Abstract
Active cytoplasmic RNA localization depends on the attachment of RNA-binding proteins that dictate the destination of the RNA molecule. In this study, we used an electrophoretic mobility-shift assay in combination with equilibrium and kinetic analyses to characterize the assembly of the human zipcode-binding protein IMP1 on targets in the 3′-UTR from Igf-II mRNA and in H19 RNA. In both cases, two molecules of IMP1 bound to RNA by a sequential, cooperative mechanism, characterized by an initial fast step, followed by a slow second step. The first step created an obligatory assembly intermediate of low stability, whereas the second step was the discriminatory event that converted a putative RNA target into a ‘locked’ stable RNP. The ability to dimerize was also observed between members of the IMP family of zipcode-binding proteins, providing a multitude of further interaction possibilities within RNP granules and with the localization apparatus.
INTRODUCTION
RNA localization is a way of achieving asymmetric protein synthesis, which is an important feature of cell migration during the development of multicellular organisms. Somewhat paradoxically, the paradigm in terms of molecular details of RNA localization is still provided by Ash1 mRNA in the unicellular Saccharomyces cerevisiae. Here, the synthesis of Ash1p at the bud tip is governed by localization of the corresponding mRNA via intracellular transport as an RNP by a myosin motor on microfilaments [reviewed in (1)]. Our knowledge of the molecular details behind mRNA transport in other systems is more rudimentary, although evidence for molecular motors such as kinesins and dynein transporting mRNA cargo on microtubules is emerging, especially in Drosophila [reviewed in (2)]. Elucidation of the trans-acting factors connecting the RNA cargo with the molecular motors is in progress, but the mechanistic details which provide selectivity and stability for long-distance transport of the appropriate RNA are virtually unknown.
The VICKZ family of zipcode-binding proteins [acronym suggested in (3)] has members such as Vg1RBP/Vera in Xenopus laevis, IMP1-3 in human, CRD-BP in mouse, KOC in human and ZBP1 in chicken, all of which contain two RNA recognition motifs and four hnRNP K homology domains (4–9). The functional entity, at least in terms of high-affinity RNA-binding and granule formation, is provided by the four KH domains (10), which constitute the phylogenetically conserved part in invertebrates (11). This family of zipcode-binding proteins displays asymmetric cytoplasmic localization and attaches to several mRNAs involved in cell migration and growth, of which β-actin mRNA has been shown to localize to lamellipodia in chicken embryo fibroblasts and to growth cones in developing neurons via a so-called zipcode in the 3′-untranslated region (3′-UTR) [reviewed in (12)]. In addition, the untranslated H19 RNA is directed to lamellipodia in NIH 3T3 cells by cis-elements in its 3′ part (13). Although cytoplasmic at steady state, members of the family such as chicken ZBP1 and human IMP1 have been shown to enter the nucleus (14,15), which has led to the proposal that mRNA cargo is picked up in the nucleus and—by inference—its subsequent fate determined by this nuclear event (16,17). Regardless of whether selection of cargo mRNA takes place in the nucleus or in the cytoplasm, the selection has to be specific and, once attached, stable; otherwise, the cytoplasmic transport of the assembled RNP granules will be jeopardized.
In this study, we provide insight into how IMP1 attaches to two different RNA targets, one being a cDNA SELEX target identified in mouse Igf-II 3′-UTR and the other a previously identified target in human H19 RNA. The details of the binding mechanism are obtained by applying both equilibrium studies and kinetic analysis of association and dissociation of RNPs and reveal cooperative, sequential dimerization of IMP1 on the RNA. The salient feature of the mechanism is the formation of an obligate assembly intermediate, before a stable RNP is produced.
MATERIALS AND METHODS
In vitro transcription of RNA targets
Radiolabelled and unlabelled RNA were generated by T7 RNA-polymerase-directed in vitro transcription from templates inserted downstream from a T7 RNA polymerase promoter, and transcripts were purified by denaturing 5% PAGE. Radiolabelled RNA was synthesized to a specific activity of 33 Ci/mmol uridine by including 20 μCi [α-32P]UTP and 600 pmol unlabelled UTP in a 10 μl transcription reaction.
A target originally obtained by a cDNA SELEX analysis of E12.5 mice corresponding to positions 760–892 in the mouse IGF-II 3′-UTR was reduced in size to 86 nt and contained positions 788–874 downstream from the translational stop codon of the IGF-II prepropeptide. The sequence is CACUUUUCCCUACCUACCCGAAAAAGCACAUGAUAUACUUCACAUCCACAGGCAUUAACACACACAUACACAUAACACAUAUGCAC.
The H segment originally identified in human H19 RNA (13) was reduced in size to 101 nt and contained positions 1891–1989. The sequence is CUCCCUCUUCUUCUUUUUCAUCCUUCUGUCUCUUUGUUUCUGAGCUUUCCUGUCUUUCCUUUUUUCUGAGAGAUUCAAAGCCUCCACGACUCUGUUUCCCC. The examined in vitro transcripts contained the above wild-type sequences with two extraneous guanosines at the 5′ terminus.
Recombinant proteins
Untagged versions of IMP proteins were obtained by inserting the reading frames into NdeI- and XhoI-cleaved pET42 vector (Novagen) followed by expression in RNAse-E-deficient Escherichia coli cells (Invitrogen) containing plasmid-encoded transfer RNAs (tRNAs) for rare Arg, Ile and Leu codons. Following ultrasonication of bacterial cell pellets in 20 mM Tris–HCl, pH 7.8, 5 mM MgCl2, 100 mM KCl, 1 mM DTT and 1.4 μg/ml aprotinin, Triton X-100 was added to 0.4%, and the debris was removed by centrifugation at 8000 r.p.m. at 4°C for 10 min. The supernatant was made up to 10% in glycerol, and layered on a sucrose cushion consisting of 1.1 M sucrose, 20 mM Tris–HCl, pH 7.8, 5 mM MgCl2, 100 mM KCl, 1 mM DTT and 0.1% Triton X-100 and centrifuged for 2 h at 4°C at 40 000 r.p.m. The resulting pellet was washed in 20 mM Tris–HCl, pH 7.8, 5 mM MgCl2, 100 mM KCl, 1 mM DTT and 0.1% Triton X-100, and resuspended in 20 mM Tris–HCl, pH 7.8, 5 mM MgCl2, 650 mM KCl, 1 mM DTT and 0.1% Triton X-100. The suspension was centrifuged for 1 h at 4°C at 40 000 r.p.m., and the supernatant adjusted to 200 mM KCl and 10% glycerol, before it was applied to a 2 ml Heparin–Sepharose (Amersham Biosciences) column equilibrated in 20 mM Tris–HCl, pH 7.8, 5 mM MgCl2, 200 mM KCl, 1 mM DTT, 0.1% Triton X-100 and 10% glycerol. After washing with the equilibration buffer, the protein was eluted by the same solution containing 350 mM KCl.
Electrophoretic mobility-shift analysis
Radiolabelled RNA (26–28 pM) were incubated with recombinant IMP proteins at concentrations in the 0.17–4.5 nM range and 100 ng E.coli tRNA for different time periods at 30°C in 10 μl of 20 mM Tris–HCl, pH 7.8, 140 mM KCl, 2 mM MgCl2 and 0.1% Triton X-100. After addition of Ficoll (2%) and bromophenol blue at 30°C, samples were applied directly to a cold 1 mM 5% polyacrylamide gel (19:1) in 90 mM Tris–borate, pH 8.3, running at 80 V for 5 h.
Off-rates were estimated at 30°C by adding 40 nM unlabelled RNA to preformed radiolabelled complexes, and the decay of the RNP complex was followed by gel electrophoresis as above.
After electrophoresis, gels were dried and 32P was measured in an InstantImager (Packard).
Pull-down of endogenous IMP1
Segments corresponding to KH1-4, KH12 and KH34 (Figure 1) were inserted in the BamHI and EcoRI sites of pET28 (Novagen) and expressed in E.coli BL21/DE3 cells containing pRI952 (10). Cells were lysed under native conditions as described in the Novagen pET manual, and recombinant protein was attached to paramagnetic nickel beads (Qiagen) in 300 mM NaCl, 50 mM sodium phosphate, 15 mM imidazole, 2 μg/ml aprotinin, 15 μM leupeptin, pH 8.0. The beads were washed three times in 15 mM Tris–HCl, pH 8.0, 140 mM KCl, 1.5 mM MgCl2, 10 mM imidazole, 0.1% Triton X-100 and used directly for affinity purification.
Figure 1.
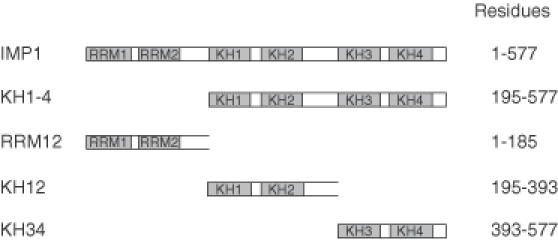
Modular architecture of IMP1 and the constructs used in the present study.
A frozen cell-pellet of ∼5 × 107 human rhabdomyosarcoma (RD) cells was resuspended in 800 μl 15 mM Tris–HCl, pH 8.0, 140 mM KCl, 1.5 mM MgCl2, 0.3% Triton X-100, 6 μg/ml aprotinin, 45 μM leupeptin, 3 μM pepstatin, before the lysate was incubated with 2500 U of RNAse T1 for 15 min on ice and centrifuged at 10 000 g for 3 min at 4°C. The supernatant was adjusted to a total protein concentration of 3 μg/μl with 15 mM Tris–HCl, pH 8.0, 140 mM KCl, 1.5 mM MgCl2, 15 mM imidazole, before 800 μl was added to 70 μl washed paramagnetic nickel beads covered with either His-tagged KH1-4, KH12 or KH34. Incubation was for 30 min at room temperature followed by two washes with 15 mM Tris–HCl, pH 8.0, 140 mM KCl, 1.5 mM MgCl2, 10 mM imidazole and 0.1% Triton X-100. Bound proteins were eluted with 45 μl of 8 M urea in 20 mM Tris–HCl, pH 8,0, 50 mM NaCl, 0.2% Triton X-100 for 2 min at 42°C followed by 5 min at room temperature, which leaves the majority of the His-tagged protein on the beads. Western analysis with an anti-IMP1 antibody was carried out as described previously.
Protein boundary mapping
A heart muscle kinase (HMK) phosphorylation site was inserted into the C-terminus of KH1-4 by exchanging five amino acids, aa569–573 (SNQAQ) with the HMK site RRASV by PCR as described previously (10). The PCR fragment was cloned into EcoRI and XhoI sites in pET28 (Novagen), and KH1-4HMK was expressed and attached to Ni-beads (Sigma). HMK-catalysed 32P labelling of KH1-4HMK was performed as described in (18), and the end-labelled protein was eluted with 250 mM imidazole. End-labelled KH1-4HMK of 100 ng was prespun for 1 min at 10 000 g to remove aggregates, before the supernatant was digested with 0.5 ng chymotrypsin or 1.5 ng LysC in 10 mM HEPES–KOH, pH 7.9, 100 mM KCl, 2 mM MgCl2, 0.05% NP40, 0.5 μg tRNA and 0.5 μg BSA by incubation at 37°C for 15 min.
Packed glutathion–Sepharose beads (15 μl) with attached glutathione S-transferase (GST)–KH1-4 or GST–KH34 were incubated in a total volume of 60 μl of 20 mM Tris–HCl, pH 8.0, 150 mM KCl, 1.5 mM MgCl2 and 0.1% Triton X-100 with partially digested, end-labelled KH1-4HMK for 20 min at 20°C. Beads were pelletted for 2 min at 1000 g, the supernatant recovered and the pellet washed with 500 μl buffer before it was resuspended in SDS–PAGE load buffer.
Pellets and supernatants from the pull-down were analysed in 7% stacking/20% separation polyacrylamide–SDS–tricin gels (18) and subjected to autoradiography. The intensity of the bands was analysed by a Fujix Bas 2000 imager.
Surface plasmon resonance
His-tagged versions of segments corresponding to KH1-4, KH12 and KH34 (Figure 1) were purified under native conditions according to the manufacturer's instructions (Novagen pET manual), and ∼6000 resonance units of His–KH1-4 were immobilized on a CM5 sensorchip by carbodiimide coupling. One microlitre of each of the protein preparations were diluted in 100 μl of 10 mM HEPES–KOH, pH 7.4, 150 mM NaCl, 3 mM EDTA and 0.005% (v/v) polysorbate 20. Samples were injected at a flow of 20 μl/min over 5 min. The surface was reconstituted by washing in 0.2 M glycine for 30 s.
Data analysis
Binding constants were obtained by fits to an equation for successive binding of IMP1 to two different sites:
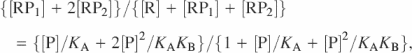
assuming that [Pfree] ≈ [Ptot]; R, RP1 and RP2 represent radioactively labelled RNA obtained by electrophoretic mobility-shift experiments performed as described above, corresponding to free RNA and RNA with one or two bound protein molecules, P, respectively. KA and KB are the equilibrium dissociation constants for the first and second bound step, respectively.
Kinetic constants for the second step of attachment were obtained from fits to [RP2]/{[RP1] + [RP2]} = N1e−[P]kt + N2 for the on-rate and to [RP2]/{[R] + [RP2]} = N0e−kt for the off-rate, where k is the rate constant, t is the time, N1, N2 and N0 are fitting parameters (N0 is the fraction of RNA in complex with two protein molecules at time zero).
Fits were carried out using the UltraFit (3.01) software package from Biosoft, and the estimated constants presented with standard errors from fitting the equation to the data derived from an individual experiment. Variations between experiments are shown as the mean ± SEM from the indicated number of independent experiments.
RESULTS
IMP isoforms heterodimerize on a target in the 3′-UTR of Igf-II mRNA
Initial titration experiments revealed that two molecules of IMP1 bound to an RNA target from the 3′-UTR of mouse Igf-II mRNA (positions 781–866 downstream of the translational stop codon), which was originally obtained by a cDNA SELEX analysis of E12.5 mice. The target was further trimmed in size to 86 nt by nested PCR (the sequence is shown in Materials and Methods) to improve the resolution of the subsequent gel electrophoretic mobility-shift analysis.
Since the functional entity of IMP1, at least in terms of RNA binding and granule formation, resides in its four KH domains (10), it was examined whether the KH1-4 entity from IMP1 was able to dimerize with the full-length IMP1 on the RNA target. The autoradiograph in Figure 2A shows the result from the non-denaturing gel electrophoretic analysis. Mixtures of KH1-4 and IMP1 (tracks 4–6) contain a new molecular species that is absent in solutions containing either KH1-4 (track 2) or IMP1 (track 3). The new species is a dimer between KH1-4 and IMP1 exhibiting an intermediate mobility between pure KH1-4 and IMP1 homodimers.
Figure 2.
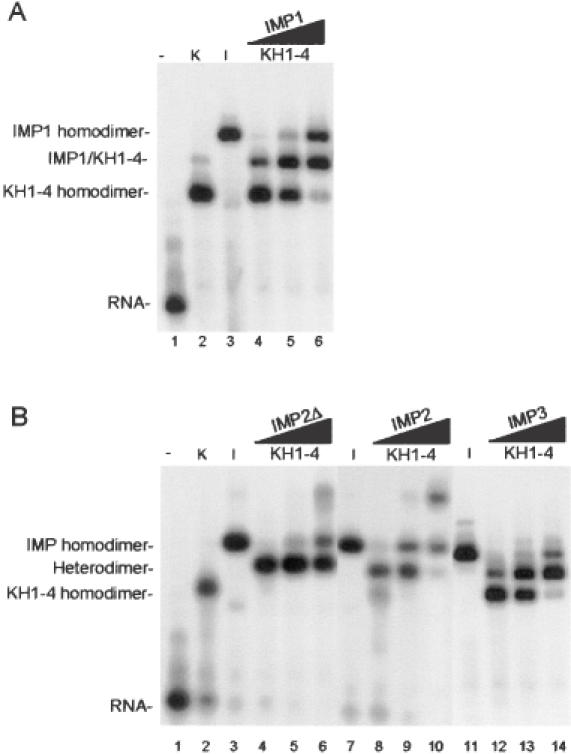
Dimerization of IMP isoforms examined by gel mobility-shift analysis. (A) Track 1, radiolabelled RNA target (28 pM) on its own; track 2, in the presence of 1.5 nM KH1-4; track 3, in the presence of 1.5 nM IMP1; tracks 4–6, in the presence of both 1.5 nM KH1-4 and 0.5, 1.5 or 4.5 nM IMP1, respectively. (B) Similar to (A) but IMP1 is substituted with IMP2Δ in tracks 3–6, with IMP2 in tracks 7–10 and IMP3 in tracks 11–14. The slow migrating species in track 2 in (A) and in tracks 6 and 10 in (B) are oligomers of KH1-4, IMP2Δ and IMP2, respectively.
In previous studies, we observed a similar ability of IMP isoforms to attach to various RNA targets (6,13); so, the electrophoretic assay was exploited to examine whether full-length IMP2, a splice variant of IMP2 (IMP2Δ), and IMP3 were able to form heterodimers with the functionally independent KH1-4 entity of IMP1. The autoradiograph in Figure 2B shows that IMP2Δ, IMP2 and IMP3 all are able to form heterodimers (tracks 4–6, 8–10 12–14, respectively) with the IMP1 KH1-4 entity on the RNA target. In addition, the IMP2Δ and the IMP2 isoforms exhibit higher-order complexes at 4.5 nM (tracks 6 and 10).
IMP1 binds in a sequential manner to RNA
To establish whether the dimerization of protein molecules occurred prior or subsequent to RNA attachment, an ‘order-of-addition’ time course experiment was carried out. Pre-incubation of a concentrated solution of IMP1 (37 nM) was followed by dilution to 1.0 nM in the absence of RNA, which was added subsequently (IMP1 first), or the concentrated IMP1 solution was diluted directly into 28 pM RNA (RNA-target first). The rationale behind the experiment is that a mechanism wherein dimerization of protein molecules prior to RNA attachment is rate limiting will be favoured by dilution into an RNA-containing solution, whereas a sequential mechanism is insensitive to the order-of-addition. The resulting autoradiograph is shown in Figure 3A, where the tracks at the left exhibit the association kinetics following incubation of dilute IMP1 before the addition of RNA, whereas the tracks at the right reveal the kinetics after concentrated IMP1 was diluted directly into an RNA-containing solution. The β-emission detected by the Instantimager from three independent experiments is shown in Figure 3B, in which the concentration of RNA in the complex with two molecules of protein (Complex II) is plotted against the time of incubation. The data show that dilution of concentrated IMP1 into an RNA-containing solution does not facilitate the formation of Complex II, indicating that the two molecules of IMP1 bind in a sequential manner to its RNA target. The sequential addition is also apparent directly from the autoradiograph, where it is observed that Complex I is a precursor for Complex II.
Figure 3.
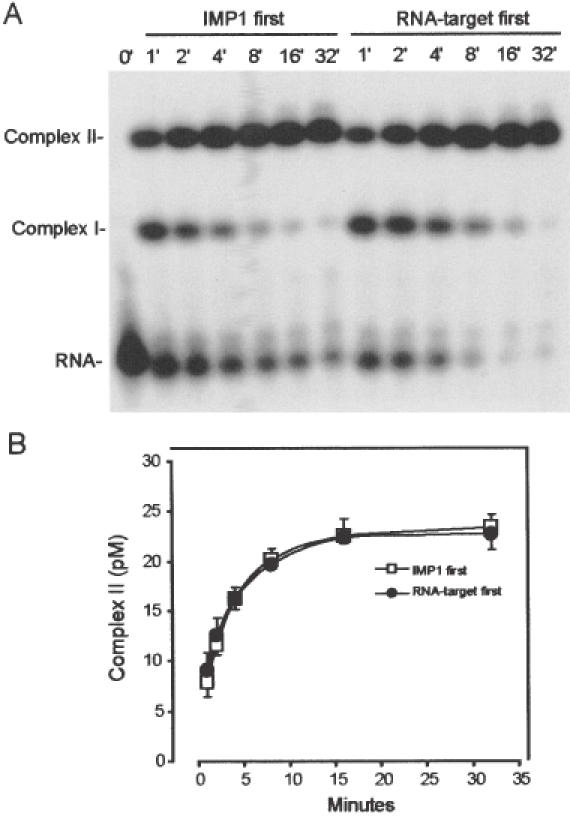
Sequential attachment of IMP1 to the radiolabelled RNA target. (A) Autoradiograph of the time course of the binding of 1.0 nM IMP1, pre-incubated in the absence of 28 pM RNA (left part), or diluted directly into the RNA-containing solution (right part). Complex I contains one IMP1 molecule and complex II contains two protein molecules. (B) InstantImager analysis of the formation of complex II as a function of time derived from three independent experiments.
IMP1 binds cooperatively to RNA
To examine whether the sequential addition of the two protein molecules to RNA occurred in a cooperative manner, the distribution between free RNA, RNA with one molecule attached (Complex I), and RNA with two molecules attached (Complex II), was analysed by non-denaturing gel electrophoresis. Incubation was carried out for 30 min with 0.17–1.64 nM IMP1. Provided that each sample has reached equilibrium before gel electrophoresis and that the latter reflects the distribution of RNP complexes prior to electrophoresis (see below), the data from the autoradiograph in Figure 4A can be fitted to an equation for the binding of IMP1 to two non-identical sites A and B (see Materials and Methods for data analysis). The dissociation constants for the two sites are estimated to KA = 1.1 ± 0.3 nM and KB = 0.31 ± 0.09 nM, respectively, reflecting a positive cooperativity in the presented experiment. The binding experiment was repeated six times, and the entire dataset plotted as the mean ± SEM in Figure 4B. Based on estimates from seven experiments, KA and KB values of 2.1 ± 0.4 and 0.58 ± 0.09 nM were obtained, respectively.
Figure 4.
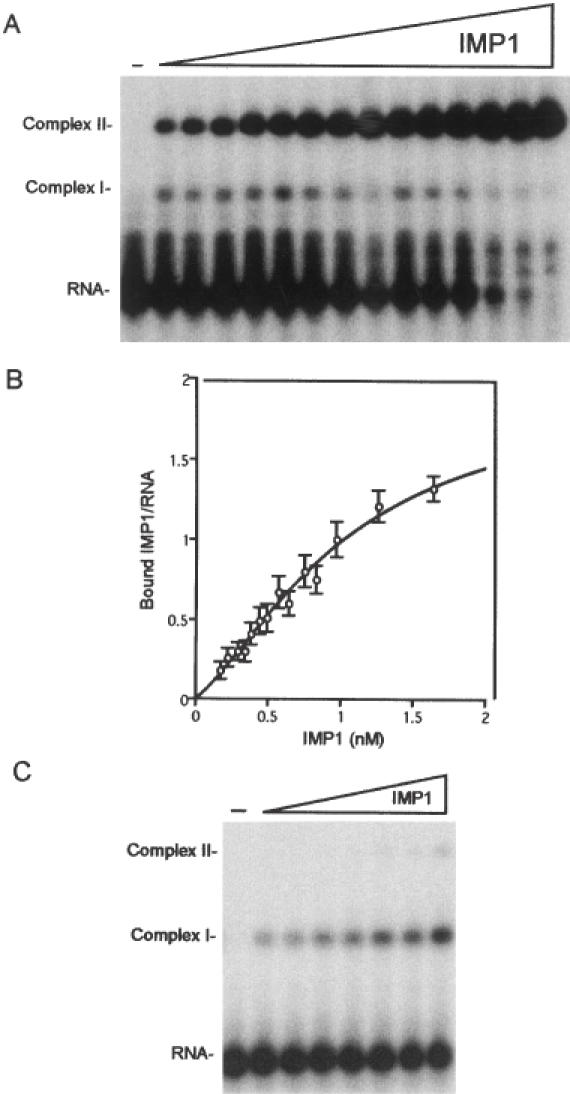
Cooperative binding of IMP1 to the radiolabelled RNA target. (A) Twenty-eight picomolar RNA was incubated for 30 min with IMP1 concentrations in the 0.17–1.64 nM range and subsequently analysed by gel mobility-shift analysis. Complex I contains one, and Complex II contains two IMP1 molecules. (B) InstantImager data from seven independent experiments plotted as the number of bound IMP1 molecules versus the protein concentration, and the curve is a fit to the equation shown in Materials and Methods. (C) Twenty-eight picomolar RNA was incubated for 2 min with 1.3-fold increments of IMP1 in the 0.17–0.83 nM range and subsequently analysed by gel mobility-shift analysis.
Due to the limitations of handling samples fast enough, it is impossible to estimate the fast on- and off-rates of the first step of attachment, but their magnitudes imply that equilibrium is attained readily. Therefore, we repeated the experiment in Figure 4A, but this time incubation with IMP1 in the 0.17–0.83 nM range was only for 2 instead of 30 min before gel electrophoresis to favour formation of Complex I. The aim was to obtain an independent estimate of KA, and, in doing so, examining the suitability of the electrophoretic assay if applied to a transient molecular species. The result of the gel electrophoretic analysis is shown in Figure 4C, and the dissociation constant for the simple equilibrium R + P ⇔ RP1 was extrapolated to 2.1 nM.
The association and dissociation rate constants of the second step of attachment
Since Figure 3 showed that the first step is considerably faster than the second step, an attempt to estimate the association rate of the second step was carried out. Five independent association experiments between 28 pM RNA and 1 nM IMP1, similar to the approach outlined in Figure 3A, were performed, and the time course of each individual experiment was fitted to the equation [RP2]/([RP1] + [RP2]) = N1e−[P]kt + N2 (see Materials and Methods for details). Based on the association kinetics of the five individual experiments, the association rate constant for the second step of attachment was estimated to be 2.73 ± 0.34 × 106 M−1 s−1.
The autoradiograph in Figure 5A shows the time course of the dissociation of preformed Complex II in the presence of 40 nM unlabelled RNA added at time 0′ to trap dissociating protein. The dissociation experiment was carried out five times, and Figure 5B depicts the average decay observed in these experiments. Since the dissociation of Complexes II to I is considerably slower than the subsequent dissociation of Complex I to RNA, as revealed by the absence of Complex I in the autoradiograph, the dissociation rate constant of Complex II of each individual experiment was fitted to the equation [RP2]/([R] + [RP2]) = N0e−kt. Based on these individual fits, the dissociation rate was estimated to 1.92 ± 0.38 × 10−3 s−1. Using the rate constants derived from the experiments presented in this section, we infer that the dissociation constant KB = koff/kon for the second step is 0.70 nM, which should be compared with the 0.58 nM obtained from the equilibrium analysis above.
Figure 5.
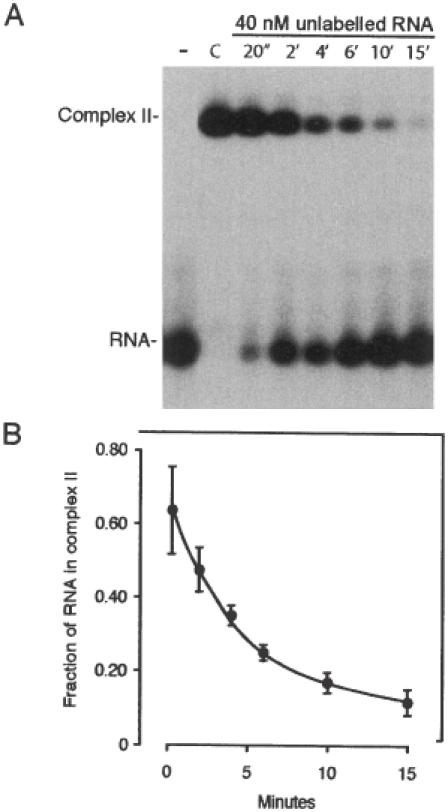
Dissociation kinetics of Complex II. (A) Autoradiograph of the time course of dissociation of Complex II following the addition of 40 nM unlabelled RNA target. Complex II was formed initially by incubating 28 pM RNA with 1.5 nM IMP1 for 30 min under standard conditions (track C), and the unlabelled RNA was added at time 0. (B) InstantImager analysis of data from five independent dissociation experiments plotted as the fraction of RNA in Complex II versus time of incubation with unlabelled RNA.
The KH34 didomain encompasses the protein dimerization motif
Since Figure 2 revealed that KH1-4 is able to dimerize with full-length IMP1 in the presence of an RNA target, it was examined whether KH1-4 was able to associate with endogenous IMP1 in an RNA-depleted cytoplasmic extract. The western analysis in Figure 6A (track 1) shows that immobilized His–KH1-4 is able to bind endogenous IMP1 in the absence of cytoplasmic RNA. The experiment was repeated with immobilized His–KH12 (track 2) and His–KH34 (track 3), which revealed that KH34 was sufficient for the pull-down of endogenous IMP1.
Figure 6.
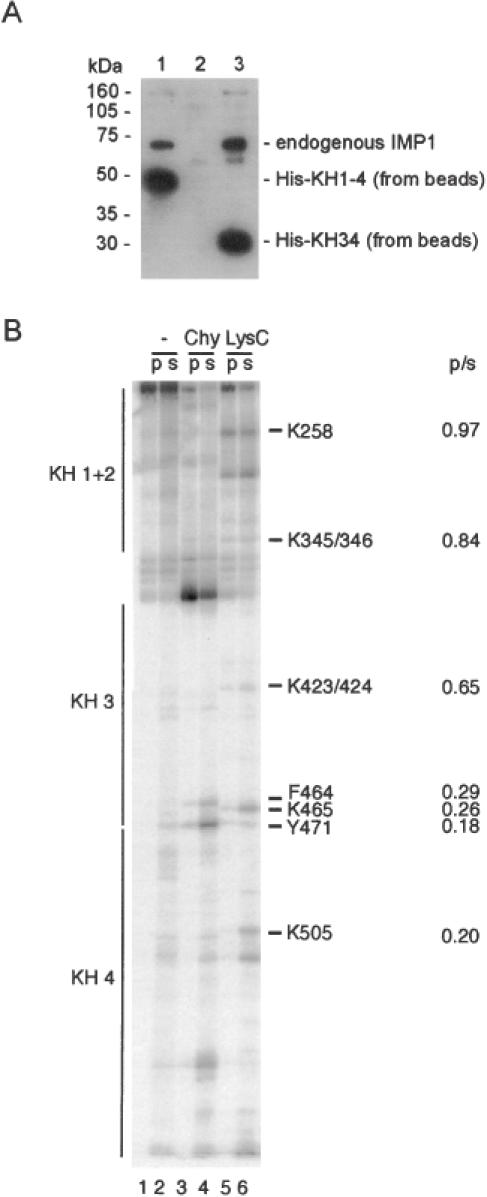
The dimerization motif is located in the KH34 didomain. (A) Western analysis of the ability of endogenous IMP1 from an RNAse T1-treated cytoplasmic extract of RD-cells to associate with His–KH1-4 (track 1), His–KH12 (track 2) or His–KH34 (track 3) on nickel beads. The anti-IMP1 antibody is raised towards the C-terminal 10 amino acids (6), thus being unable to detect His–KH12 in track 2. (B) C-terminally labelled KH1-4 was partially cleaved with either chymotrypsin or LysC and then bound to immobilized GST–KH1-4. The pellet (p) or the supernatant (s) was analysed by polyacrylamide–SDS–tricin gel electrophoresis. The positions of the four KH domains are shown at the left, and amino acid positions and p/s ratios at the right of the autoradiograph.
To rule out the possibility of an indirect effect via cytoplasmic components and to obtain a clearer picture of the protein–protein dimerization interface, a high resolution GST pull-down experiment was carried out with a GST-tagged version of KH1-4 bound to glutathion beads as bait. In this experiment, the prey was a collection of protein fragments obtained by partial proteolysis of C-terminally 32P-labelled KH1-4 by either chymotrypsin or Lys-C. The rationale behind the approach is that only C-terminally labelled fragments encompassing an intact dimerization motif will bind to immobilized KH1-4 and consequently be precipitated by centrifugation. Fragments without a dimerization motif can be recovered in the supernatant, providing an identification of the N-terminal boundary of the motif by SDS–tricine–PAGE. The autoradiograph in Figure 6B from the electrophoretic analysis of bound- and free protein fragments shows that the fragments encompassing KH34 are able to bind to immobilized KH1-4, whereas the fragments containing only the KH4 module are not. The N-terminal boundary is situated in the vicinity of lysine-424, so the N-terminal part of KH3 is dispensable for dimerization. The experiment was repeated with immobilized GST–KH34 with a similar result (data not shown), so we conclude that KH34 in IMP1 is sufficient for dimerization.
To obtain an estimate of the affinity of protein–protein interaction, a His-tagged version of KH1-4 was immobilized on a CM5 sensorchip, and the ability to interact with KH12, KH34 or KH1-4 domains in solution was examined by surface plasmon resonance. While KH12 was unable to bind, both KH34 and KH1-4 exhibited a dissociation constant of 2.0 ± 0.4 × 10−8 M (n = 3) (see Supplementary Material for representative sensorgrams), thus reinforcing the conclusion drawn from the pull-down experiments.
Excess of the dimerization motif does not reduce the rate of formation of dimerized IMP1 on the RNA target
Since the experiment in Figure 6 showed that KH34 contains the dimerization motif in IMP1, the association kinetics of 28 pM RNA and 1.5 nM full-length IMP1 in the presence of 15 nM KH34 or RRM12 was measured. Neither KH34 nor RRM12 binds IMP1 targets at concentrations upto 100 nM (10). The aim of the experiment is to examine whether the second IMP1 molecule enters the preformed obligate assembly intermediate via an RNA or a protein interface. In the latter, the presence of a 10-fold molar excess of the dimerization motif would be expected to decrease the rate of formation of a stable Complex II. The autoradiograph in Figure 7A shows that the molar excess of KH34 gives rise to an IMP1/KH34 dimer with full-length IMP1 on the RNA target, confirming that the dimerization interface on the assembly intermediate is at least partially blocked. However, the presence of the blocked intermediate is transient and has no effect on the rate of formation of dimerized full-length IMP1 on RNA, when compared to either a parallel experiment with an identical molar excess of RRM12, which is unable to dimerize and therefore acts as a negative control, or to experiments in the absence of a didomain (Figure 3). Since KH34 is unable to bind RNA on its own (Figure 7, right panel), and since the level of free Complex I is only 40% of that observed in the presence of RRM12, we conclude that the second full-length molecule of IMP1 is likely to enter both Complex I and the IMP1–KH34 dimer via an RNA interface rather than via the dimerization motif.
Figure 7.
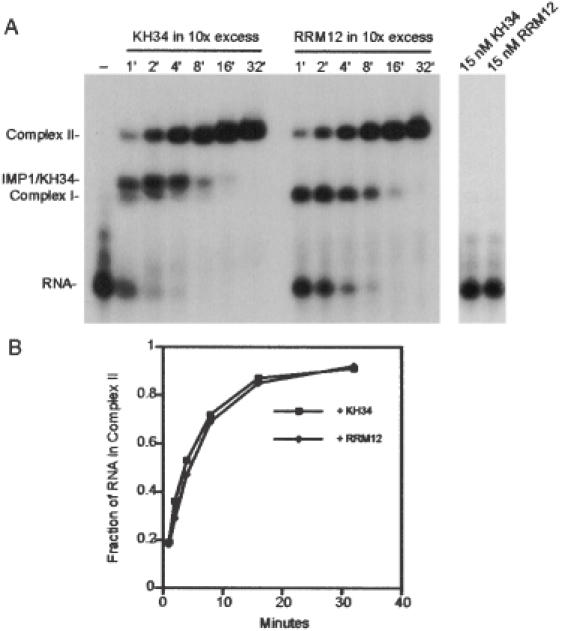
Formation of Complex II in the presence of a 10-fold molar excess of the dimerization motif. (A) Left panel, autoradiograph of the time course of the formation of Complex II from 28 pM radiolabelled RNA and 1.5 nM IMP1 in the presence of either 15 nM KH34 or 15 nM RRM12. Complex I contains one, and Complex II contains two IMP1 molecules. IMP1–KH34 is a dimer between one molecule of full-length IMP1 and one molecule of the KH34 didomain from IMP1. (A) Right panel, autoradiograph showing that neither 15 nM KH34 nor 15 nM RRM12 is able to bind the radiolabelled RNA after 32 min. (B) InstantImager analysis of the formation of Complex II as a function of time derived from the data in (A).
Stability on other RNA targets
To establish that the sequential, cooperative mechanism described for the above-mentioned target in the 3′-UTR of mouse Igf-II mRNA is representative, a kinetic analysis was carried out on a similarly sized target from human H19 RNA (the sequence is shown in Materials and Methods). A qualitatively similar picture emerged in terms of a sequential, cooperative dimerization mechanism (autoradiographs not shown). The equilibrium constants were estimated to KA = 0.84 ± 0.09 nM and KB = 0.76 ± 0.08 nM, and the association rate and dissociation rate constants for the second step of attachment are fitted to 1.7 ± 0.1 × 106 M−1 s−1 and 1.6 ± 0.1 × 10−3 s−1, respectively.
The Igf-II and H19 RNA targets subjected to kinetic analysis are smaller (86 and 101 nt, respectively) than the RNA targets we have employed in previous studies (10,15). This was a prerequisite in order to obtain autoradiographs that were amenable to kinetic analysis. However, the stability of larger targets can be analysed by electrophoresis essentially as described in Figure 5A. The t1/2 values obtained from such experiments are shown in Table 1 together with the values for the smaller targets. In general, the complexes between the RNA targets and IMP1 are exceedingly stable, and the t1/2 values for two previously published targets were greater than the 2 h time span of the experiment.
Table 1. Half-lives of IMP1 RNP complexes at 30°C.
| RNA target | Length (nt) | Half-life | Apparent Kda |
|---|---|---|---|
| mIGF-II 3′-UTR/781–866 | 86 | 6 min | 0.7 nM |
| mIGF-II 3′-UTR/753–884 | 148 | >2 h | 0.2 nMb |
| H19/1891–1989 | 101 | 7 min | 1.0 nM |
| H19/1836–2008 | 173 | 43 min | 0.4 nM (13) |
| IGF-II leader 3/729–890 | 162 | >2 h | 0.1 nM (6) |
aThe apparent Kd is derived from the protein concentration where 50% of total RNA is in the form of RP2.
bUnpublished data.
DISCUSSION
In this study, the dynamics behind the recognition of RNA targets by the human zipcode-binding protein IMP1 was examined. The RNA-binding protein employs a cooperative, sequential mechanism of dimerization on RNA that may be summarized by the equation R + 2P ⇔ RP1 + P ⇔ RP2. The first step is fast and creates an obligatory assembly intermediate RP1 of low stability, whereas the second step is the discriminatory event that dictates whether a putative RNA target with one protein attached is converted into a ‘locked’ stable RP2 with two bound protein molecules. The process may serve two interrelated functions crucial for a protein involved in RNA trafficking over considerable distances; namely selectivity during the assembly of the RNP and stability once formed.
The human branch of the VICKZ family consists of three members plus a splice variant, that show overlapping spatiotemporal expression and appear to have similar RNA target specificities as homodimers (M. A. Kristensen, J. Nielsen, F. C. Nielsen and J. Christiansen, unpublished data). As depicted in Figure 2, the KH1-4 entity of IMP1 can heterodimerize with the three other isoforms, and the IMP2Δ splice variant [also known as p62 (19)] actually exhibited a preponderance to heterodimerize. This may provide an additional level of complexity in RNP formation and function. Since the protein interaction partners of the four members are unknown, it is uncertain whether heterodimerization is physiologically important in providing additional interaction possibilities that are commonly observed with transcription factors.
No simple consensus sequence in IMP1 RNA targets have yet been identified, but the common characteristic of IMP1 targets is the lack of guanosines (7 and 11% in the two examined targets), which indicates a low structuring potential facilitating dynamic flexibility during assembly. The requirement of two protein molecules for a stable interaction provides the possibility of upto 12 RNA-binding modules per RNA target; so, even if each module recognizes a small motif such as has been reported for the KH3 domain from Nova-1 (20), the total RNA interface could still be quite extensive. Our data support this notion, because increases of the size of the examined targets from 86 to 148 nt and from 101 to 173 nt, respectively, yielded more stable RNP complexes (see Table 1). With the exception of the target in the Igf-II leader 3, the examined RNAs exhibit 2:1 stoichiometry, so the increased stability cannot be rationalized in terms of the formation of a higher-order RNP complex, but is likely to be a consequence of additional contacts in the larger targets. In light of a recent report of a 100-fold increase in the levels of all Igf-II mRNA isoforms in mammary epithelial cells in CRD-BP (identical to mouse IMP1) transgenic mice (21), it should be emphasized that the employed SELEX target resides in the 3′-UTR, which is identical in the Igf-II mRNA isoforms, and thus may be involved in the CRD-BP-induced increase.
We propose the following model for sequential, cooperative addition of two IMP1 molecules on RNA. One IMP1 molecule binds to the RNA in an unstable complex with high on- and off-rates. The second IMP1 molecule then enters the initial complex through interaction with another site on the RNA followed by stabilization of the final RNP complex by protein–protein interaction between the two protein molecules. Based on the values of the equilibrium dissociation constants, the two IMP1 molecules bind to the examined RNA targets with positive cooperativity, since KB < 4 × KA (KB = 4 × KA for two identical non-interacting sites). Figures 3 and 4C strongly support a sequential mechanism of attachment with fast formation of an unstable Complex I followed by slow formation of a stable Complex II. An alternative model with monomer to preformed dimer exchange is only compatible with the order-of-addition experiment in Figure 3 under special circumstances and is incompatible with Figure 7, where KH34 would be expected to lower the level of preformed IMP1 dimer and thus the association rate in the latter model. The pull-down experiments in Figure 6 and the surface plasmon resonance study reveal that KH34 encompasses a dimerization motif, which is not the case for KH12. A similar motif was also identified in the homologous Xenopus Vg1RBP and shown to self-associate transiently in the absence of RNA (22). Therefore, it is likely that the transient association of KH34 with Complex I (Figure 7A) is due to a protein–protein interaction, since KH34 on its own cannot bind RNA at concentrations upto 100 nM (10). However, this protein–protein interaction does not decrease the rate of formation of Complex II, implying that the second full-length protein molecule initially enters Complex I via an RNA interface and subsequently dimerizes once bound to the RNA target. Such an interpretation would put demands on the geometry of the protein molecule in relation to Complex I and provide a rationale for a sequential mechanism of attachment in terms of obtaining selectivity. This ‘proofreading’ step is important, because once formed Complex II is ‘locked’ as revealed by the long half-lives in Table 1. IMP1 is present in RNP granules of 200–700 nm optical diameter (10), so the initial two-step cooperative mechanism of IMP1 attachment to an RNA target is a nucleating event that presumably determines the further cellular fate of a particular mRNA in terms of intracellular localization, stability and/or translatability.
A few examples of kinetic analysis of RNA–protein interactions are available for comparison, and they vary considerably. The bacteriophage MS2 translational operator complex is a paradigm for RNA–protein interactions, and in this case stopped-flow fluorescence measurements provided an on-rate of 2 × 109 M−1 s−1 (23), which is three orders of magnitude greater than our estimate of 2.73 × 106 M−1 s−1 and similar to the diffusion-controlled limit (24). The U1A-hairpin II interaction with an on-rate constant of 1.1 × 107 M−1 s−1 (25) and the association of HuD with a 13 nt target occurring at 6.4 × 105 M−1 s−1 (26), both measured by surface plasmon resonance, are closer to the rate constant observed for the second step of IMP1 attachment. In fact, we envisage that HuD may employ a similar mechanism of attachment to the one identified in the present study, since HuD is also reported to be involved in granule formation and long-distance RNA trafficking (27). In the same vein, it would be interesting to elucidate the molecular details behind the dimerization mechanism of other RNA-transporting proteins such as FMRP, Staufen and She2p.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Lena B. Johansson for skilful technical assistance and to the Danish Natural Science and Medical Research Councils for support.
REFERENCES
- 1.Chartrand P., Singer,R.H. and Long,R.M. (2001) RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol., 17, 297–310. [DOI] [PubMed] [Google Scholar]
- 2.Tekotte H. and Davis,I. (2002) Intracellular mRNA localization: motors move messages. Trends Genet., 18, 636–642. [DOI] [PubMed] [Google Scholar]
- 3.Yaniv K. and Yisraeli,J.K. (2002) The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene, 287, 49–54. [DOI] [PubMed] [Google Scholar]
- 4.Havin L., Git,A., Elisha,Z., Oberman,F., Yaniv,K., Schwartz,S.P., Standart,N. and Yisraeli,J.K. (1998) RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev., 12, 1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshler J.O., Highett,M.I., Abramson,T. and Schnapp,B.J. (1998) A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol., 8, 489–496. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen J., Christiansen,J., Lykke-Andersen,J., Johnsen,A.H., Wewer,U.M. and Nielsen,F.C. (1999) A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol., 19, 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyle G.A., Betz,N.A., Leeds,P.F., Fleisig,A.J., Prokipcak,R.D. and Ross,J. (1998) The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res., 26, 5036–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueller-Pillasch F., Pohl,B., Wilda,M., Lacher,U., Beil,M., Wallrapp,C., Hameister,H., Knochel,W., Adler,G. and Gress,T.M. (1999) Expression of the highly conserved RNA binding protein KOC in embryogenesis. Mech. Dev., 88, 95–99. [DOI] [PubMed] [Google Scholar]
- 9.Ross A.F., Oleynikov,Y., Kislauskis,E.H., Taneja,K.L. and Singer,R.H. (1997) Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol., 17, 2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen F.C., Nielsen,J., Kristensen,M.A., Koch,G. and Christiansen,J. (2002) Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J. Cell Sci., 115, 2087–2097. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen J., Cilius Nielsen,F., Kragh Jakobsen,R. and Christiansen,J. (2000) The biphasic expression of IMP/Vg1-RBP is conserved between vertebrates and Drosophila. Mech. Dev., 96, 129–132. [DOI] [PubMed] [Google Scholar]
- 12.Bassell G.J., Oleynikov,Y. and Singer,R.H. (1999) The travels of mRNAs through all cells large and small. FASEB J., 13, 447–454. [DOI] [PubMed] [Google Scholar]
- 13.Runge S., Nielsen,F.C., Nielsen,J., Lykke-Andersen,J., Wewer,U.M. and Christiansen,J. (2000) H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J. Biol. Chem., 275, 29562–29569. [DOI] [PubMed] [Google Scholar]
- 14.Oleynikov Y. and Singer,R.H. (2003) Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr. Biol., 13, 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen J., Adolph,S.K., Rajpert-De Meyts,E., Lykke-Andersen,J., Koch,G., Christiansen,J. and Nielsen,F.C. (2003) Nuclear transit of human zipcode-binding protein IMP1. Biochem. J., 376, 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dreyfuss G., Kim,V.N. and Kataoka,N. (2002) Messenger-RNA-binding proteins and the messages they carry. Nature Rev. Mol. Cell Biol., 3, 195–205. [DOI] [PubMed] [Google Scholar]
- 17.Farina K.L. and Singer,R.H. (2002) The nuclear connection in RNA transport and localization. Trends Cell Biol., 12, 466–472. [DOI] [PubMed] [Google Scholar]
- 18.Jensen T.H., Jensen,A. and Kjems,J. (1995) Tools for the production and purification of full-length, N- or C-terminal 32P-labeled protein, applied to HIV-1 Gag and Rev. Gene, 162, 235–237. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J.Y., Chan,E.K., Peng,X.X. and Tan,E.M. (1999) A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J. Exp. Med., 189, 1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis H.A., Musunuru,K., Jensen,K.B., Edo,C., Chen,H., Darnell,R.B. and Burley,S.K. (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell, 100, 323–332. [DOI] [PubMed] [Google Scholar]
- 21.Tessier C.R., Doyle,G.A., Clark,B.A., Pitot,H.C. and Ross,J. (2004) Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res., 64, 209–214. [DOI] [PubMed] [Google Scholar]
- 22.Git A. and Standart,N. (2002) The KH domains of Xenopus Vg1RBP mediate RNA binding and self-association. RNA, 8, 1319–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lago H., Parrott,A.M., Moss,T., Stonehouse,N.J. and Stockley,P.G. (2001) Probing the kinetics of formation of the bacteriophage MS2 translational operator complex: identification of a protein conformer unable to bind RNA. J. Mol. Biol., 305, 1131–1144. [DOI] [PubMed] [Google Scholar]
- 24.Gutfreund H. (1995) Kinetics for the Life Sciences: Receptors, Transmitters and Catalysts. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 25.Katsamba P.S., Myszka,D.G. and Laird-Offringa,I.A. (2001) Two functionally distinct steps mediate high affinity binding of U1A protein to U1 hairpin II RNA. J. Biol. Chem., 276, 21476–21481. [DOI] [PubMed] [Google Scholar]
- 26.Park-Lee S., Kim,S. and Laird-Offringa,I.A. (2003) Characterization of the interaction between neuronal RNA-binding protein HuD and AU-rich RNA. J. Biol. Chem., 278, 39801–39808. [DOI] [PubMed] [Google Scholar]
- 27.Kasashima K., Sakashita,E., Saito,K. and Sakamoto,H. (2002) Complex formation of the neuron-specific ELAV-like Hu RNA-binding proteins. Nucleic Acids Res., 30, 4519–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


