Abstract
Available evidence suggests that Mg2+ ions are involved in reactions catalyzed by hammerhead ribozymes. However, the activity in the presence of exclusively monovalent ions led us to question whether divalent metal ions really function as catalysts when they are present. We investigated ribozyme activity in the presence of high levels of Mg2+ ions and the effects of Li+ ions in promoting ribozyme activity. We found that catalytic activity increased linearly with increasing concentrations of Mg2+ ions and did not reach a plateau value even at 1 M Mg2+ ions. Furthermore, this dependence on Mg2+ ions was observed in the presence of a high concentration of Li+ ions. These results indicate that the Mg2+ ion is a very effective cofactor but that the affinity of the ribozyme for a specific Mg2+ ion is very low. Moreover, cleavage by the ribozyme in the presence of both Li+ and Mg2+ ions was more effective than expected, suggesting the existence of a new reaction pathway—a cooperative pathway—in the presence of these multiple ions, and the possibility that a Mg2+ ion with weak affinity for the ribozyme is likely to function in structural support and/or act as a catalyst.
INTRODUCTION
Naturally existing catalytic RNAs include hammerhead, hairpin, hepatitis delta virus (HDV) and Varkud Satellite (VS) ribozymes, group I and II introns, and the RNA subunit of RNase P (1–7). In addition, recent structural and chemical analyses strongly suggest that ribosomal RNA might also be a ribozyme (8–11). In addition, the possibility exists that the RNA component of the spliceosome might be a ribozyme too (12). The earliest research on ribozymes suggested that all ribozymes might be metalloenzymes that require divalent metal ions, in particular Mg2+ ions, for catalysis, and that all might operate by a basically similar mechanism. However, subsequent, extensive studies revealed that the catalytic activity of hairpin ribozymes is independent of divalent metal ions (7,13–18). Thus, the various types of ribozymes appear to exploit different cleavage mechanisms, which depend upon the architecture of the individual ribozyme. Even hammerhead ribozymes, which have generally been characterized as typical metalloenzymes, can no longer be categorized unambiguously.
Naturally existing hammerhead ribozymes were originally identified in some RNA viruses, and it was demonstrated that they act in cis during viral replication by the rolling circle mechanism (3). In the laboratory, ribozymes have been engineered such that they act in trans against other RNA molecules and catalyze the cleavage of phosphodiester bonds at specific sites to generate specific products, each of which has a 2′,3′-cyclic phosphate and a 5′-hydroxyl group (19–22). The transesterification mechanism includes deprotonation of the 2′-hydroxyl moiety of a ribose group, nucleophilic attack of the 2′-oxygen on the adjacent phosphorus atom, and protonation of the 5′-oxyanion leaving group (Figure 1A). A large body of evidence also indicates that the P9/G10.1 site binds a metal ion with high affinity, with other metal ion-binding sites being located around the G5 nucleobase and A13 phosphate near the site of cleavage (23–36). Thus, the idea that ribozymes are metalloenzymes has been generally accepted. However, it was reported recently that hammerhead ribozymes are active in the presence of very high concentrations of monovalent cations, such as Li+ or NH4+ ions, in the absence of divalent metal ions (37). This finding raises the possibility that hammerhead ribozymes should not be classified as metalloenzymes. Therefore, we decided to investigate ribozyme activity in the presence of Mg2+ ions and the effects of Mg2+ ions in the presence of Li+ ions on ribozyme activity, using a well-studied model hammerhead ribozyme (R32) and its substrate (S11), both of which are shown in Figure 1B (5–7,25,28,32–34,38–41).
Figure 1.
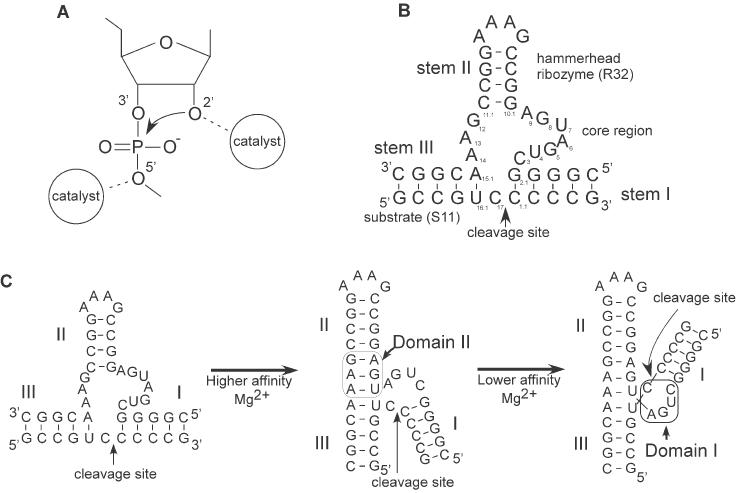
(A) Schematic representation of the proposed mechanism of the hammerhead ribozyme reaction. The 2′-hydroxyl moiety is activated by the catalyst and attacks the adjacent phosphate nucleophilically, with subsequent cleavage of the bond at the 5′-oxygen. The developing negative charge on the leaving 5′-oxygen is stabilized by another catalyst. (B) The sequences and secondary structures of the hammerhead ribozyme (R32) and substrate (S11) used in this study. (C) The proposed two-stage folding scheme for the hammerhead ribozyme–substrate complex. The higher-affinity Mg2+ ion(s) drives the formation of domain II, which contains non-Watson–Crick pairings and the lower-affinity Mg2+ ion(s) rotates around helix I, forming the catalytic core.
We investigated the dependence on the concentration of Mg2+ and Li+ ions of ribozyme activity over a range from 5 mM to ∼1 M Mg2+ ions and 1 to 5 M Li+ ions, respectively. Although several research groups have reported similar analyses (42–50), the concentrations of Mg2+ ions used in most of these studies were <100 mM, and little information is available about activity at a higher concentration, such as 1 M. The activity at high concentrations of Mg2+ ions, as compared to physiological concentrations, is useful for studies of the chemistry of the hammerhead ribozyme and it allowed us to estimate the intrinsic cleavage rate constant and to compare an enzymatic reaction to a non-enzymatic reaction. We were also able to calculate acceleration energy more precisely and to investigate the dependence on Mg2+ ions in the presence of high concentrations of Li+ ions. Considering the results of such analyses, we propose the existence of a new cooperative pathway that involves divalent metal ions, such as Mg2+, and monovalent ions, such as Li+, in the reaction catalyzed by a hammerhead ribozyme. We also discuss the nature of the catalysts.
MATERIALS AND METHODS
Preparation of the hammerhead ribozyme and its substrate
The ribozyme (R32) and its substrate (S11) were synthesized chemically on a DNA/RNA synthesizer (model 394; PE Applied Biosystems, Foster City, CA) using phosphoramidic chemistry with 2′-tert butyldimethylsilyl (TBDMS) protection as described previously (25). Chemically synthesized oligonucleotides were deprotected in a mixture of 28% ammonia and ethanol (3:1) at 55°C for 8 h. The mixture was evaporated to dryness and the residue was allowed to dissolve in 1 ml of 1 M tetrabutylammonium fluoride (TBAF; Sigma-Aldrich, Japan K.K., Tokyo, Japan) at room temperature for 12 h. After the addition of 1 ml of water, the mixture was desalted on a gel-filtration column (Bio-Gel P-4; Bio-Rad Laboratories, Hercules, CA). Fully deprotected oligonucleotides were purified by gel electrophoresis on a 20% polyacrylamide gel that contained 7 M urea, the respective bands were excised from the gel, and oligonucleotides were extracted in water. The oligonucleotides were recovered by ethanol precipitation and then solutions were desalted on a gel-filtration column (TSK-GEL G3000PW; TOSOH, Tokyo, Japan) by high-performance liquid chromatography (HPLC) with ultrapure water. All of the RNA oligomers were quantitated in terms of absorbance at 260 nm.
The substrate S11 was labeled with [γ-32P]ATP by T4 polynucleotide kinase (Takara Bio, Inc., Shiga, Japan). Radiolabeled S11 was purified on a 20% polyacrylamide gel that contained 7 M urea and then purified by the standard procedure, as described above, with desalting on a gel-filtration column (NAP™-10 column; Amersham Biosciences, K.K., Tokyo, Japan).
Quantification of the ribozyme reaction
All ribozyme reactions were performed under single-turnover conditions to ensure that conversion of the ribozyme–substrate complex to the ribozyme–product complex could be monitored kinetically without complications due to complex formation and slow release of products. The solution for the ribozyme reaction contained a trace amount of 5′-32P-labeled S11 and 25 mM Bis-Tris buffer at pH 6.0 and 25°C. The pH values of all 1.25× pre-stock Bis-Tris buffers that contained appropriate metal ions (metal-ion–buffer) were adjusted appropriately with HCl and we confirmed that each buffer had the appropriate pH under the chosen reaction conditions. Each reaction was initiated by addition of the substrate to a mixture of metal-ion–buffer and ribozyme, and aliquots were removed from the reaction mixture at appropriate intervals. Each aliquot was mixed with more than three volumes of a stop solution that contained 100 mM MES (pH 6), 100 mM EDTA, 7 M urea, xylene cyanol (0.1%) and bromophenol blue (0.1%), and then it was stored at −80°C prior to analysis. Since EDTA does not chelate Mg2+ and Li+ ions efficiently at lower pH values, we confirmed that reactions did not continue in the stop solution and that quenching was effective due to high concentration of urea in this solution. Uncleaved substrate and 5′-cleaved products were separated on a 20% polyacrylamide gel that contained 7 M urea. The extent of each cleavage reaction was quantitated with an imageanalyzer (Storm 830; Molecular Dynamics, Sunnyvale, CA). For each reaction, an observed rate constant was determined by non-linear least-squares fitting of the time course of reaction using a pseudo-first-order equation.
RESULTS AND DISCUSSION
The dependence of the activity of the hammerhead ribozyme on the concentration of Li+ ions
We examined the dependence of the activity of the ribozyme on the concentration of Li+ ions to confirm that Li+ ions affect the ribozyme's activity, as reported previously by others (51). We performed reactions under single-turnover conditions at pH 7.5 and 25°C. The results are shown in Figure 2A. Although the activity reached a plateau at ∼3 M Li+ ions, the dependence on the concentration of Li+ ions was observed (with a slope of three) below the plateau. In the study by O'Rear et al. (51), the hammerhead ribozyme reaction exhibited second-order dependence on Li+ ions up to 4 M at pH 7.5. Although our curve is steeper than the curve that they obtained under the same conditions, with the exception of the concentrations of Li+ ions, the slopes of both profiles obtained with Li+ ions are clearly steeper than those of profiles obtained with Mg2+ ions. Our profile is unique insofar as we observed saturation of the ribozyme reaction in the range of 3–5 M (Figure 2A). This plateau suggests that the ribozyme reaction might involve the binding of Li+ ions to the ribozyme–substrate complex.
Figure 2.
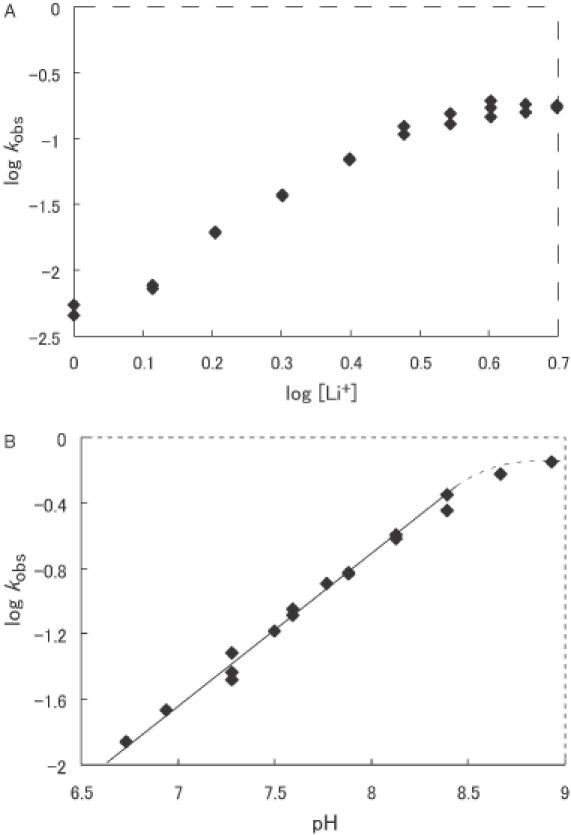
(A) Dependence of cleavage activity on the concentration of Li+ ions. The activity increased linearly with the concentration of Li+ ions until ∼3 M. Reaction conditions: 25 mM Bis-Tris, pH 7.5, and varying concentrations of Li+ ions as indicated. (B) Dependence on pH of the activity in 2 M Li+ ions. The Li+-containing buffer used in this experiment included Bis-Tris propane and Tris. The cleavage rate increased linearly with pH, yielding a slope of 0.9.
We also confirmed the dependence on pH, with a slope of unity, of the activity in the presence of a high concentration of Li+ ions, as shown in Figure 2B. This result supports the hypothesis that the cleavage step is the rate-limiting step even at such a high ionic strength. The pH profile is also consistent with the previous report by Curtis and Bartel (52). As shown in Figure 2A, the observed rate constant at a saturating concentration of Li+ ions (3–5 M) at pH 7.5 was ∼0.17 min−1. The estimated observed rate constant in the presence of 800 mM Mg2+ ions was 32 min−1 at pH 7.5, as determined from the value of 1 min−1 at pH 6 at the same temperature as that at which the Li+ experiment was performed (see later). The difference was, thus, approximately 200-fold under similar conditions. The true difference might be even greater because, in contrast to the results with Li+, the rate at 800 mM Mg2+ does not reach a plateau value (see later). These data indicate clearly that Mg2+ ions are more suitable for an effective hammerhead ribozyme reaction than any other monovalent ions (Li+ ions have the highest activity of all monovalent ions in the ribozyme reaction).
The dependence of the activity of the hammerhead ribozyme on the concentration of Mg2+ ions
We attempted first to determine how many Mg2+ ions might be involved in our model ribozyme reaction and the saturating concentration of Mg2+ ions in the reaction, beyond which the cleavage rate constant no longer increases. We examined the dependence on the concentration of Mg2+ ions of the activity of the R32 hammerhead ribozyme up to ∼1 M Mg2+ ions. We performed reactions with R32-S11 (Figure 1B) under single-turnover conditions with a saturating amount of ribozyme with respect to the amount of S11 for the same reasons as noted above. The reaction in the presence of Mg2+ ions is accelerated with increases in pH, with a slope of unity (7,40,43,44,47,53). We adjusted the pH of reactions to 6.0 to slow down the reaction so that we could determine the rate constants of rapid reactions precisely. As shown by closed circles in Figure 3, the dependence on Mg2+ ions was approximately first-order. However, no plateau was reached under our conditions, even above 800 mM Mg2+ ions. The continuous increase in rate constant upon the addition of more and more Mg2+ ions indicates the involvement of one Mg2+ ion that has low affinity for the hammerhead ribozyme–substrate complex. At 800 mM Mg2+ ions, the rate constant approached 1.1 min−1 at pH 6.0, and this is the limit of detection of a rapid cleavage reaction under standard laboratory conditions.
Figure 3.
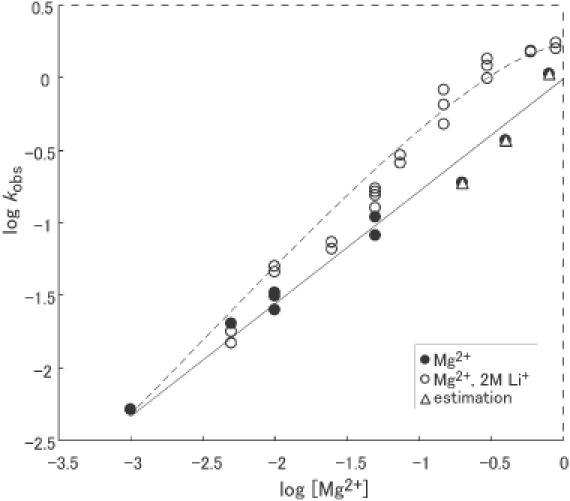
The Mg2+-titration curve for cleavage activity in the presence (open circles) and absence (filled circles) of Li+ ions. In the absence of Li+ ions, the line drawn with the linear best fit had a slope of 0.7. The activity increased with increasing concentrations of Mg2+ ions and did not reach a plateau value even at 800 mM Mg2+ ions. In the presence of 2 M Li+ ions, the activity increased with increases in the concentration of Mg2+ ions up to ∼600 mM. Triangles are the calculated rates in the presence of Mg2+ ions plus 2 M Li+ ions, which were determined simply by adding the respective reaction rates together. Since the observed rate in the presence of 2 M Li+ ions only at pH 6 was <0.01 min−1, triangles are at almost the same positions as the filled circles. The rates observed in the presence of Mg2+ ions and Li+ ions together (open circles) were clearly higher than the calculated values (triangles).
Analyses of the structure of hammerhead ribozymes and of the conformational changes caused by interactions with Mg2+ ions have indicated that two major conformational changes occur: the formation of domain II, which is followed by the formation of domain I, as shown in Figure 1C (38,45,48,50,54). The formation of domain II results in coaxial stacking of helices II and III, induced by the binding of a higher-affinity Mg2+ ion(s) to P9 phosphate and N7 of G10.1 (P9/G10.1) of the ribozyme–substrate complex (38,45,48,50,54–58). The second transition is the formation of the catalytic domain with movement of stem I toward stem II, which is induced by the binding of a lower-affinity Mg2+ ion(s). The Kd values of the two domains have been determined by various methods to be several hundred micromolar and several millimolar, respectively (45,46,48,50,54). Thus, at several hundred millimolar Mg2+ ions, the formation of domains II and I should be complete.
Taking this structural information into consideration, we can reasonably conclude that the very-low-affinity Mg2+ ion that we detected in the present study might be involved in some step other than the formation of domains I and II. This step might be a conformational change or the binding of a catalytic species to the ribozyme–substrate complex. Walter and coworkers recently reported that a third and previously undetected metal ion at rather high concentrations might play a role in the induction of a minor conformational adjustment that leads to the formation of the active state after the formation of domains I and II (59). The Mg2+ ion that we detected had very low affinity, and the relative level of truly active ribozyme species at a concentration of several millimolar Mg2+ ions corresponded to <1% of all the ribozyme–substrate complexes in the reaction mixture [compare 1 min−1 in 10 mM MgCl2 at pH 8 and 25°C (60) with 100 min−1 in 800 mM MgCl2 at the same pH and the same temperature (see later)].
One might agree that Figure 3 does not support the existence of a very-low-affinity binding site for an Mg2+ ion on the ribozyme–substrate complex since the binding curve does not reach a plateau. If the dependence that we observed here were due only to ionic strength, no plateau would be observed. However, we did observe a plateau in the case of Li+ ions (Figure 2A). Because reactions were so rapid, even at low pH, we could not monitor the kinetics at even higher concentrations of Mg2+ ions, such as 3 M. However, it is likely that, in ribozyme reactions, ionic strength is less important than the ion radius and/or the electron density of metal ions since (i) a correlation between the ion radius of Group I monovalent metal ions and ribozyme activity has been observed (52,57) and (ii) an apparent plateau was reached in the presence of high concentrations of Mg2+ ions plus 2 M Li+ ions (Figure 3). Although we cannot calculate a Hill constant for Mg2+ ions and estimate the number of binding sites for Mg2+ ions from our current data, all the available data strongly support the possibility of the existence of a very-low-affinity metal-binding site(s).
Misra and Draper (61) proposed a model for the stabilization of RNA by Mg2+ ions that arises from two distinct binding modes, diffuse binding and site binding. Diffusely bound Mg2+ ions are defined as fully solvated ions that interact with RNA only through long-range electrostatic interactions. Site-bound Mg2+ ions are defined as partially desolvated ions that are attracted to electronegative pockets. In general, the affinity of diffusely bound Mg2+ ions appears to be lower than that of site-bound Mg2+ ions. Thus, it is possible that the very-low-affinity Mg2+ ions that we detected here might be involved in diffuse binding. Although diffuse binding can sometimes play a dominant role in stabilizing the tertiary structures of small RNAs (61), we cannot exclude the possibility that such diffuse binding Mg2+ ion might play a role in catalysis in the ribozyme reaction at a specific site in the ribozyme–substrate complex.
In 800 mM Mg2+ ions, the observed rate constant of the reaction catalyzed by the hammerhead ribozyme can be estimated to be ∼100 min−1 at pH 8 and 25°C, from the dependence on pH with a slope of unity. The estimated rate constant at concentrations of Mg2+ ions >800 mM should be much higher than that at 800 mM since the cleavage reaction did not reach saturation at 800 mM Mg2+ and approached the estimated observed rate constant of a ‘kissing ribozyme’, under saturating conditions with respect to Mg2+ ions and pH (62).
The dependence of the activity of the hammerhead ribozyme on Mg2+ ions in the presence of a high concentration of Li+ ions
In order to investigate the properties of Mg2+ ions in the ribozyme reaction in further detail, we examined the dependence on Mg2+ ions in the presence of a high concentration (2 M) of Li+ ions. We chose the Li+ ion as the monovalent cation because this ion is the most active of a large variety of monovalent cations (57). At 2 M Li+ ions at pH 6.0, the hammerhead ribozyme had detectable activity in the absence of Mg2+ ions. We varied the concentration of Mg2+ ions from 5 to 900 mM and measured the rate constant. As shown by open circles in Figure 3, we observed approximately first-order dependence on the concentration of Mg2+ ions up to 600 mM, and then the rate constant started to reach a plateau from 600 to 900 mM Mg2+ ions. These data indicate that, even in the presence of a very high concentration of Li+ ions, an Mg2+ ion with very low affinity plays an important role in the hammerhead ribozyme reaction. The plateau that seemed to appear in the presence of high concentrations of Li+ ions (Figure 3, open circles) was not observed in the absence of Li+ ions (closed circles). This apparent plateau was not due to misfolding of RNA at such a high ionic concentration because the extent of cleavage at the end of the reaction was always >90% under these conditions. The plateau might indicate that Li+ ions help the ribozyme–substrate complex to fold into a more active form, contributing to the enhanced binding of the very-low-affinity Mg2+ ion(s) to the complex.
We examined reactions in the presence of Mg2+ ions only (Figure 3, closed circles) and in the presence of Li+ ions only (57). We then calculated rate constants (Figure 3, triangles) on the assumption that Mg2+ and Li+ ions function independently. It should be noted that the observed rate constant in the presence of both Mg2+ ions and 2 M Li+ ions together (Figure 3, open circles) was definitely higher than the calculated rates (Figure 3, triangles) at pH 6 (compare open circles with triangles in Figure 3). One might expect that, in such an experiment (i) the activities in the presence of both metal ions together would be lower than the estimated activities (triangles) because of the lower ability of Mg2+ ions to bind to RNA in the presence of high ionic strength due to 2 M Li+ ions and (ii) the stimulation by Mg2+ ions in the presence of such a high concentration of Li+ ions would be smaller than in the absence of Li+ ions. It has been reported that stimulation by Cd2+ ions is reduced in a ribozyme reaction in the presence of 4 M Li+ ions (51), while stimulation by Cd2+ ions is observed in reaction mixtures that contain 10 mM Ca2+ ions instead of 2 M Li+ ions (23). However, the results in the present study were quite opposite: (i) the activities in the presence of Mg2+ ions and 2 M Li+ ions together (open circles in Figure 3) were higher than the calculated activities (triangles in Figure 3), and (ii) the stimulation by Mg2+ ions in the presence of 2 M Li+ ions (open circles in Figure 3) was even greater than that by Mg2+ ions in the absence of Li+ ions (closed circles in Figure 3).
Our observations suggest the existence of a new cooperative pathway that involves Li+ and Mg2+ ions, in which both metal ions function cooperatively in structural support and/or as the catalyst(s) to increase the rate constant of cleavage (57). In the case of cleavage of RNA by another type of ribozyme, the RNA subunit of RNase P, a similar cooperativity between the metal ions, e.g. Mg2+ and Ca2+, has been reported (63).
What is the catalyst in the hammerhead reaction?
To the best of our knowledge, this report is the first to describe the dependence of ribozyme activity on a high concentration of Mg2+ ions under single-turnover conditions (Figure 3). The reaction did not reach a plateau even at 800 mM Mg2+ ions. Under these conditions at pH 6, the observed rate constant was 1.1 min−1. The activity of the ribozyme is known to depend on pH, with a slope of unity. Thus, we can estimate an observed rate constant for the ribozyme reaction of 110 min−1 at pH 8. It is very unlikely that this rate constant of cleavage can occur without an effective catalyst(s) (64). So, what is the catalyst(s) of the reaction?
In studies of solvent isotope effects on the ribozyme reaction in the presence of deuterium and Li+, Mg2+ or NH4+ ions, we failed previously to observe any proton transfers in the transition state during the ribozyme reaction in the presence of either Li+ or Mg2+ ions but we did observe proton transfer in the presence of NH4+ ions (53,65). We interpreted such results by suggesting that metal ions, such as Mg2+ and Li+, function as a Lewis acid catalyst while NH4+ ions function as a general acid catalyst during the ribozyme reaction. Thus, the catalyst can change according to the conditions around the ribozyme. On the basis of this hypothesis and the involvement of two other kinds of Mg2+ ions in the formation of domains I and II, it is possible that the newly identified Mg2+ ion with very low affinity that we observed in this study might be the true catalyst. Our novel cooperative pathway might involve an Mg2+ ion in a catalytic role after monovalent cations and other Mg2+ ions have acted to generate the pre-active conformation just before chemical cleavage by the hammerhead ribozyme. In the presence of monovalent ions exclusively, monovalent ions can also function as the catalyst but they are not as effective. Mg2+ ions have a higher charge density than Li+ ions and would function better as catalyst in the reaction. Thus, when both Mg2+ and Li+ ions are present in the reaction mixture, the high activity of the ribozyme reaction is likely due to an Mg2+ ion catalyst. The Li+ ions act to support the formation of the domains I and II in cooperation with Mg2+ ions, as discussed above.
We are at present constructing a reaction scheme for ribozyme reactions in the presence of both Mg2+ and Li+ ions, namely, a ‘cooperative pathway’, by determining the details of the stoichiometric relationships among these metal ions (57). Examination of ribozyme-catalyzed reactions in the presence of various metal ions in combination should clarify the roles of metal ions in catalysis.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Laura Nelson for helpful comments.
REFERENCES
- 1.Cech T.R., Zaug,A.J. and Grabowski,P.J. (1981) In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell, 27, 487–496. [DOI] [PubMed] [Google Scholar]
- 2.Guerrier-Takada C., Gardiner,K., Marsh,T., Pace,N. and Altman,S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell, 35, 849–857. [DOI] [PubMed] [Google Scholar]
- 3.Symons R.H. (1992) Small catalytic RNAs. Annu. Rev. Biochem., 61, 641–671. [DOI] [PubMed] [Google Scholar]
- 4.Carola C. and Eckstein,F. (1999) Nucleic acid enzymes. Curr. Opin. Chem. Biol., 3, 274–283. [DOI] [PubMed] [Google Scholar]
- 5.Warashina M., Zhou,D.M., Kuwabara,T. and Taira,K. (1999) Ribozyme structure and function. In Söll,D., Nishimura,S. and Moore,P.B. (eds), Comprehensive Natural Products Chemistry. Elsevier Science Ltd, Oxford, Vol. 6, pp. 235–268. [Google Scholar]
- 6.Warashina M., Takagi,Y., Stec,W.J. and Taira,K. (2000) Differences among mechanisms of ribozyme-catalyzed reactions. Curr. Opin. Biotechnol., 11, 354–362. [DOI] [PubMed] [Google Scholar]
- 7.Takagi Y., Warashina,M., Stec,W.J., Yoshinari,K. and Taira,K. (2001) Recent advances in the elucidation of the mechanisms of action of ribozymes. Nucleic Acids Res., 29, 1815–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noller H.F., Hoffarth,V. and Zimniak,L. (1992) Unusual resistance of peptidyl transferase to protein extraction procedures. Science, 256, 1416–1419. [DOI] [PubMed] [Google Scholar]
- 9.Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 10.Muth G.W., Ortoleva-Donnelly,L. and Strobel,S.A. (2000) A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science, 289, 947–950. [DOI] [PubMed] [Google Scholar]
- 11.Cech T.R. (2000) Structural biology. The ribosome is a ribozyme. Science, 289, 878–879. [DOI] [PubMed] [Google Scholar]
- 12.Collins C.A. and Guthrie,C. (2000) The question remains: is the spliceosome a ribozyme? Nature Struct. Biol., 10, 850–854. [DOI] [PubMed] [Google Scholar]
- 13.Hampel A. and Cowan,J.A. (1997) A unique mechanism for RNA catalysis: the role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol., 4, 513–517. [DOI] [PubMed] [Google Scholar]
- 14.Nesbitt S., Hegg,L.A. and Fedor,M.J. (1997) An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol., 4, 619–630. [DOI] [PubMed] [Google Scholar]
- 15.Young K.J., Gill,F. and Grasby,J.A. (1997) Metal ions play a passive role in the hairpin ribozyme catalysed reaction. Nucleic Acids Res., 25, 3760–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowrira B.M., Berzal-Herranz,A. and Burke,J.M. (1993) Ionic requirements for RNA binding, cleavage, and ligation by the hairpin ribozyme. Biochemistry, 32, 1088–1095. [DOI] [PubMed] [Google Scholar]
- 17.Earnshaw D.J. and Gait,M.J. (1998) Hairpin ribozyme cleavage catalyzed by aminoglycoside antibiotics and the polyamine spermine in the absence of metal ions. Nucleic Acids Res., 26, 5551–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seyhan A.A. and Burke,J.M. (2000) Mg2+-independent hairpin ribozyme catalysis in hydrated RNA films. RNA, 6, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlenbeck O.C. (1987) A small catalytic oligoribonucleotide. Nature, 328, 596–600. [DOI] [PubMed] [Google Scholar]
- 20.Haseloff J. and Gerlach,W.L. (1988) Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature, 334, 585–591. [DOI] [PubMed] [Google Scholar]
- 21.Hutchins C.J., Rathjen,P.D., Forster,A.C. and Symons,R.H. (1986) Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res., 14, 3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koizumi M., Hayase,Y., Iwai,S., Kamiya,H., Inoue,H. and Ohtsuka,E. (1989) Design of RNA enzymes distinguishing a single base mutation in RNA. Nucleic Acids Res., 17, 7059–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S., Karbstein,K., Peracchi,A., Beigelman,L. and Herschlag,D. (1999) Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry, 38, 14363–14378. [DOI] [PubMed] [Google Scholar]
- 24.Maderia M., Hunsicker,L.M. and DeRose,V.J. (2000) Metal–phosphate interactions in the hammerhead ribozyme observed by 31P NMR and phosphorothioate substitutions. Biochemistry, 39, 12113–12120. [DOI] [PubMed] [Google Scholar]
- 25.Yoshinari K. and Taira,K. (2000) A further investigation and reappraisal of the thio effect in the cleavage reaction catalyzed by a hammerhead ribozyme. Nucleic Acids Res., 28, 1730–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peracchi A., Beigelman,L., Scott,E.C., Uhlenbeck,O.C. and Herschlag,D. (1997) Involvement of a specific metal ion in the transition of the hammerhead ribozyme to its catalytic conformation. J. Biol. Chem., 272, 26822–26826. [DOI] [PubMed] [Google Scholar]
- 27.Knöll R., Bald,R. and Fürste,J.P. (1997) Complete identification of nonbridging phosphate oxygens involved in hammerhead cleavage. RNA, 3, 132–140. [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamatsu Y., Warashina,M., Kuwabara,T., Tanaka,Y., Yoshinari,K. and Taira,K. (2000) Significant activity of a modified ribozyme with N7-deazaguanine at G10.1: the double-metal-ion mechanism of catalysis in reactions catalysed by hammerhead ribozymes. Genes Cells, 5, 603–612. [DOI] [PubMed] [Google Scholar]
- 29.Peracchi A., Beigelman,L., Usman,N. and Herschlag,D. (1996) Rescue of abasic hammerhead ribozymes by exogenous addition of specific bases. Proc. Natl Acad. Sci. USA, 93, 11522–11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peracchi A., Karpeisky,A., Maloney,L., Beigelman,L. and Herschlag,D. (1998) A core folding model for catalysis by the hammerhead ribozyme accounts for its extraordinary sensitivity to abasic mutations. Biochemistry, 37, 14765–14775. [DOI] [PubMed] [Google Scholar]
- 31.Murray J.B. and Scott,W.G. (2000) Does a single metal ion bridge the A-9 and scissile phosphate groups in the catalytically active hammerhead ribozyme structure? J. Mol. Biol., 296, 33–41. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka Y., Morita,E.H., Hayashi,H., Kasai,Y., Tanaka,T. and Taira,K. (2000) Well-conserved tandem G•A pairs and the flanking C•G pair in hammerhead ribozymes are sufficient for capture of structurally and catalytically important metal ions. J. Am. Chem. Soc., 122, 11303–11310. [Google Scholar]
- 33.Suzumura K., Warashina,M., Yoshinari,K., Tanaka,Y., Kuwabara,T., Orita,M. and Taira,K. (2000) Significant change in the structure of a ribozyme upon introduction of a phosphorothioate linkage at P9: NMR reveals a conformational fluctuation in the core region of a hammerhead ribozyme. FEBS Lett., 473, 106–112. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka Y., Kojima,C., Morita,E.H., Kasai,K., Ono,A., Kainosho,M. and Taira,K. (2002) Identification of the metal ion binding site on an RNA motif from hammerhead ribozymes using 15N-NMR spectroscopy. J. Am. Chem. Soc., 124, 4595–4601. [DOI] [PubMed] [Google Scholar]
- 35.Hansen M.R., Simorre,J.P., Hanson,P., Mokler,V., Bellon,L., Beigelman,L. and Pardi,A. (1999) Identification and characterization of a novel high affinity metal-binding site in the hammerhead ribozyme. RNA, 5, 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feig A.L., Panek,M., Horrocks,W.D. and Uhlenbeck,O.C. (1999) Probing the binding of Tb(III) and Eu(III) to the hammerhead ribozyme using luminescence spectroscopy. Chem. Biol., 6, 801–810. [DOI] [PubMed] [Google Scholar]
- 37.Murray J.B., Seyhan,A.A., Walter,N.G., Burke,J.M. and Scott,W.G. (1998) The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol., 5, 587–595. [DOI] [PubMed] [Google Scholar]
- 38.Zhou J.-M., Zhou,D.-M., Takagi,Y., Kasai,Y., Inoue,A., Baba,T. and Taira,K. (2002) Existence of efficient divalent metal ion-catalyzed and inefficient divalent metal ion-independent channels in reactions catalyzed by a hammerhead ribozyme. Nucleic Acids Res., 30, 2374–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasai Y., Shizuku,H., Takagi,Y., Warashina,M. and Taira,K. (2002) Measurements of weak interactions between truncated substrates and a hammerhead ribozyme by competitive kinetic analyses: implications for the design of new and efficient ribozymes with high sequence specificity. Nucleic Acids Res., 30, 2383–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou D.-M. and Taira,K. (1998) The hydrolysis of RNA: from theoretical calculations to the hammerhead ribozyme-mediated cleavage of RNA. Chem. Rev., 98, 991–1026. [DOI] [PubMed] [Google Scholar]
- 41.Takagi Y. and Taira,K. (1995) Temperature-dependent change in the rate-determining step in a reaction catalyzed by a hammerhead ribozyme. FEBS Lett., 361, 273–276. [DOI] [PubMed] [Google Scholar]
- 42.Koizumi M. and Ohtsuka,E. (1991) Effects of phosphorothioate and 2-amino groups in hammerhead ribozymes on cleavage rates and Mg2+ binding. Biochemistry, 30, 5145–5150. [DOI] [PubMed] [Google Scholar]
- 43.Dahm S.C., Derrick,W.B. and Uhlenbeck,O.C. (1993) Evidence for the role of solvated metal hydroxide in the hammerhead cleavage mechanism. Biochemistry, 32, 13040–13045. [DOI] [PubMed] [Google Scholar]
- 44.Hendry P. and McCall,M.J. (1995) A comparison of the in vitro activity of DNA-armed and all-RNA hammerhead ribozymes. Nucleic Acids Res., 23, 3928–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bassi G.S., Murchie,A.I.H., Walter,F., Clegg,R.M. and Lilley,D.M.J. (1997) Ion-induced folding of the hammerhead ribozyme: a fluorescence resonance energy transfer study. EMBO J., 16, 7481–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horton T.E., Clardy,D.R. and DeRose,V.J. (1998) Electron paramagnetic resonance spectroscopic measurement of Mn2+ binding affinities to the hammerhead ribozyme and correlation with cleavage activity. Biochemistry, 37, 18094–18101. [DOI] [PubMed] [Google Scholar]
- 47.Peracchi A. (1999) Origins of the temperature dependence of hammerhead ribozyme catalysis. Nucleic Acids Res., 27, 2875–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bassi G.S., Møllegaard,N.E., Murchie,A.I.H. and Lilley,D.M.J. (1999) RNA folding and misfolding of the hammerhead ribozyme. Biochemistry, 38, 3345–3354. [DOI] [PubMed] [Google Scholar]
- 49.Hunsicker L.M. and DeRose,V.J. (2000) Activities and relative affinities of divalent metals in unmodified and phosphorothioate-substituted hammerhead ribozymes. J. Inorg. Biochem., 80, 271–281. [DOI] [PubMed] [Google Scholar]
- 50.Hammann C., Norman,D.G. and Lilley,D.M.J. (2001) Dissection of the ion-induced folding of the hammerhead ribozyme using 19F-NMR. Proc. Natl Acad. Sci. USA, 98, 5503–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Rear J.L., Wang,S., Feig,A.L., Beigelman,L., Uhlenbeck,O.C. and Herschlag,D. (2001) Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA, 7, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Curtis E.A. and Bartel,D.P. (2001) The hammerhead cleavage reaction in monovalent cations. RNA, 7, 546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takagi Y. and Taira,K. (2002) Detection of a proton-transfer process by kinetic solvent isotope effects in NH4+-mediated reactions catalyzed by a hammerhead ribozyme. J. Am. Chem. Soc., 124, 3850–3852. [DOI] [PubMed] [Google Scholar]
- 54.Bassi G.S., Møllegaard,N.E., Murchie,A.I.H., von Kitzing,E. and Lilley,D.M.J. (1995) Ionic interactions and the global conformations of the hammerhead ribozyme. Nature Struct. Biol., 2, 45–55. [DOI] [PubMed] [Google Scholar]
- 55.Schiemann O., Fritscher,J., Kisseleva,N., Sigurdsson,S.T. and Prisner,T.F. (2003) Structural investigation of a high-affinity MnII binding site in the hammerhead ribozyme by EPR spectroscopy and DFT calculations. Effects of neomycin B on metal-ion binding. ChemBioChem, 4, 1057–1065. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka Y., Kasai,Y., Mochizuki,S., Wakisaka,A., Morita,E.H., Kojima,C., Toyozawa,A., Kondo,Y., Taki,M., Takagi,Y., Inoue,A., Yamasaki,K. and Taira,K. (2004). Nature of the chemical bond formed with the structural metal ion at the A9/G10.1 motif derived from hammerhead ribozymes. J. Am. Chem. Soc., 126, 744–752. [DOI] [PubMed] [Google Scholar]
- 57.Takagi Y., Inoue,A. and Taira,K. (2004). Analysis of on a cooperative pathway involving multiple cations in hammerhead reactions. J. Am. Chem. Soc., in press. [DOI] [PubMed] [Google Scholar]
- 58.Warashina M., Kuwabara,T., Nakamatsu,Y., Takagi,Y., Kato,Y. and Taira,K. (2004) Analysis of the conserved P9-G10.1 metal binding motif in hammerhead ribozymes with extra nucleotide inserted between A9 and G10.1 residues. J. Am. Chem. Soc., in press. [DOI] [PubMed] [Google Scholar]
- 59.Rueda D., Wick,K., McDowell,S.E. and Walter,N.G. (2003) Diffusely bound Mg2+ ions slightly reorient stems I and II of the hammerhead ribozyme to increase the probability of formation of the catalytic core. Biochemistry, 42, 9924–9936. [DOI] [PubMed] [Google Scholar]
- 60.Stage-Zimmermann T.K. and Uhlenbeck,O.C. (1998) Hammerhead ribozyme kinetics. RNA, 4, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Misra V.K. and Draper,D.E. (2002) The linkage between magnesium binding and RNA folding. J. Mol. Biol., 317, 507–521. [DOI] [PubMed] [Google Scholar]
- 62.Khvorova A., Lescoute,A., Westhof,E. and Jayasena,S.D. (2003) Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nature Struct. Biol., 10, 708–712. [DOI] [PubMed] [Google Scholar]
- 63.Brännvall M. and Kirsebom,L.A. (2001) Metal ion cooperativity in ribozyme cleavage of RNA. Proc. Natl Acad. Sci. USA, 98, 12943–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y. and Breaker,R.R. (1999) Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc., 121, 5364–5372. [Google Scholar]
- 65.Sawata S., Komiyama,M. and Taira,K. (1995) Kinetic evidence based on solvent isotope effects for the nonexistence of a proton-transfer process in reactions catalyzed by a hammerhead ribozyme—implication to the double-metal-ion mechanism of catalysis. J. Am. Chem. Soc., 117, 2357–2358. [Google Scholar]


