (ORF) with non-compatible 3′ overhanging ends of 1 and 4 bases, respectively. The linearized substrate was subsequently gel-purified. Transformation using 100 ng of linear- or circular DNA was performed as described above. After the transformation process cells were diluted and plated onto SC-ura-met and SC-ura plates. The gap repair frequency was calculated as the number of URA+ MET+ transformants obtained from transformation with linear DNA divided by the number of URA+ MET+ transformants obtained from transforming circular DNA as described (55).
For the genetic analysis, representative URA+ transformants with no size or shape bias, were picked to SC-ura master plates, and further incubated for 2 days to ensure that they are URA+. The URA+ transformants were replica-plated onto SC-met and YPD plates, to determine the percentage of the URA+ transformants that were also MET+, and to allow non-selective growth, respectively. The plates were incubated at 30°C for 2–3 days, after which the YPD plates were again replica-plated onto 5-FOA and lead plates, to assess the mitotic stability of URA3 and MET17 markers, respectively. After 3 days of growth at 30°C, plates were scored as follows: URA+ MET+ cells that were 5-FOA resistant and grew as black/brown patches on lead plates represented unstable (no crossover), URA+u MET+u, transformants. URA+ MET+ cells that were 5-FOA sensitive and grew as white patches on lead plates represented stable (with crossover), URA+s MET+s, transformants (see Figure 1). The statistical significance of the data was evaluated in a chi-square test.
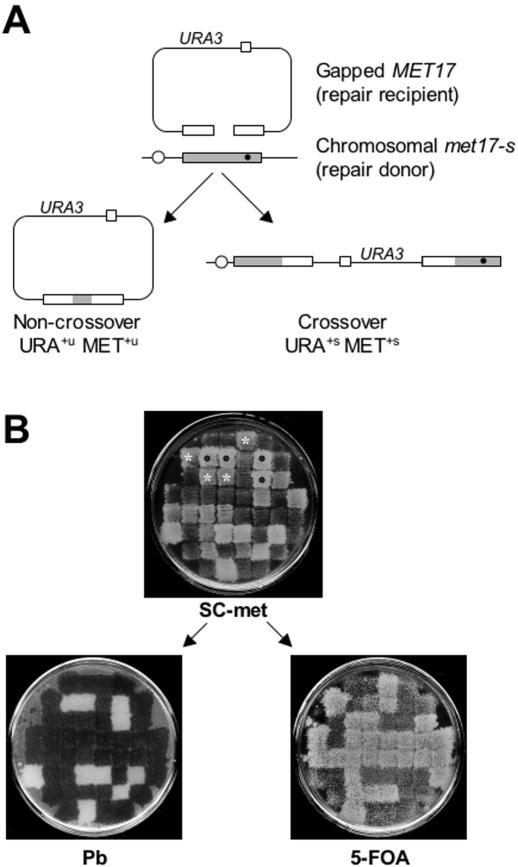
Figure 1.
The double-stranded DNA gap repair system. (A) The ars plasmid containing the URA3 and MET17 markers is digested with BspEI and EcoNI to create a 238 bp gap leaving 1177 and 1021 bp of homology to the chromosomal met17-s gene on either sides of the gap, respectively. The chromosomal met17-s mutation is 216 bp downstream of the break site and is indicated as a black dot. The repaired plasmid can remain episomal (non-crossover) resulting in unstable transformants (MET+u URA+u) or integrate in the chromosome (crossover) resulting in stable transformants (MET+s URA+s). The figure is not drawn to scale. (B) Genetic analysis of the repair events. URA+ transformants are replica-plated onto SC-met media to determine, which transformants are also MET+. After several generations of non-selective growth, the transformants are replica-plated on 5-FOA plates and YPD plates containing lead nitrate (labeled Pb). Black dots represent URA+ MET+ transformants that are stable integrants indicative of crossover (MET+, white on lead plates; URA+, no growth on 5-FOA plates) and the white stars represent URA+ MET+ transformants that are unstable indicative of non-crossover events (MET−, black on lead plates; URA−, confluent growth on 5-FOA plates due to concomitant loss of both markers through plasmid loss under non-selective growth).
Southern analysis
Genomic DNA was isolated from representative URA+u MET+u and URA+s MET+s transformants, doubly digested with BamHI and SnaBI, separated by electrophoresis on a 0.7% TBE gel, and subjected to Southern hybridization using a DIG-labeled BamHI/SnaBI MET17 (1.5 kb) probe.
Survival experiment
Wild-type and mec1 cells were transformed as described above. Aliquots of cells were removed at each point during the transformation process: after LiOAc wash, after 1 h incubation in LiOAc solution, after addition of DNA, after addition of PEG and after the 15 min heat shock. Cells were diluted and plated onto YPD plates to determine survival during transformation. Averages of the two experiments are presented.
Rad53 immunoblot
Total protein extracts were prepared using trichloroacetic acid precipitation (66) from the cells used in the survival experiment. Approximately 15 μg of whole cell protein extract per well were electrophoresed on a 9% polyacrylamide gel and analyzed by western blotting using anti-Rad53 antibodies (Santa Cruz Biotechnology).
RESULTS
Experimental system
To examine the effect of the DNA damage checkpoints on recombinational DNA repair, we utilized an elegant plasmid-based, double-strand gap repair assay developed by Symington and colleagues (55) (see Figure 1). The system employs the MET17 gene as a selectable marker. MET17 serves also as a color marker, as met17 mutants form dark brown colonies on lead-containing media, whereas MET17 wild-type cells form white colonies. A plasmid carrying a 238 bp double-stranded DNA gap in the MET17 gene is repaired by homologous recombination using the chromosomal mutant copy of the gene, met17-s, as a donor [(55); Table 2]. The plasmid also carries a URA3 marker to allow selection for URA+ and counter-selection for URA− on 5-FOA plates. Gap repair events by homologous recombination are selected for as MET+ URA+ transformants (Figure 1) and a mutation in the pivotal recombination gene RAD52 virtually eliminates gap repair leading to a 250 fold reduction (Table 2), consistent with the original analysis (55). The gap repair substrate plasmid contains an ars sequence allowing it to either integrate into the chromosome or remain extrachromosomal after gap repair. Integration of the plasmid during gap repair is indicative of an associated crossover and results in stable transformants (URA+s MET+s). Gap repair not associated with a crossover results in unstable transformants (URA+u MET+u) that can easily be identified by genetic means (Figure 1 and Materials and methods). Both types of transformants were also verified by physical means [(55); see Figure 4]. Gap repair efficiency is determined as the number of URA+ MET+ transformants using the gapped plasmid divided by the number of URA+ MET+ transformants with the circular plasmid. Therefore, the gap repair frequency is independent of the transformation efficiency of the individual strains. Hence, this system allows one to determine the efficiency of recombinational repair and its crossover/non-crossover outcome.
Table 2. Transformation efficiencies and repair frequencies.
| Relevant genotype | Transformation efficiency (×1000)a | Repair frequencyb | Fold reduction |
|---|---|---|---|
| WTc (LSY697) | 346 | 26.3 | 1 |
| WT (WDHY1800) | 387 | 24.6 | 1 |
| mec1 | 90 | 5.0 | 5 |
| rad53 | 12 | 5.4 | 5 |
| dun1 | 134 | 24.8 | 1 |
| chk1 | 213 | 30.7 | 1 |
| rad53 chk1 | 31 | 2.6 | 9 |
| rad51 | 157 | 0.8 | 31 |
| rad52 | 31 | 0.1 | 250 |
| mec1 rad51 | 10 | 0.8 | 31 |
aTransformation efficiencies (URA+ MET+ colonies) per 1 μg of circular plasmid DNA.
bRepair frequency (%): number of URA+ MET+ transformants obtained from the gapped plasmid divided by the number of URA+ MET+ transformants obtained from the circular plasmid × 100. Averages of at least three transformation experiments are presented.
cThis strain does not contain the RNR1 overexpression cassette TRP1::GAP-RNR1 (see Table 1). WT = wild type.
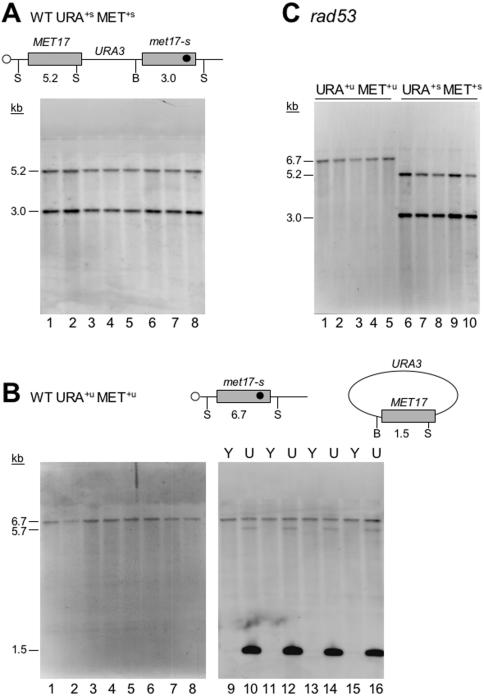
Figure 4.
Physical analysis of transformants. (A) Total DNA isolated from eight YPD-grown URA+s MET+s wild-type transformants was doubly digested with BamHI (B) and SnaBI (S) and subjected to Southern analysis using a MET17 probe. The digest pattern indicates a simple integration of the plasmid as shown schematically above the Southern blot. (B) Total DNA isolated from eight YPD-grown URA+u MET+u wild-type transformants was doubly digested with BamHI (B) and SnaBI (S) and subjected to Southern analysis using a MET17 probe. The 6.7 kb band is indicative of the chromosomal met17-s as shown schematically above the Southern blots (lanes 1 to 8). When DNA was isolated from SC-ura-grown (U) cultures to select for the plasmid (lanes 10, 12, 14, 16), a prominent 1.5 kb band appeared, which represents the repaired MET17 ORF on the plasmid. The 1.5 kb band is absent in total DNA from YPD-grown (Y) transformants (lanes 9, 11, 13, 15), indicative of plasmid loss. The faint 5.7 kb band observed in SC-ura-grown lanes most likely represents non-specific hybridization of the probe with the abundant digested plasmid (7.2 kb plasmid − 1.5 kb MET17 ORF = 5.7 kb). (C) Total DNA isolated from rad53 transformants was doubly digested with BamHI and SnaBI and subjected to Southern analysis using a MET17 probe. All URA+s MET+s and URA+u MET+u transformants from each class exhibited the expected digest pattern.
DNA damage checkpoints are activated and required for survival during transformation
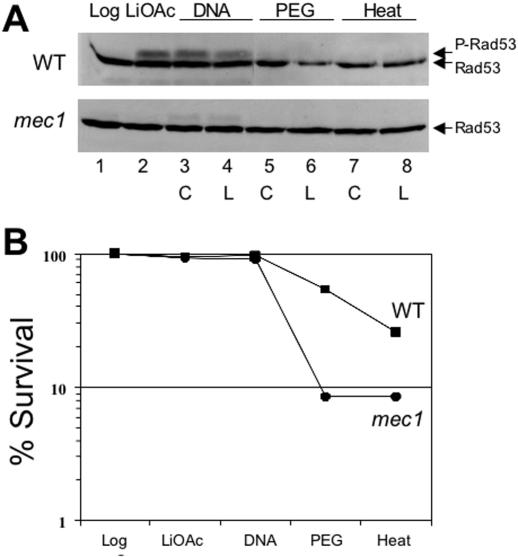
Transformation-based assays measuring homologous recombination between plasmid-borne and chromosomal DNA sequences have been pivotal in the development of the DSBR model for homologous recombination (33). The LiOAc-based transformation protocol is slightly mutagenic in yeast (unpublished data) and the transformation procedure introduces DNA with double-stranded ends that potentially could elicit a DNA damage checkpoint response during transformation. To monitor if the DNA damage checkpoint response is activated during the transformation procedure, we used immunoblots to determine the phosphorylation status of the Rad53 checkpoint kinase as a convenient marker of checkpoint activation (67). An aliquot of cells was removed at each step of the transformation protocol and analyzed for the Rad53 phosphorylation status. As shown in Figure 2A, Rad53 kinase is transiently phosphorylated during the early steps of the transformation procedure, after the 1 h incubation in 100 mM LiOAc solution and prior to the addition of DNA (Figure 2A upper panel, lane 2). There is no greater induction of Rad53 phosphorylation after the addition of the transforming DNA, after a further 30 min of incubation. The transforming DNA with double-stranded DNA ends did not lead to increased checkpoint induction compared with circular DNA (Figure 2A upper panel, lanes 3 and 4), which was expected because the transformation protocol includes an excess of carrier DNA. Rad53 phosphorylation is only transient and is turned off during the following 1 h incubation with PEG (Figure 2A upper panel, lanes 5 and 6). The heat shock did not cause Rad53 activation. Consistent with the response to DNA damage, Rad53 phosphorylation was largely dependent on Mec1 kinase, as Rad53 phosphorylation was strongly diminished in Mec1-deficient cells (Figure 2A, lower panel).
Figure 2.
Transient activation of DNA damage checkpoints during transformation. (A) Phosphorylation status of Rad53 kinase was monitored at different stages of the transformation protocol by direct immunoblotting of whole cell extracts. Log = logarithmic cells washed once with 100 mM LiOAc solution; LiOAc = after 1 h incubation in 100 mM LiOAc; DNA = after 30 min incubation with either circular (C) or linear (L) transforming DNA; PEG = after 1 h incubation in PEG; Heat = after 15 min heat shock at 42°. WT, wild-type cells WDHY1800; mec1 WDHY1814. P-Rad53 indicates the phosphorylated form of Rad53 protein. (B) Survival of wild type and mec1 cells was determined at the identical time points as in (A). The averages of two experiments are presented.
The activation of the DNA damage checkpoints during transformation might be reflective of a possible function. Hence, we analyzed the survival of wild-type and mec1 cells during the transformation procedure. Aliquots of cells were removed at the identical time points as in Figure 2A, diluted and plated on YPD plates to determine cell survival. Since there was no difference in checkpoint activation between circular and linear DNA (Figure 2A), we performed the survival experiment only with circular DNA. The transformation process is cytotoxic; only 26% of the wild type and 9% of mec1 cells survive the procedure (Figure 2B). As shown in Table 2, the transformation efficiency of wild-type cells is about four times greater than that of mec1 cells, suggesting that the low transformation efficiency of mec1 cells is largely due to their survival defect during transformation.
Mec1 and Rad53 kinases are required for efficient gap repair
Our previous work has shown that MEC1-deficient cells [esr1-1 allele of (51)] are significantly compromised in carrying out DNA damage-induced recombination between heteroalleles in vegetative diploid cells (41). In the original description of the plasmid gap repair assay used here, Symington and colleagues (55) noted that rad53-21 cells exhibited a 2-fold reduction in gap repair frequency. Using a set of isogenic strains with null mutations in the major DNA damage checkpoint kinase genes MEC1, RAD53, DUN1 and CHK1, we identified a significant contribution of the Mec1 and Rad53 kinases to the efficiency of double-strand gap repair (Table 2). Both mec1 and rad53 cells exhibited a highly significant 5-fold reduction in the gap repair frequency compared to wild type (p ≤ 0.0003), showing that 80% of the gap repair events depend on a functioning DNA damage checkpoint. It is important to stress that this reduction in gap repair frequency is independent of the reduced transformation efficiency in these mutants, as the gap repair efficiency is normalized by the transformation efficiency of the circular plasmid. The gap repair frequencies of dun1 and chk1 cells were not significantly different from wild-type cells (p ≥ 0.42). The relatively small difference in repair efficiency between the rad53 chk1 double mutant (2.6%) and the rad53 (5.4%) or mec1 (5%) single mutants was found to be statistically insignificant (p > 0.51).
MEC1 and RAD53 are essential genes and the lethality of the deletion mutants was suppressed by the constitutive overexpression of RNR1, encoding the largest subunit of ribonucleotide reductase (59). Since all strains for these experiments contain the overexpression cassette TRP1::GAP-RNR1, we wanted to establish whether the increased level of Rnr1 protein would affect the gap repair assay. As shown in Table 2, the two wild-type strains with (WDHY1800) or without (LSY697) the RNR1 overexpression construct exhibited near identical transformation efficiencies and gap repair frequencies (p = 0.92). Thus, the RNR1 overexpression does not appear to affect gap repair in this assay, consistent with previous analyses that also suggested that suppression of the mec1 or rad53 lethality does not suppress other DNA checkpoint defects in these mutants (68).
DNA damage checkpoints modulate gap repair efficiency through the Rad51 pathway
To study further the involvement of Mec1 kinase in recombinational repair, we constructed the double mutant mec1 rad51. rad52 cells are severely defective in gap repair (250-fold reduction), whereas rad51 cells show a lesser reduction in repair efficiency of 31-fold (Table 2), consistent with earlier determinations (55,69). We reasoned that analysis of the mec1 rad51 double mutant would be more informative than that of the mec1 rad52 strain, because the gap repair frequency in rad51 cells is not maximally reduced leaving room for an additive/synergistic phenotype in the double mutant. The gap repair frequency of the mec1 rad51 double mutant was found to be identical to the frequency of the rad51 single mutant (Table 2). This result suggests that mec1 and rad51 are epistatic for their effect on the efficiency of gap repair. This view is supported by the analysis of the crossover/non-crossover data (see below; Figure 3).
Figure 3.
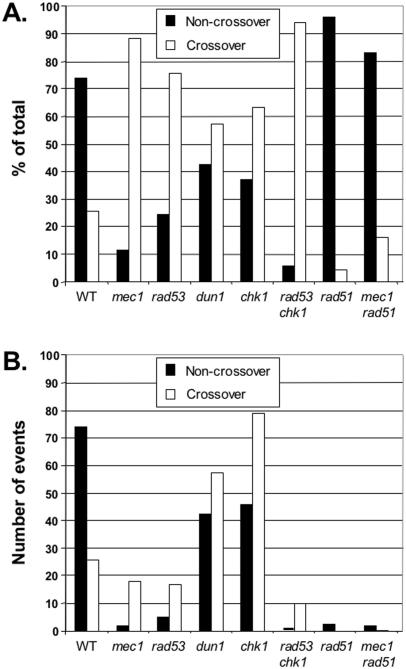
Distribution of crossover to non-crossover events. Graphic representation of the distribution of crossover (URA+s MET+s transformants) and non-crossover (URA+u MET+u transformants) events in wild-type and mutant S.cerevisiae strains. (A) Normalized data. The raw data are from Table 3 but were normalized to (URA+u MET+u events) + (URA+s MET+s events) as 100%. Except for the rad51 mec1 double mutant strain, these two classes represent 91% or more of the total URA+ MET+ transformants (see text). (B) Absolute data. The fractions of crossover and non-crossover events were multiplied by the gap repair efficiency (Table 2) and the data are plotted in comparison to 100 events in wild-type cells.
DNA damage checkpoint kinases suppress mitotic crossovers
The plasmid-based gap repair assay (55) allows for the genetic (Table 3, Figure 3) and physical analysis (Figure 4) of the URA+ MET+ transformants to determine if the gap repair event was associated (URA+s MET+s) or was not associated (URA+u MET+u) with a crossover event (Figure 1). Wild-type S.cerevisiae cells repair DSBs during mitotic growth using homologous recombination with little associated crossovers (4,6,55,57,70). This is also reflected in the gap repair data, where only 26% of the URA+ MET+ transformants were associated with a crossover (Figure 3A), which is in excellent agreement with the originally reported value (55). Crossover association with gap repair is dramatically changed in mec1 cells, where 88% of the events were associated with crossovers (Figure 3A). The difference to wild-type cells is highly significant (p < 0.0001). A similarly dramatic increase in crossover association was seen in the rad53 chk1 double mutant with 94%. The difference between the mec1 and the rad53 chk1 strains is not statistically significant (p = 0.49). Crossover association was also found to have increased to 76% in the rad53 single mutant but to a lower degree than in the mec1 or rad53 chk1 strains. The difference between the rad53 single mutant and the rad53 chk1 double mutant was found to be statistically significant (p = 0.01). Although gap repair efficiency was not affected in dun1 or chk1 single mutant strains (Table 2), crossover association is significantly increased in both strains compared to wild type (p ≤ 0.0002). It is unclear, why we found a lower crossover association (4%) than previously reported [21%; (55)] in the rad51 strain. However, it is important to note that the bias in favor of non-crossover outcome during gap repair remained as in wild-type cells. Importantly, the mec1 rad51 double mutant showed 17% crossover association. This resembles the rad51 and wild-type bias in favor of non-crossover outcome significantly more than the bias in favor of crossover association found in the mec1 single mutant. The difference in crossover association between wild-type and mec1 rad51 cells was statistically insignificant (p = 0.86). This suggests that rad51 is not only epistatic to mec1 for the gap repair efficiency phenotype but also for the crossover association phenotype.
Table 3. Phenotypes of URA+ transformants to determine crossover and non-crossover events.
| Relevant genotype | URA+a | % of URA+ transformants that were | ||||||
|---|---|---|---|---|---|---|---|---|
| URA+ MET+b | URA+s MET+s | URA+u MET+u | Othersc | URA+ MET−d | URA+s MET− | URA+u MET− | ||
| WTe | 226 | 50 | 13 | 36 | 1 | 50 | 24 | 26 |
| mec1 | 250 | 26 | 21 | 3 | 2 | 74 | 45 | 29 |
| rad53 | 200 | 43 | 31 | 10 | 2 | 57 | 35 | 22 |
| dun1 | 210 | 52 | 27 | 20 | 5 | 48 | 30 | 18 |
| chk1 | 250 | 51 | 29 | 17 | 5 | 49 | 37 | 12 |
| rad53 chk1 | 164 | 32 | 28 | 2 | 2 | 68 | 46 | 22 |
| rad51 | 260 | 27 | 1 | 25 | 1 | 73 | 20 | 53 |
| rad51 mec1 | 139 | 13 | 1 | 5 | 7 | 87 | 54 | 33 |
aNumber of URA+ transformants analyzed.
bThis column represents the sum total of all URA+ MET+ events.
cOther transformants include URA+u MET+s and URA+s MET+u.
dThis column represents the sum total of all URA+ MET− events.
eWDHY1800 was used as the wild type (WT).
The checkpoint mutants affected the relative frequency of crossovers but some mutants also had an effect on gap repair efficiency. Hence, we plotted in Figure 3B the absolute numbers of crossover and non-crossover events compared to 100 total events in wild-type cells (73 non-crossovers, 27 crossovers). From the data with mec1, rad53 and the rad53 chk1 double mutant it might appear as if the effect on crossover frequency is entirely accounted for by a specific loss of non-crossover events. However, the relative frequency and absolute number of crossover events in dun1 and chk1 single mutants is increased (Figure 3B), showing that the increase in crossover is not only due to the loss of non-crossover events.
To confirm the genetic results we analyzed representative transformants from each class by Southern analysis. All wild-type transformants representing stable recombinants (integration), URA+s MET+s, and all transformants representing unstable recombinants (no integration), URA+u MET+u, analyzed showed the expected pattern (Figure 4). The digest pattern in Figure 4A indicates a simple integration of the plasmid, with the wild-type copy of MET17 upstream of met17-s and the URA3 marker in between the two alleles. As for the URA+u MET+u transformants (Figure 4B), the digest pattern confirms the presence of only one chromosomal allele of met17-s. Since the genomic DNA was prepared from YPD-grown cultures, as described previously (55), we decided to analyze four of these transformants further by isolating genomic DNA from SC-ura-grown cultures to select for the plasmid and compare the digest pattern with YPD-grown cultures. As shown in the right panel of Figure 4B, when grown under selective conditions, the expected 1.5 kb band appears, which is indicative of the repair of the plasmid-borne MET17 ORF. Representative rad53 transformants were also examined, and all transformants from each class exhibited the expected Southern profile (Figure 4C). We conclude that the results from the physical analysis (Figure 4) fully confirm the genetic results (Figure 3), as has been extensively demonstrated in the original publication of the gap repair system (55).
In all strains, with the exception of the rad51 mec1 double mutant, 91% or more of the URA+ MET+ transformants were classified either as URA+s MET+s (indicative of crossover) or as URA+u MET+u (indicative of non-crossover) transformants (Table 3). In rad51 mec1 cells, 50% of the URA+ MET+ transformants were found to be URA+s MET+u. We were unable to determine the exact molecular nature of these transformants, because these cells were very sick and ceased to grow. It is possible that reversion of the chromosomal ura3 marker contributes to this class, as the transformation efficiency of this strain is very low (Table 2).
The analysis focused on the URA+ MET+ events, because they allow classification into crossover and non-crossover events. About one half of the URA+ transformants were MET− in wild-type cells (Table 3). These URA+ MET− events fall into two classes (55). URA+s MET− transformants result when the chromosomal met17-s marker has co-converted with the gap and a crossover led to plasmid integration, or by conversion/reversion of the ura3-1 allele. URA+u MET− transformants result when the chromosomal met17-s marker has co-converted in the absence of an associated crossover or by NHEJ. The proportion of URA+ MET− events was increased in mec1 and rad53 chk1 cells, whereas rad53, dun1, chk1 mutants showed the same distribution of MET+ to MET− events amongst the URA+ transformants as wild type (Table 3). Amongst the URA+ MET− events in mec1 and rad53 chk1 cells a bias toward crossover outcomes was evident (URA+s MET− transformants) compared with wild type cells. The strong increase in URA+u MET− transformants in rad51 cells was expected and identified before as being due to NHEJ events (55). In conclusion, the analysis of the URA+ MET− events does not suggest a dramatic increase in co-conversion of the DSB gap with met17-s marker in checkpoint mutants.
DISCUSSION
Using a plasmid-based, double-strand gap repair assay (55) we demonstrate that the DNA damage checkpoints control the efficiency and the crossover/non-crossover outcome of recombinational repair in the yeast S.cerevisiae. Three important conclusions can be drawn as follows. First, the efficiency of gap repair is reduced 5-fold in checkpoint-deficient cells, meaning that 80% of the gap repair events depend on a functional DNA damage checkpoint. Second, the DNA damage checkpoints suppress the formation of crossovers during gap repair. Third, the analysis of individual mutants suggests that the DNA damage checkpoints have multiple terminal targets in the recombinational repair pathway, including one or more major targets controlling efficiency and at least three targets controlling crossover/non-crossover outcome.
DNA damage checkpoints modulate the efficiency of recombinational DNA repair
DNA damage checkpoint mutants exhibit pleiotropic phenotypes in response to DNA damage, and their contribution to survival after genotoxic stress may result from several different effector pathways including cell cycle regulation, transcriptional regulation, regulation of DNA replication and DNA repair. The plasmid-based gap repair assay allows measuring the effect of DNA checkpoints on recombinational repair of linear plasmids independent of survival, as the repair efficiency is normalized to the transformation efficiency of the circular plasmid. Thus, the loss of viability of mec1 cells during transformation, which accounts for the low transformation efficiency, does not influence the measurements of gap repair efficiency. Double-strand gap repair efficiency is reduced 5-fold in deletion mutants of the MEC1 and RAD53 genes, which encode checkpoint kinases. A 2-fold reduction in the same gap repair system was reported for a hypomorphic RAD53 mutation, rad53-21 (55). This difference is explained by the fact that we are using a null allele for RAD53, which requires suppression by RNR1 overexpression for viability. Instead the hypomorphic rad53-21 allele [originally isolated as sad1-1 (45)] is viable and apparently retains some residual function.
Mec1 kinase is known to control the activation of Rad53 kinase in the checkpoint pathway (Figure 5A) suggesting that Mec1 exerts its effect via its control of Rad53 kinase. Presently, it is unclear how many targets Rad53 might have to regulate gap repair efficiency. It is unlikely that Chk1 kinase makes a contribution to gap repair efficiency, as the single mutant cells repair with the same efficiency as wild-type cells. Moreover, the difference in gap repair efficiency between rad53 single mutants and rad53 chk1 double mutants was found to be statistically insignificant.
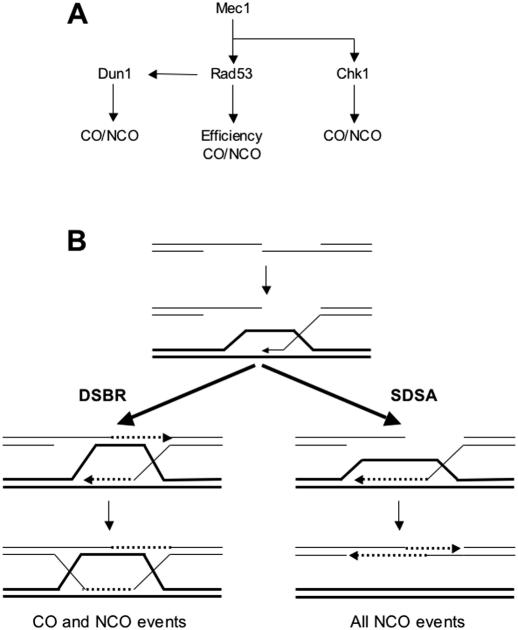
Figure 5.
Model for checkpoint kinase-mediated control of DSB repair in S.cerevisiae. (A) DNA damage checkpoint pathways. Schematic representation of the organization of the Mec1, Rad53, Chk1 and Dun1 kinases in the DNA damage checkpoints in S.cerevisiae and the pathways to control efficiency and outcome of recombinational DNA repair. CO, crossover; NCO, non-crossover. For details see text. (B) Model for recombinational repair of DSBs. After DSB formation, the 5′ ends are resected to form 3′ ending single-stranded DNA tails. Rad51 protein-mediated DNA strand invasion leads to the formation of a D-loop. In DSBR, the second end of the DSB is captured by the D-loop leading to the formation of double Holliday junction, which can be resolved to crossover and non-crossover products. In SDSA, the invading strand reverses the D-loop and anneals with the second end to result in non-crossover products only. Dashed lines indicate newly synthesized DNA. CO: crossover, NCO: non-crossover. For details see text.
How does the checkpoint pathway regulate gap repair efficiency? Double-strand gaps are repaired by a recombinational pathway in wild-type yeast cells. In rad51 mutants, gap repair is reduced 30- to 100-fold [Table 2; (55)], whereas rad52 mutants exhibit a more extreme reduction [250- to 550-fold; Table 2; (55)]. This difference between the rad51 and the rad52 mutant is also found in many other recombination assays (4,6). It reflects the roles of Rad51 and Rad52 proteins in the formation and functioning of the Rad51 filament during DSBR and SDSA (Figure 5B) and the Rad51-independent role of Rad52 protein in SSA (4,6). By analyzing the mec1 rad51 double mutant, we have shown that rad51 is epistatic to mec1, as the double mutant exhibits the same repair efficiency as the rad51 single mutant. This result suggests that Mec1 kinase modulates only the Rad51-dependent recombination pathway but probably not the SSA pathway. Checkpoint control of the Rad52-dependent SSA pathway would have predicted a more severe phenotype in the rad51 mec1 double mutant than in the rad51 single mutant, because a RAD52 mutation leads to significantly lower gap repair efficiency than a RAD51 mutation. Epistasis between mutations in the ATM kinase gene and RAD51 has been reported in chicken DT40 cells (71) and inhibitor studies also suggested a role of ATM in regulation of homologous recombination in hamster cells (72). Although the budding yeast Tel1 kinase appears to be more related in primary sequence to the vertebrate ATM kinase, it appears that Mec1 has occupied many of the functions that are split between ATM and ATR in higher eukaryotes (42–44). The possible targets of Mec1 kinase in the Rad51 pathway to regulate repair efficiency have not been identified yet. A candidate is the Rad55–Rad57 heterodimer, which was found to be phosphorylated after DNA damage (41) and likely functions in the nucleation of the Rad51 filament (10). Regulation at the Rad51 filament formation step would equally affect the DSBR and SDSA sub-pathways of recombinational DNA repair (Figure 5B). A positive role of DNA damage checkpoints in regulating DSB repair by homologous recombination has also been inferred from the cell cycle arrest-independent DNA damage sensitivity and defects in recombinational DNA repair of checkpoint-deficient cells. Among these are mutants in the budding yeast RAD24 gene, which encodes a checkpoint-specific alternative large RFC subunit (73), in budding yeast MEC1 (41) and in mammalian ATM (47,74,75).
DNA damage checkpoint kinases suppress crossover association during recombinational DNA repair in vegetative cells
In wild-type cells a significant bias toward non-crossover during recombination in vegetative (somatic) cells has been identified for chromosomal recombination (4,6). A similar bias is found in plasmid–chromosome recombination systems [(55,57,58,76) and this study]. This has led to the proposal that DSB repair in vegetative cells proceeds primarily by synthesis-dependent strand annealing (SDSA) (77) or migrating D-loop models (58), in which no Holliday junction intermediate is formed that could be processed to a crossover outcome (Figure 5B). Importantly, the 3:1 bias in favor of non-crossovers in wild-type cells is dramatically reversed to 9:1 in favor of crossovers in checkpoint-deficient mec1 cells. Thus, the DNA damage checkpoint kinases actively suppress crossovers during recombinational repair.
We considered the possibility that the increase in crossover association in checkpoint-deficient cells was due to the specific loss of non-crossover events leading to MET− transformants. These events would be URA+ MET− transformants, where the chromosomal met17-s marker had co-converted with the gap. First, it is important to note that in rad53, chk1 and dun1 cells the increase in crossover association was not accompanied by an increase in URA+ MET− transformants (Table 3). Second, in mec1 (and rad53 chk1) cells the proportion of URA+ MET− events increased from 50 to 75%. In mec1 cells, 60% of these URA+ MET− events were indicative of crossover association (URA+s MET−) and only 40% were consistent with non-crossover or NHEJ events (URA+u MET−) (Table 3). This compares to an equal distribution of crossover and non-crossover/NHEJ events in URA+ MET− wild-type transformants. Hence, we conclude that the increase in apparent co-conversions cannot fully explain the dramatic shift to crossover outcome in checkpoint-deficient cells. Increased conversion tract length leading to more co-conversion of the met17-s marker may account for the loss of some non-crossover events in mec1 and rad53 chk1 cells, suggesting that the 9:1 bias in favor of crossover in these cells is a slight overestimate.
How does the checkpoint pathway regulate gap repair outcome? The results of the epistasis analysis suggest Mec1-mediated control acts on a Rad51-dependent recombinational repair. It appears possible that the DNA damage checkpoints specifically affect the non-crossover pathway, as the absolute numbers of crossovers remained rather constant in mec1 cells (Tables 2 and 3). However, the significant increase in crossovers in chk1 and dun1 single mutants is not accompanied by decreased gap repair efficiency, making this explanation unlikely (Figure 3B).
Crossovers associated with recombination between non-allelic homologous sequences (e.g. interspersed DNA repeats) may lead to inversion, deletions or translocations, events that contribute to genomic instability. This is consistent with the genomic instability phenotype in mammalian ATM-deficient cells, where evidence for these types of genome rearrangements were found (47), and with the dramatic increase of gross chromosomal rearrangements identified in checkpoint-deficient cells in budding yeast (78–81).
DNA damage checkpoints have multiple targets in the recombinational repair pathway
The regulation of crossover/non-crossover outcome appears to involve multiple independent targets, as suggested by the analysis of individual checkpoint kinase mutant strains. The deletion mutant in MEC1 essentially eliminates the DNA damage checkpoints in budding yeast. Mec1 controls at least three downstream kinases, Rad53, Dun1 and Chk1 (Figure 5A). The mutant data suggest at least three targets of the checkpoint kinases in the regulation of crossover outcome. The mec1 mutant shows a 9:1 bias in favor of crossovers, which is dramatically shifted from the 3:1 bias in favor of non-crossovers in wild-type cells. Strains deficient in the Rad53, Dun1 or Chk1 kinases result in significantly more crossover association compared to wild type (Figure 3), but their bias in favor of crossover is not as extreme as in mec1 cells. Importantly, the rad53 chk1 double mutant shifts the bias in favor of crossover as much as a mec1 mutant, suggesting that Mec1 kinase controls crossover outcome through two signaling branches controlled by Rad53 and Chk1. The rad53 mutation affected crossover outcome significantly more than the dun1 mutation, suggesting that Rad53 kinase controls crossover outcome by at least two pathways: one dependent on Dun1 and the other independent of Dun1. Rad53 is known to control the activation of Dun1 kinase but there is also evidence for Rad53-independent function of Dun1 (45,46,81). The observation that the rad53 chk1 double mutant affects crossover outcome as much as a mec1 mutant suggests that Dun1 functions in crossover control in an exclusively Rad53-dependent manner. Thus, we propose that Dun1, Rad53 and Chk1 perform independent phosphorylation events to modulate crossover outcome. The simplest interpretation of these genetic data is that the kinases target three different proteins but it is also possible that they perform independent phosphorylation events with distinct mechanistic consequences on the same protein(s).
The direct targets of checkpoint-mediated control of gap repair outcome are presently unknown. The balance between crossover and non-crossover outcome could be tipped in multiple ways. Symington and colleagues (55) calculated that 40% of the gap repair events are shunted into DSBR (Figure 5B), which may lead to crossover or non-crossover outcome, whereas 60% of the events were handled by SDSA, which leads to non-crossovers exclusively. Increased usage of DSBR will increase the likelihood for crossover association. The mechanisms involved in the usage of the DSBR versus SDSA sub-pathways are unknown. However, even shunting all events to DSBR fails to explain the extreme 9:1 bias in mec1 or rad53 chk1 cells, as random resolution of the double Holliday junction would result in maximally 50% crossover association. This suggests that proteins acting late in recombinational repair and involved in crossover formation or crossover suppression are potential targets. One potential target is the Sgs1–Top3 complex, which suppresses crossover formation in double-strand break repair (24). Another potential target is the Mus81–Mms4 structure-specific DNA endonuclease. Mus81 has been implicated in crossover formation in meiotic cells (22,27,82,83) and DNA damage-induced sister chromatid exchange, likely to represent crossovers between sister chromatids, in mammalian cells (84). In fission yeast, Mus81 was originally identified through its interaction with Cds1 kinase, the fission yeast Rad53 homolog (85). Fission yeast Mus81 was also found to be phosphorylated in response to genotoxic stress but the biological significance and mechanistic consequence remain to be determined (85). The interaction between Mus81 and Cds1 appears to be conserved in budding yeast, as a Rad53–Mus81 interaction was reported in a large-scale protein interaction screen (86).
In summary, our results demonstrate that 80% of the recombinational repair of a double-stranded DNA gap is dependent on a functioning DNA damage checkpoint system and that DNA damage checkpoint kinases suppress crossover formation during recombinational repair in vegetative budding yeast cells. The mutant analysis provided clear evidence for multiple independent checkpoint targets in the Rad51-dependent recombination pathway to modulate repair efficiency and outcome. It will be of great interest to determine the checkpoint targets in recombinational repair and the mechanisms involved in this regulation.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to Dr Lorraine Symington for kindly providing us with strains and plasmids for the gap repair assay and to Dr Stephan Bärtsch for his help optimizing the transformation protocol. We thank Vladimir Bashkirov, Kirk Ehmsen, Kristina Herzberg, Xuan Li, Michael Rolfsmeier and Xiao-Ping Zhang for their helpful discussions and comments on the manuscript. This work was supported by a grant from the NIH (CA92276) to W.-D.H.
REFERENCES
- 1.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 2.Gerard F., Dri,A.M. and Moreau,P.L. (1999) Role of Escherichia coli RpoS, LexA and H-NS global regulators in metabolism and survival under aerobic, phosphate-starvation conditions. Microbiology, 145, 1547–1562. [DOI] [PubMed] [Google Scholar]
- 3.Michel B., Ehrlich,S.D. and Uzest,M. (1997) DNA double-strand breaks caused by replication arrest. EMBO J., 16, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symington L.S. (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev., 66, 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gellert M. (2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem., 71, 101–132. [DOI] [PubMed] [Google Scholar]
- 6.Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasior S.L., Wong,A.K., Kora,Y., Shinohara,A. and Bishop,D.K. (1998) Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev., 12, 2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis A.P. and Symington,L.S. (2001) The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics, 159, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petukhova G., Stratton,S.A. and Sung,P. (1999) Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J. Biol. Chem., 274, 33839–33842. [DOI] [PubMed] [Google Scholar]
- 10.Sung P. (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. [DOI] [PubMed] [Google Scholar]
- 11.Sung P. (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem., 272, 28194–28197. [DOI] [PubMed] [Google Scholar]
- 12.New J.H., Sugiyama,T., Zaitseva,E. and Kowalczykowski,S.C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature, 391, 407–410. [DOI] [PubMed] [Google Scholar]
- 13.Shinohara A. and Ogawa,T. (1998) Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature, 391, 404–407. [DOI] [PubMed] [Google Scholar]
- 14.Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science, 265, 1241–1243. [DOI] [PubMed] [Google Scholar]
- 15.Petukhova G., Stratton,S. and Sung,P. (1998) Catalysis of homologous pairing by yeast Rad51 and Rad54 proteins. Nature, 393, 91–94. [DOI] [PubMed] [Google Scholar]
- 16.Van Komen S., Petukhova,G., Sigurdsson,S., Stratton,S. and Sung,P. (2000) Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell, 6, 563–572. [DOI] [PubMed] [Google Scholar]
- 17.Mazin A.V., Bornarth,C.J., Solinger,J.A., Heyer,W.-D. and Kowalczykowski,S.C. (2000) Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell, 6, 583–592. [DOI] [PubMed] [Google Scholar]
- 18.Solinger J.A., Lutz,G., Sugiyama,T., Kowalczykowski,S.C. and Heyer,W.-D. (2001) Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol., 307, 1207–1221. [DOI] [PubMed] [Google Scholar]
- 19.Solinger J.A. and Heyer,W.-D. (2001) Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc. Natl Acad. Sci. USA, 98, 8447–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solinger J.A., Kiianitsa,K. and Heyer,W.-D. (2002) Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell, 10, 1175–1188. [DOI] [PubMed] [Google Scholar]
- 21.Kiianitsa K., Solinger,J.A. and Heyer,W.D. (2002) Rad54 protein exerts diverse modes of ATPase activity on duplex DNA partially and fully covered with Rad51 protein. J. Biol. Chem., 277, 46205–46215. [DOI] [PubMed] [Google Scholar]
- 22.Heyer W.D., Ehmsen,K.T. and Solinger,J.A. (2003) Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends in Biochem. Sci., 10, 548–557. [DOI] [PubMed] [Google Scholar]
- 23.Wu L.J. and Hickson,I.D. (2003) The Bloom's syndrome helicase suppresses crossing-over during homologous recombination. Nature, 426, 870–874. [DOI] [PubMed] [Google Scholar]
- 24.Ira G., Malkova,A., Liberi,G., Foiani,M. and Haber,J.E. (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell, 115, 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rockmill B., Fung,J.C., Branda,S.S. and Roeder,G.S. (2003) The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol., 13, 1954–1962. [DOI] [PubMed] [Google Scholar]
- 26.Fabre F., Chan,A., Heyer,W.D. and Gangloff,S. (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mus81, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA, 99, 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osman F., Dixon,J., Doe,C.L. and Whitby,M.C. (2003) Generating crossovers by resolution of nicked Holliday junctions: A role of Mus81–Eme1 in meiosis. Mol. Cell, 12, 761–774. [DOI] [PubMed] [Google Scholar]
- 28.Gaillard P.-H., Noguchi,E., Shanahan,P. and Russell,P. (2003) The endogenous Mus81–Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell, 12, 747–759. [DOI] [PubMed] [Google Scholar]
- 29.Yildiz O., Majumder,S., Kramer,B. and Sekelsky,J.J. (2002) Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell, 10, 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y.L., Masson,J.Y., Shah,R., O'Regan,P. and West,S.C. (2004) RAD51C is required for Holliday junction processing in mammalian cells. Science, 303, 243–246. [DOI] [PubMed] [Google Scholar]
- 31.Hollingsworth N.M. and Brill,S.J. (2004) The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev., 18, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de los Santos T., Loidl,J., Larkin,B. and Hollingsworth,N. (2001) A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics, 159, 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orr-Weaver T., Szostak,J. and Rothstein,R. (1981) Yeast transformation: a model system for the study of recombination. Proc. Natl Acad. Sci. USA, 78, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strathern J.N., Weinstock,K.G., Higgins,D.R. and McGill,C.B. (1991) A novel recombinator in yeast based on gene II protein from bacteriophage f1. Genetics, 127, 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morimatsu K. and Kowalczykowski,S.C. (2003) RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol. Cell, 11, 1337–1347. [DOI] [PubMed] [Google Scholar]
- 36.Chanet R., Heude,M., Adjiri,A., Maloisel,L. and Fabre,F. (1996) Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol., 16, 4782–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aboussekhra A., Chanet,R., Adjiri,A. and Fabre,F. (1992) Semi-dominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA protein. Mol. Cell. Biol., 12, 3224–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiestl R.H., Prakash,S. and Prakash,L. (1990) The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics, 124, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krejci L., Van Komen,S., Li,Y., Villemain,J., Reddy,M.S., Klein,H., Ellenberger,T. and Sung,P. (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature, 423, 305–309. [DOI] [PubMed] [Google Scholar]
- 40.Veaute X., Jeusset,J., Soustelle,C., Kowalczykowski,S.C., Le Cam,E. and Fabre,F. (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature, 423, 309–312. [DOI] [PubMed] [Google Scholar]
- 41.Bashkirov V.I., King,J.S., Bashkirova,E.V., Schmuckli-Maurer,J. and Heyer,W.D. (2000) DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol., 20, 4393–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou B.B.S. and Elledge,S.J. (2000) The DNA damage response: putting checkpoints in perspective. Nature, 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 43.Nyberg K.A., Michelson,R.J., Putnam,C.W. and Weinert,T.A. (2002) Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet., 36, 617–656. [DOI] [PubMed] [Google Scholar]
- 44.Melo J. and Toczyski,D. (2002) A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol., 14, 237–245. [DOI] [PubMed] [Google Scholar]
- 45.Allen J.B., Zhou,Z., Siede,W., Friedberg,E.C. and Elledge,S.J. (1994) The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev., 8, 2401–2415. [DOI] [PubMed] [Google Scholar]
- 46.Bashkirov V.I., Bashkirova,E.V., Haghnazari,E. and Heyer,W.D. (2003) Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase. Mol. Cell. Biol., 23, 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeggo P.A., Carr,A.M. and Lehmann,A.R. (1998) Splitting the ATM: distinct repair and checkpoint defects in ataxia-telangiectasia. Trends Genet., 14, 312–316. [DOI] [PubMed] [Google Scholar]
- 48.Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- 49.D'Amours D. and Jackson,S.P. (2002) The MRE11 complex: at the crossroads of DNA repair and checkpoint signalling. Nature Rev. Mol. Cell. Biol., 3, 317–327. [DOI] [PubMed] [Google Scholar]
- 50.Liberi G., Chiolo,I., Pellicioli,A., Lopes,M., Plevani,P., Muzi-Falconi,M. and Foiani,M. (2000) Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J., 19, 5027–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato R. and Ogawa,H. (1994) An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res., 22, 3104–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szostak J.W., Orr-Weaver,T.L., Rothstein,R.J. and Stahl,F.W. (1983) The double-strand-break repair model for recombination. Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- 53.Glaser V.M., Glasunov,A.V., Tevzadze,G.G., Perera,J.R. and Shestakov,S.V. (1990) Genetic control of plasmid DNA double-strand gap repair in yeast, Saccharomyces cerevisiae. Curr. Genet., 18, 1–5. [DOI] [PubMed] [Google Scholar]
- 54.Perera J.R., Glasunov,A.V., Glaser,V.M. and Boreiko,A.V. (1988) Repair of double-strand breaks in plasmid DNA in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet., 213, 421–424. [DOI] [PubMed] [Google Scholar]
- 55.Bärtsch S., Kang,L.E. and Symington,L.S. (2000) RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol., 20, 1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orr-Weaver T.L. and Szostak,J.W. (1983) Yeast transformation: the association between double-strand gap repair and crossover. Proc. Natl Acad. Sci. USA, 80, 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plessis A. and Dujon,B. (1993) Multiple tandem integrations of transforming DNA sequences in yeast chromosomes suggest a mechanism for integrative transformation by homologous recombination. Gene, 134, 41–50. [DOI] [PubMed] [Google Scholar]
- 58.Ferguson D.O. and Holloman,W.K. (1996) Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc. Natl Acad. Sci. USA, 93, 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desany B., Alcasabas,A.A., Bachant,J.B. and Elledge,S.J. (1998) Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes. Dev., 12, 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wach A. (1996) PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S.cerevisiae. Yeast, 12, 259–265. [DOI] [PubMed] [Google Scholar]
- 61.McDonald J.P., Levine,A.S. and Woodgate,R. (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics, 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherman F., Fink,G.R. and Hicks,J.B. (1982) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 63.Cost G.J. and Boeke,J.D. (1996) A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast, 12, 939–941. [DOI] [PubMed] [Google Scholar]
- 64.Ono B.-I., Ishii,N., Fujino,S. and Aoyama,I. (1991) Role of hydrosulfie ions (HS-) in methylmercury reisstance in Saccharomyces cerevisiae. Appl. Environ. Microbiol., 57, 3183–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact cells treated with alkali cations. J. Bacteriol., 153, 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foiani M., Liberi,G., Piatti,S. and Plevani,P. (1999) In Cotteril,S. (ed.), Eukaryotic DNA Replication. A Practical Approach. Oxford University Press, Oxford, pp. 185–200. [Google Scholar]
- 67.Sanchez Y., Desany,B.A., Jones,W.J., Liu,Q.H., Wang,B. and Elledge,S.J. (1996) Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science, 271, 357–360. [DOI] [PubMed] [Google Scholar]
- 68.Zhao X.L., Muller,E.G.D. and Rothstein,R. (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell, 2, 329–340. [DOI] [PubMed] [Google Scholar]
- 69.Ira G. and Haber,J.E. (2002) Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol., 22, 6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kostriken R., Strathern,J.N., Klar,A.J., Hicks,J.B. and Heffron,F. (1983) A site-specific endonuclease essential for mating-type switching in Saccahromyces cerevisiae. Cell, 35, 167–174. [DOI] [PubMed] [Google Scholar]
- 71.Morrison C., Sonoda,E., Takao,N., Shinohara,A., Yamamoto,K. and Takeda,S. (2000) The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J., 19, 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Asaad N.A., Zeng,Z.C., Guan,J., Thacker,J. and Iliakis,G. (2000) Homologous recombination as a potential target for caffeine radiosensitization in mammalian cells: reduced caffeine radiosensitization in XRCC2 and XRCC3 mutants. Oncogene, 22, 5788–5800. [DOI] [PubMed] [Google Scholar]
- 73.Aylon Y. and Kupiec,M. (2003) The checkpoint protein Rad24 of Saccharomyces cerevisiae is involved in processing double-strand break ends and in recombination partner choice. Mol. Cell. Biol., 23, 6585–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Golding S.E., Rosenberg,E., Khalil,A., McEwen,A., Holmes,M., Neill,S., Povirk,L.F. and Valerie,K. (2004) Double strand break repair by homologous recombination is regulated by cell cycle-independent signaling via ATM in human glioma cells. J. Biol. Chem., 279, 15402–15410. [DOI] [PubMed] [Google Scholar]
- 75.Kuhne M., Riballo,E., Rief,N., Rothkamm,K., Jeggo,P.A. and Lobrich,M. (2004) A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res., 64, 500–508. [DOI] [PubMed] [Google Scholar]
- 76.Paques F., Leung,W.Y. and Haber,J.E. (1998) Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol., 18, 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nassif N., Penney,J., Pal,S., Engels,W.R. and Gloor,G.B. (1994) Efficient copying of nonhomolgous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol., 14, 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Myung K. and Kolodner,R.D. (2002) Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 99, 4500–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Myung K., Datta,A. and Kolodner,R.D. (2001) Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell, 104, 397–408. [DOI] [PubMed] [Google Scholar]
- 80.Kolodner R.D., Putnam,C.D. and Myung,K. (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science, 297, 552–557. [DOI] [PubMed] [Google Scholar]
- 81.Myung K., Chen,C. and Kolodner,R.D. (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature, 411, 1073–1076. [DOI] [PubMed] [Google Scholar]
- 82.de los Santos T., Hunter,N., Lee,C., Larkin,B., Loidl,J. and Hollingsworth,N.M. (2003) The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics, 164, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith G.R., Boddy,M.N., Shanahan,P. and Russell,P. (2003) Fission yeast Mus81•Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics, 165, 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abraham J., Lemmers,B., Hande,M.P., Moynahan,M.E., Chahwan,C., Ciccia,A., Essers,J., Hanada,K., Chahwan,R., Khaw,A.K. et al. (2003) Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J., 22, 6137–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boddy M.N., Lopez-Girona,A., Shanahan,P., Interthal,H., Heyer,W.D. and Russell,P. (2000) Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol., 20, 8758–8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ho Y., Gruhler,A., Heilbut,A., Bader,G.D., Moore,L., Adams,S.L., Millar,A., Taylor,P., Bennett,K., Boutilier,K. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]