Abstract
Increased agricultural production is an urgent issue. Projected global population is 9 million people by mid of this century. Estimation projects death of 1 million people for lack of food quality (micronutrient deficit) and quantity (protein deficit). Majority of these people will be living in developing countries. Other global challenges include shrinking cultivable lands, salinity, and flooding due to climate changes, new emerging pathogens, and pests. These affect crop production. Furthermore, they are major threats to crop genetic resources and food security. Genetic diversity in cultivated crops indicates gene pool richness. It is the greatest resource for plant breeders to select lines that enhance food security. This study was conducted by Masinde Muliro University to evaluate genetic diversity in 19 cowpea accessions from Kenya national gene bank. Accessions clustered into two major groups. High divergence was observed between accessions from Ethiopia and Australia and those from Western Kenya. Upper Volta accessions were closely related to those from Western Kenya. Low variation was observed between accessions from Eastern and Rift Valley than those from Western and Coastal regions of Kenya. Diversity obtained in this study can further be exploited for the improvement of cowpea in Kenya as a measure of food security.
1. Introduction
Cowpea, Vigna unguiculata (L.) Walp. (2n = 22), is one of the most ancient human food sources [1]. It is one of the most important legume crops in the world and it is a major food crop in Africa, Latin America, and India because of its high protein content [2, 3]. As a result, cowpea is referred to as a poor man's meat [4]. Cowpea is primarily a self-pollinating crop and its genetic base is considered to be narrow [5–7].
The world's cowpea production as at 2013 was estimated at 5,718,144.66 tonnes of which 5,421,561 tonnes were from Africa, with East Africa contributing 532,901 tonnes [8]. In Kenya, yields remain extremely low, ranging from 150 to 500 kg/ha which is attributed to abiotic and biotic stresses, lack of high yielding cultivars, and poor crop management practices [9]. The area under cowpea in Kenya is estimated at 1800 ha excluding the area under the crop in home gardens [10].
Traditionally, diversity in cowpea is estimated by measuring variation in phenotypic or qualitative traits such as flower colour, growth habit, or quantitative agronomic traits such as yield potential and stress tolerance [11] that do not necessarily reflect real genetic relationships [12]. Furthermore, the expression of quantitative traits is subject to strong environmental influence and therefore limits knowledge of the germplasm structure for development of hybrids with specific ecological adaptations [11]. DNA markers are the most popular and widely used techniques to differentiate among genotypes at species and subspecies level [13]. Comparative studies in plants have shown that simple sequence repeat (SSR) markers, which are single locus markers with multiple alleles, are more valuable and provide an effective means for discriminating between genotypes [14, 15]. This study characterized 19 cowpea accessions from different sources that had been preserved at the national gene bank of Kenya using two SSR markers (Table 2) that had highest polymorphic amplification of both local and inbred lines of cowpea as reported by Badiane et al. [16].
Table 2.
Primer sequences used.
| Primer code | Primer sequence 5′ to 3′ |
|---|---|
| SSR-6540 | 5′-GGACATTTAGGATTGGGTGG-3′ |
| 5′-CCATAGGTTAAACTTATTGTACTC-3′ | |
| SSR-6652 | 5′-CAAAATTCCACGGTCACC-3′ |
| 5′-CGGGACTTGAGGTAGCGCG-3′ |
2. Materials and Methods
2.1. DNA Extraction
Seedlings of each cowpea accession (Table 1) were grown in pots of sterile soil in a greenhouse with 3 plants per accession. Leaf samples were purposively sampled from three plants per accession from 15-day-old seedlings [16] and frozen in liquid nitrogen and genomic DNA extracted according to the prescribed protocol of the DNeasy Plant Mini Kit (Qiagen).
Table 1.
List of the names of all 19 cowpea accessions used in this study and their geographic distributions (locations).
| Sample number | Accession name | Genebank accession number | District and locality of collection | Latitude | Longitude | Date of collection |
|---|---|---|---|---|---|---|
| 1 | Rift Valley 040539 | GBK-040539 | Turkana; Nadoto | 2.7333°N | 35.11667°E | 18.9.1994 |
| 2 | Australia 016157 | GBK-016157 | Uasin Gishu | — | — | 5.1.1989 |
| 3 | Coast 032338 | GBK-032338 | Kwale; Mwachanda | — | — | 11.2.1992 |
| 4 | Coast 032344 | GBK-032344 | Kwale; Marenje village | 4.46167°S | 39.12833°E | 11.3.1992 |
| 5 | Coast 032723 | GBK-032723 | — | — | — | 1.1.1976 |
| 6 | Rift Valley 040472 | GBK-040472 | Kabarnet | — | — | 9.2.1994 |
| 7 | Coast 031913 | GBK-031913 | Busia | 0.45694°N | 34.191389°E | 7.10.1992 |
| 8 | Western 047102 | GBK-047102 | Kakamega; Bunyala East | 0.44172°N | 034.68136°E | 22.11.2004 |
| 9 | Western 047111 | GBK-047111 | Vihiga; Mudete | 0.11785°N | 034.76527°E | 24.11.2004 |
| 10 | Eastern 046585 | GBK-046585 | Mwingi; Nzelune-Makilungi | 1.284167°N | 38.258611°E | 29.8.2003 |
| 11 | Western 044082 | GBK-044082 | Meru; Nkubu market | 0.066667°S | 37.666667°E | 2.11.1997 |
| 12 | Eastern 033061 | GBK-033066 | Embu; Embu research station | 3.508889°S | 37.454722°E | 1.12.1992 |
| 13 | Eastern 033066 | GBK-033061 | Embu; Embu research station | 3.508889°S | 37.454722°E | 1.12.1992 |
| 14 | Ethiopia 015141 | GBK-015141 | Siaya; Kigilo | — | — | 5.1.1989 |
| 15 | Eastern 033060 | GBK-033060 | Embu; Embu research station | 3.508889°S | 37.454722°E | 1.12.1992 |
| 16 | Upper Volta 022436 | GBK-022436 | Kilifi | — | — | 10.1.1975 |
| 17 | Western 047048 | GBK-047048 | Busia; Ageng'a | 0.22152°N | 034.08540°E | 19.11.2004 |
| 18 | Western 047119 | GBK-047119 | Vihiga; Serem Tiriki East | 0.07745°N | 034.8548°E | 24.11.2004 |
| 19 | Rift Valley 032108 | GBK-032108 | Nandi; Kaptumo location | 0.067500°N | 35.067500°E | 27.8.1992 |
| 20 | Western 047082 | GBK-047082 | Busia; Bumala | 0.30394°N | 034.20103°E | 19.11.2004 |
Data obtained from the national gene bank of Kenya.
2.2. PCR Amplification of DNA and Electrophoresis
PCR was carried out in 0.2 mL tubes with a reaction volume of 25 μL containing 2.5 μL 10x PCR buffer, 1 μL of both primers, 1 mM of each dNTPs, 0.5 U Taq DNA polymerase, and 50 ng DNA. The tubes were placed in an Eppendorf Mastercycler Gradient thermocycler programmed for initial denaturation at 94°C for 1 minute, followed by 35 cycles for 30 seconds at 94°C, 30 seconds at 55°C, 1 minute at 75°C, and final extension for 10 minutes at 72°C.
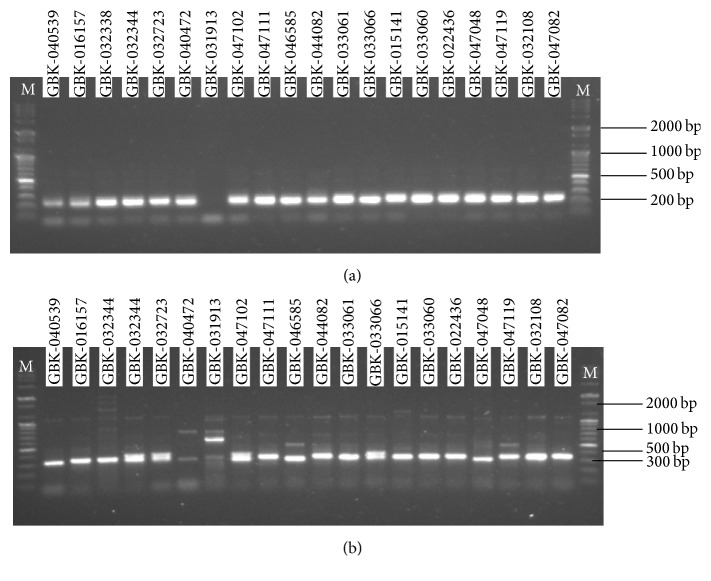
The PCR products were resolved on a agarose gel (1%) using 0.5x TBE containing 1 mg/mL ethidium bromide with a vertical electrophoresis apparatus. The gel was photographed using Alphaimager 2200 under UV transilluminator (Figure 1). The resolved products were extracted from the gel and purified using the Qiagen DNA purification kit according to the prescribed protocol. DNA quantification was done by using a DNA NanoDrop 2000/2000c Spectrophotometer.
Figure 1.
(a) PCR products of the 20 accessions of cowpea amplified using SSR primer 6652: M indicates the ladder. (b) PCR products of the 20 accessions of cowpea amplified using SSR primer 6540: M indicates the ladder.
3. Phylogenetic Analysis
The sequences obtained were first edited by BioEdit version (version 7) and then nucleotide alignments were generated using ClustalW software. The evolutionary history was inferred using the Neighbor-Joining method. Analyses were conducted using the Jukes-Cantor model [17]. The analysis involved 19 nucleotide sequences. Codon positions included were the 1st, 2nd, 3rd, and noncoding positions. All ambiguous positions were removed for each sequence pair. Evolutionary analyses were conducted in MEGA6. The confidence of the branches was measured by bootstrapping with 1,000 replicates [18].
4. Results
4.1. PCR Products
Nineteen cowpea accessions gave PCR products of ~300 bp (Figure 1(b)) for the SSR-6540 marker, while SSR-6652 gave PCR products of ~200 bp (Figure 1(a)) and these were not able to give any significant similarity from the NCBI and hence were not considered useful for this study.
4.2. Phylogenetic Analysis
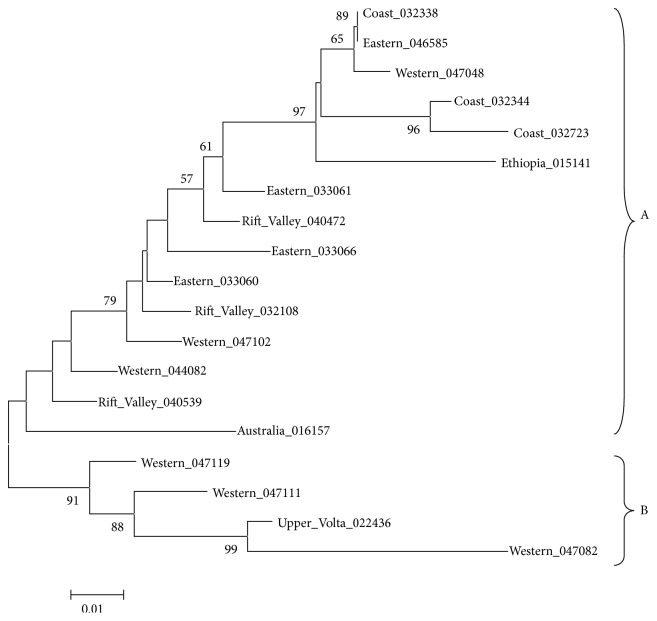
These accessions were clustered into two main groups: A and B, as indicated in Figure 2. Cluster A is comprised of 16 accessions most diverged that form seven subclusters with bootstrap support of 79 (Eastern_033066, Rift_Valley_032108, Eastern_033060, and Western_047102), 59 (Rift_Valley_040472), 62 (Eastern_033061), 96 (Ethiopia_015141), 98 (Coast_032344 and Coast_032373), 63 (Western_047048), and 88 (Eastern_046585, Coast_032338). Three accessions in this cluster (Western_044082, Rift_Valley_040539, and Australia_016157) had their bootstrap less than 50; hence, their branches were not reliable.
Figure 2.
Phylogenetic tree of cowpea varieties: varieties having similar distances are genetically similar.
Cluster B is comprised of four accessions that exhibit moderate level of divergence, forming four subclusters with bootstrap support of 91 (Western_047119), 88 (Western_047111), and 99 (Western_047082 and Upper_Volta_022436).
5. Discussion
Genetic diversity is the extent to which material differs within a group of plants [19]. The low genetic variability in the cowpea accessions used in this study is consistent with the findings of previous studies due to the fact that a single domestication event is involved in the origin of this crop [7, 9, 20–24].
The low genetic divergence observed in this study is in agreement with the findings of Padulosi and Ng [25], who attributed it to the self-pollination nature of this crop. Given that the accessions were from different regions, it could also indicate high-gene flow within regions and limited time for significant genetic differentiation along geographical lines as indicated by Karuma et al. 2008 [9]. Highest levels of divergence between the accession from Western Kenya, Australia, and Ethiopia could be attributed to the fact that the accessions have been popularly cultivated in the respective regions over time giving enough time for significant genetic differentiation along geographical lines [9]. At the same time, it could indicate genetic evidence of cowpea being a very diverse taxon as reported by Huynh et al. 2013 [26]. This therefore would mean that the studied germplasm from Australia and Ethiopia has some amount of diversity that can be used for cowpea improvement in Western Kenya. In the same manner, it can be argued that Upper_Volta_022436 has some amount of diversity that can be used for improvement of cowpea at the Rift Valley and Eastern and Coastal region of Kenya. Eastern Kenya constitutes 85% of the total production of cowpea in Kenya [27]. The comparison of the genetic distances between accessions from Western Kenya (Western_047111, Western_047102, Western_044082, Western_047119, Western_047048, and Western_047082) to those from Eastern Kenya revealed a closer genetic relationship with Eastern_046585 than all other accessions from the same region (Eastern_033060, Eastern_033061, and Eastern_033066). It could be argued that these accessions could have common origin with accession Eastern_046585 and the observed variability is attributed to hybridization as indicated by Adewale et al. [28].
Similarly, two Western Kenya accessions (Western_047102 and Western_044082) exhibited a closer relationship with Rift Valley_032108. The low genetic distances observed between these accessions could possibly reflect the initial bottleneck during domestication maintained by the inherent self-pollination mechanism of cowpea crop [7]. On the contrary higher genetic distance observed between Western_047082 and Rift Valley_040539 and between Western_047048 and both Rift_Valley_040539 and Rift Valley_040472 could possibly indicate that the diversity in these accessions could be used for the improvement of cowpea crop in these two regions [26].
On the other hand, the genetic distances of the accessions from Western Kenya to the accessions from Australia, Ethiopia, and Upper Volta revealed that there was greater divergence between Western_047082 and the accession from Australia and Ethiopia, but this accession was genetically closely related with the accession from Upper Volta which is in agreement with Ba et al. [23]. This could mean that these two accessions (Western_047082 and Upper_Volta_022436) are actually the same accession, despite being collected from different regions. This study has also observed that there is higher diversity between Australia_016157 and the accessions from the Coastal region of Kenya which could be exploited to improve the cowpea germplasm in the Coastal region of Kenya. This study has proved that although the genetic base of cowpea is narrow, there exists some level of diversity between cowpea accessions in the Kenya gene bank that can be exploited for the improvement of cowpea crop as a measure for food security in Kenya.
Acknowledgments
This study was made possible through the financial support of ASARECA. The cowpea accessions were provided by the gene bank in Kenya. The authors acknowledge Mary Silingi for assisting them in accessing the gene bank data for the accessions used in this work.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Summerfield R. I., Huxley P. A., Steele W. Cowpea (Vigna unguiculata (L) Walp) Field Crop Abstracts. 1974;27:301–312. [Google Scholar]
- 2.Diouf D., Hilu K. W. Microsatellites and RAPD markers to study genetic relationships among cowpea breeding lines and local varieties in Senegal. Genetic Resources and Crop Evolution. 2005;52(8):1057–1067. doi: 10.1007/s10722-004-6107-z. [DOI] [Google Scholar]
- 3.Kareem K. T., Taiwo M. A. Interactions of viruses in cowpea: effects on growth and yield parameters. Virology Journal. 2007;4:15–21. doi: 10.1186/1743-422x-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharmar S., Upadhyaya H. D., Rootkiwal M., Varshney R. K., Gowda C. L. L. Chickpea. In: Singh M., Upadhyaya H. D., Bisht I. S., editors. Genetic and Genomic Resources of Grain Legume Improvement. London, UK: Elsevier; 2013. pp. 81–104. [Google Scholar]
- 5.Sharawy W. M., El-Fiky Z. A. Characterization of cowpea (Vigna unguiculata L.) accessions based on yield traits and RAPD-PCR analyses. Arab Journal of Biotechnology. 2002;6:67–78. [Google Scholar]
- 6.Fang J., Devanand P. S., Chao C. C. T. Genetic Diversity of Cowpea [Vigna unguiculata (L.) Walp] Breeding Lines from Different Countries. Las Vegas, Nev, USA: AFLP Markers; 2005. [Google Scholar]
- 7.Asare A. T., Gowda B. S., Galyuon I. K. A., Aboagye L. L., Takrama J. F., Timko M. P. Assessment of the genetic diversity in cowpea (Vigna unguiculata L. Walp.) germplasm from Ghana using simple sequence repeat markers. Plant Genetic Resources. 2010;8(2):142–150. doi: 10.1017/s1479262110000092. [DOI] [Google Scholar]
- 8. FAO 2014 FAOSTAT, http://faostat3.fao.org/download /Q/QC/E 1/04/2014.
- 9.Karuma R. W., Kiplagat O., Ateka E., Owuoche G. Genetic diversity of Kenyan cowpea accessions based on morphological and microsatellite markers. East African Agricultural and Forestry Journal. 2008;76:3–4. [Google Scholar]
- 10.Muthamia J. G. N., Kanampiu F. K. Focus on Agricultural Research for Sustainable Development in a Changing Economic Environment: Proceedings of the 5th KARI Scientific Conference, 14th to 16th October, 1996, KARI Headquarters, Kaptagat Road, Loresho, Nairobi, Kenya. Kenya Agricultural Research Institute; 1996. On-farm cowpea evaluation in the marginal areas of eastern Kenya. [Google Scholar]
- 11.Kameswara R. N. Biotechnology for Plant Resources Conservation and Use. Kampla, Uganda: Principles of Seed Handling in Genebanks Training Course; 2004. [Google Scholar]
- 12.Patil D. M., Sawardekar S. V., Gokhale N. B., et al. Genetic diversity analysis in cowpea [VignaUnguiculata (L.) Walp.] by using RAPD markers. International Journal of Innovative Biotechnology and Biochemistry. 2013;1:15–23. [Google Scholar]
- 13.Kumar L. S. DNA markers in plant improvement: an overview. Biotechnology Advances. 1999;17(2-3):143–182. doi: 10.1016/s0734-9750(98)00018-4. [DOI] [PubMed] [Google Scholar]
- 14.Powell W., Morgante M., Andre C., et al. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding. 1996;2(3):225–235. doi: 10.1007/bf00564200. [DOI] [Google Scholar]
- 15.Li C.-D., Fatokun C. A., Ubi B., Singh B. B., Scoles G. J. Determining genetic similarities and relationships among cowpea breeding lines and cultivars by microsatellite markers. Crop Science. 2001;41(1):189–197. doi: 10.2135/cropsci2001.411189x. [DOI] [Google Scholar]
- 16.Badiane F. A., Gowda B. S., Cissé N., Diouf D., Sadio O., Timko M. P. Genetic relationship of cowpea (Vigna unguiculata) varieties from Senegal based on SSR markers. Genetics and molecular research : GMR. 2012;11(1):292–304. doi: 10.4238/2012.February.8.4. [DOI] [PubMed] [Google Scholar]
- 17.Jukes T. H., Cantor C. R. Evolution of protein molecules. In: Munro H. N., editor. Mammalian Protein Metabolism. New York, NY, USA: Academic press; 1969. pp. 21–132. [Google Scholar]
- 18.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 19.Van Hintum Th. J. L. Core Collections of Plant Genetic Resources. 1995. Hierarchical approaches to analysis of genetic diversity in crop plants; pp. 23–34. [Google Scholar]
- 20.Doebley J. Isozymic evidence and the evolution of crop plants. In: Soltis D. E., Soltis P. S., editors. Isozymes in Plant Biology. Portland, Ore, USA: Dioscorides Press; 1989. pp. 165–191. [Google Scholar]
- 21.Pasquet R. S. Allozyme diversity of cultivated cowpea Vigna unguiculata (L.) Walp. Theoretical and Applied Genetics. 2000;101(1-2):211–219. doi: 10.1007/s001220051471. [DOI] [Google Scholar]
- 22.Coulibaly S., Pasquet R. S., Papa R., Gepts P. AFLP analysis of the phenetic organization and genetic diversity of Vigna unguiculata L. Walp. reveals extensive gene flow between wild and domesticated types. Theoretical and Applied Genetics. 2002;104(2-3):358–366. doi: 10.1007/s001220100740. [DOI] [PubMed] [Google Scholar]
- 23.Ba F. S., Pasquet R. S., Gepts P. Genetic diversity in cowpea [Vigna unguiculata (L.) Walp.] as revealed by RAPD markers. Genetic Resources and Crop Evolution. 2004;51(5):539–550. doi: 10.1023/b:gres.0000024158.83190.4e. [DOI] [Google Scholar]
- 24.Magembe E. M. Genetic diversity analysis among cowpea [vignaunguiculata (l.) walp] accessions from Sub-Saharan Africa using simple sequence repeats (ssr's) University of Nairobi Publications, 2008, https://www.uonbi.ac.ke/
- 25.Padulosi S., Ng N. Q. A useful and unexploited herb, Vigna marina (Leguminosae-Papilionoideae) and the taxonomic revision of its genetic diversity. Bulletin du Jardin botanique National de Belgique. 1993;62(1–4):119–126. doi: 10.2307/3668270. [DOI] [Google Scholar]
- 26.Huynh B.-L., Close T. J., Roberts P. A., et al. Gene pools and the genetic architecture of domesticated cowpea. Plant Genome. 2013;6, article 3 doi: 10.3835/plantgenome2013.03.0005. [DOI] [Google Scholar]
- 27.Alghali A. M. On‐farm evaluation of control strategies for insect pests in cowpea with emphasis on flower thrips, Megalurothrips sjostedti Trybom (Thysanoptera: Thripidae) Tropical Pest Management. 1992;38(4):420–424. doi: 10.1080/09670879209371740. [DOI] [Google Scholar]
- 28.Adewale B. D., Adeigbe O. O., Aremu C. O. Genetic distance and diversity among some cowpea (Vigna unguiculata L. Walp)genotypes. International Journal of Research in Plant Science. 2011;1:9–14. [Google Scholar]