Abstract
Heat shock induces the transcriptional activation of large heterochromatic regions of the human genome composed of arrays of satellite III DNA repeats. A number of RNA-processing factors, among them splicing factor SF2/ASF, associate with these transcription factors giving rise to nuclear stress bodies (nSBs). Here, we show that the recruitment of SF2/ASF to these structures is mediated by its second RNA recognition motif. Amino acid substitutions in the first α-helix of this domain, but not in the β-strand regions, abrogate the association with nSBs. The same mutations drastically affect the in vivo activity of SF2/ASF in the alternative splicing of adenoviral E1A transcripts. Sequence analysis identifies four putative high-affinity binding sites for SF2/ASF in the transcribed strand of the satellite III DNA. We have verified by gel mobility shift assays that the second RNA-binding domain of SF2/ASF binds at least one of these sites. Our analysis suggests that the recruitment of SF2/ASF to nSBs is mediated by a direct interaction with satellite III transcripts and points to the second RNA-binding domain of the protein as the major determinant of this interaction.
INTRODUCTION
Exposure to a variety of physical and chemical stresses profoundly affects the gene expression programme and the organization of the cells. To cope with the deleterious effects of these changes, cells activate the expression of heat-shock proteins, a group of molecular chaperons that protect against the accumulation of non-native proteins (1,2). The key player in this response is the heat-shock transcription factor 1 (HSF1) which, upon activation, binds to specific DNA sequences, called heat-shock elements (HSE), in the promoters of heat-shock genes. The activation of HSF1 involves a complex multi-step pathway in which the inert monomer oligomerizes to a DNA-binding, transcriptionally active trimer that relocalizes within the nucleus [for a review see (3)]. In human cells, activation of HSF1 is also accompanied by the accumulation of this transcription factor in novel sub-nuclear compartments, called nuclear stress bodies (nSBs) that transiently appear in stressed cells (4,5). While the function of these bodies is still a matter of investigation, a great deal of data have been recently obtained concerning their structure and dynamics. The nSBs are assembled on to the pericentromeric heterochromatic regions of specific human chromosomes, among them the extended q12 band of chromosome 9 (6,7). Assembly of nSBs on the 9q12 band is initiated by the interaction of HSF1 with HSE-like motifs in the satellite III repeats that compose most of this heterochromatic region (6). Upon binding, HSF1 directs the recruitment of the histone acetytransferase CREB-binding protein (8) that accounts for the stress-induced accumulation of acetylated histones within nSBs (8,9). This leads to the transcriptional activation of these heterochromatic regions and to the production of RNA molecules that are complementary to the satellite III repeats by RNA polymerase II (SatIII transcripts) (8,9). The characterization of these RNAs is still not complete, however, it appears that they derive from the transcription of only one strand, and that they are apparently never exported to the cytoplasm but remain associated with the sites of transcription (8,9). Concomitant with the synthesis of SatIII transcripts, a number of RNA-binding proteins involved in pre-mRNA processing accumulates within nSBs (10). These include two hnRNP proteins (HAP and M), two members of the SR family of splicing regulators (SF2/ASF and SRp30c) and Sam68, a protein that links RNA processing to the signal transduction cascade.
Alternative splicing of pre-mRNA is a versatile regulatory mechanism of gene expression in eukaryotes (11,12). A large number of studies in the last few years have elucidated the basic elements governing alternative splice site choice. In addition to the classical splicing signals, accessory elements located within the recognized exon or the flanking introns can either stimulate (enhancer) or repress (silencer) exon definition (13). The best-characterized splicing enhancers are typically purine-rich and function by binding one or more SR proteins [for a review see (14)]. SR factors are essential splicing factors and also regulate alternative splicing of many pre-mRNAs. They are modular proteins formed by one or more RNA recognition motifs (RRMs) and by a trans-activation domain that is rich in RS dipeptides (15). High-affinity RNA binders for some SR proteins have been identified using different approaches and shown to act as splicing enhancers when introduced in the reporter genes (16–18). The level of RNA-processing factors is known to control the alternative splicing pattern of transcripts. In particular, the relative concentration of splicing factor SF2/ASF, which is recruited to nSBs (10), and of its antagonistic factor hnRNP A1, whose distribution is not affected by stress (19), controls the selection between alternative 5′ splice sites (20–22). We have previously shown that the association of specific splicing factors with nSBs is accompanied by a change in the alternative splicing pattern of the adenoviral E1A transcripts, suggesting the possibility that this redistribution of RNA-processing factors may have a functional role in the post-transcriptional regulation of gene expression during the recovery from stress (10).
Given the pivotal role of SF2/ASF in splicing, we have investigated the protein domains that control the sub-nuclear relocation of this factor in response to thermal stress. In this paper, we show that the second RRM of SF2/ASF is necessary and sufficient for the recruitment to nSBs. The first α-helix of this domain is critical for this function and for the activity of SF2/ASF in alternative splicing. Finally, we show that RRM2 is important for the association of SF2/ASF with high-affinity binding sites in the transcribed strand of the Satellite III sequence. Collectively, our analysis highlights the central role of RRM2 in the SF2/ASF function.
MATERIALS AND METHODS
Cell culture and transfection
HeLa cells were grown in Dulbecco's modified Eagles medium (Cambrex, Belgium), 10% fetal bovine serum (Sigma, USA), 50 μg/ml gentamicin and 4 mM l-glutamine. For heat-shock experiments, cells were incubated for 1 h at 42°C in a complete medium supplemented with 10 mM HEPES buffer (Sigma) and were allowed to recover for 1 h at 37°C. HeLa cells grown on cover slips were transfected with 2 μg of plasmids encoding splicing factors and, when indicated, with 1 μg of adenovirus E1A reporter plasmid. Transfections were carried out using the Lipofectin Reagent as recommended by the supplier (Invitrogen, USA).
Plasmids
The full-length cDNAs for SF2/ASF, SRp30c and SRp40 were kindly provided by Dr Caceres (15). SR proteins and SF2/ASF mutants were cloned in frame with the green fluorescent protein (GFP) into the pEGFP-C1 (Clontech, USA) vector or expressed N-terminally tagged to the T7 epitope by means of the pCGTHCFFLT7 expression vector (23). The SF2/ASF mutants described in the text were obtained by a PCR-based technique using PWO polymerase (Roche Molecular Biochemicals, Switzerland) according to the protocol described previously (24). Primers used in PCR were purchased from MWG-Biotech (Germany). All plasmids were verified by sequencing with the ‘Thermo Sequenase Cy5.5 Dye Terminator Cycle Sequencing Kit’ using the ‘SEQ4x4 Personal Sequencing System’ (Amersham Pharmacia Biotech, Sweden). The pCMVE1A plasmid containing the E1A minigene was a gift from Dr Chabot (22). The SF2/ASF full-length (GST-SF2), the deletion mutants GST-SF2-ΔRS (amino acids 1–193) and GST-SF2-ΔRRM2 (amino acids 1–248 Δ113–193), the second RRM (GST-RRM2, amino acids 108–193) and its mutated version (GST-RRM2[WQD-AAA]) were all expressed in Escherichia coli as GST fusions by sub-cloning the necessary regions of the SF2/ASF cDNA into EcoRI–SalI sites of pGEX-4T3 vector (Amersham Pharmacia Biotech).
Indirect immunofluorescence
Indirect immunofluorescence analysis of HeLa cells was performed 24 h after transfection. The cells were washed with PBS, fixed with 4% formaldehyde for 7 min and then permeabilized in 0.5% Triton X-100 in PBS for 7 min on ice. Primary affinity-purified rabbit anti-HAP polyclonal antibody (19) and monoclonal antibody anti-splicing factor SC-35 (Sigma) were used. Secondary antibodies were: rhodamine-conjugated anti-rabbit immunoglobulin (IgG) goat antibody and Cy™5 anti-mouse IgG goat antibody (Jackson ImmunoResearch Laboratories, USA). Antibodies were diluted to a working concentration in PBS containing 5% skimmed milk (Difco, USA), added to the cover slips and incubated for 1 h at 37°C in a humid chamber. After staining, the coverslips were washed three times with PBS, rinsed and mounted in 90% glycerol in PBS. Confocal microscopy was performed with a TCS-NT digital scanning confocal microscope (Leica, Germany) equipped with a 63×/NA = 1.32 oil immersion objective. We used the 488 nm laser line for the excitation of GFP (detected at 500 nm < λ < 540 nm), the 543 nm laser line for the rhodamine fluorescence (detected at λ > 590 nm) and the 633 nm laser line for the Cy™5 fluorescence (detected at λ > 650 nm). The pinhole diameter was kept at 1 μm. Images were exported to Adobe Photoshop (Adobe System, USA).
In vivo E1A alternative splicing
Total RNAs were extracted 24 h after transfection using the RNeasy Kit (Qiagen, Germany) according to the protocol recommended by the supplier. To avoid plasmid contamination, RNA samples were digested with 100 U of RNase-free DNase I (Roche Molecular Biochemicals) for 30 min at room temperature. The RT–PCR was performed as described previously (25). Total RNA (1 μg) was retro-transcribed with 50 U of MuLV reverse transcriptase (Perkin-Elmer Life Science Products, USA) in a 20 μl reaction as recommended by the supplier. Amplification reactions (50 μl) contained 2 μl of the reverse transcription reaction, 20 pmol of primers, 2 mM MgCl2, 200 μM each dNTPs and 2.5 U of Taq polymerase (Perkin-Elmer Life Science Products) in a standard buffer. To quantify the amplification products, 1 μCi of [α-32P]dCTP, 3000 Ci/mmol (Amersham Pharmacia) was added to the reactions. The primers used were E1A-569 (5′-ATTATCTGCCACGGAGGTGT-3′) and E1A-1315 (5′-GGATAGCAGGCGCCATTTTA-3′). Amplifications were performed for 30 cycles with the following profile: 1 min at 94°C, 1 min at 56°C and 1 min and 30 s at 72°C. No amplification was detectable if the reverse transcription reaction was omitted. An aliquot (10 μl) of each reaction was loaded on to a 5% polyacrylamide gel in Tris–borate–EDTA buffer (TBE). The bands were revealed and quantified with the PhosphorImager 445 SI apparatus (Molecular Dynamics, USA) using the ImageQuant version 1.0 program (Molecular Dynamics).
Yeast two-hybrid screening
Yeast strains, the human HeLa library and cloning vectors were from Clontech. Library screenings and yeast manipulation were carried out according to the protocol recommended by the manufacturer. RRM2 was PCR-amplified with suitable primers from SF2/ASF wild-type (cDNA sequence 322–579 accession no. NM_006924) or from the WQD-AAA mutant and cloned into the EcoRI–SalI sites of pAS2.1. The Saccharomyces cerevisiae Y190 strain was transformed with pAS2.1-RRM2 and used as a recipient to screen a HeLa cDNA library (Clontech, cat. no. HL4000AA). A total of 7 × 107 transformants were screened on leu−, trp− and his− synthetic medium containing 25 mM 3-amino-1,2,4-triazole (Sigma). The his+ colonies were isolated and tested in the β-galactosidase filter assay. The plasmids were isolated from colonies plated on leu−, trp+ and his+ synthetic medium containing 10 μg/ml cycloheximide (Sigma) and sequenced.
Gel mobility shift assays
The synthetic RNA oligonucleotides (16 nt) used for gel mobility shift assay were as follows: SMN1, GGUUUCAGACAAAAUC; SMN2, GGUUUUAGACAAAAUC; and SatIII, GGAAUCAACACGAGUG. The RNA oligonucleotides were provided in the stable 2′-ACE protected form; deacetylation with 2′-deprotection buffer was performed prior to use as recommended by the supplier (Dharmacon). The RNA oligonucleotides were radiolabeled with [32γ-P]ATP following the 5′ end-labeling protocol with T4 polynucleotide kinase (Promega). Unincorporated nucleotides were removed from RNA by using G-25 Sephadex Columns (Roche). Gel shift assays were performed as described previously (26) using 15 fmol of radiolabeled RNA oligonucleotides and 4 pmol of recombinant proteins in a 20 μl reaction. The samples were incubated for 20 min at room temperature. The RNA–protein complexes were separated from free RNA by electrophoresis through 5% native polyacrylamide gels in 0.5% TBE buffer. The gels were dried and analyzed with the PhosphorImager 445 SI apparatus (Molecular Dynamics, Sunnyvale, CA), using the ImageQuant version 1.0 program (Molecular Dynamics).
Expression and purification of GST fusion proteins
Expression and purification of all GST fusion proteins [GST-SF2/ASF, GST-RRM2, GST-RRM2(WQD-AAA), GST-SF2-ΔRS and GST-SF2-ΔRRM2] were performed in the same way. Briefly, 1 liter of transformed XL1Blue cultures were induced with 2 mM isopropyl-β-d-thiogalactopyranoside for 3 h, and the harvested cells were lysed by sonication in PBS buffer with 1% Triton X-100. Fusion proteins were purified from the soluble fraction by glutathione–agarose beads (Sigma) and eluted with 20 mM glutathione, 50 mM Tris–HCl, pH 8.0. The eluted proteins were dialyzed for 3 h against 10 mM Tris–HCl, pH 8.0, 100 mM NaCl and concentrated for 30 min in the same buffer containing 50% glycerol. Protein concentration was determined by the Bradford method and was monitored by Coomassie blue staining of SDS–polyacrylamide gels.
RESULTS
RRM2 is necessary and sufficient for the recruitment of SF2/ASF to nuclear stress bodies
In response to thermal stress, the splicing factor SF2/ASF is recruited to nSBs [Figure 1 and (10)]. In order to investigate the molecular mechanisms underlying this phenomenon, we sought to identify the protein domain involved. SF2/ASF is composed of three different domains: (i) a canonical RRM at the N-terminus that is followed by (ii) a protein region that is distantly related to the RRM (called pseudo-RRM or RRM2) and finally (iii) an extended tail rich in Ser-Arg dipeptides at the C-terminus that is thought to mediate protein–protein interactions (see Figure 1 for a scheme). These domains were, either individually or in different combinations, overexpressed in human HeLa cells as GFP fusions and tested for their ability to associate after heat shock (1 h at 42°C followed by 1 h of recovery at 37°C) with nSBs labeled using antibodies to hnRNP HAP. It is worth recalling that, as shown previously (10), the GFP by itself is unable to associate with nSBs. As expected, GFP-SF2/ASF was efficiently recruited to nSBs (Figure 1). Although the deletion mutants displayed a certain level of nucleolar accumulation, association with nSBs was clearly detectable with two constructs containing only two of the three SF2/ASF domains, namely RRM1-RRM2 and RRM2-SR (Figure 1). In contrast, a fusion protein (RRM1-SR) lacking RRM2 failed to associate with nSBs pointing to this domain as the one necessary for the redistribution of SF2/ASF upon stress (Figure 1).
Figure 1.
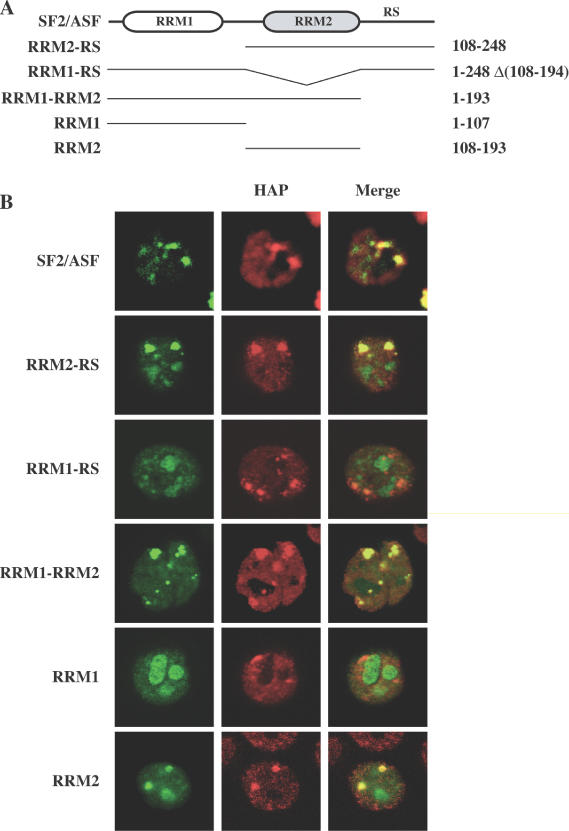
RRM2 of SF2/ASF is necessary and sufficient for the recruitment to nSBs. (A) Scheme of the SF2/ASF protein and of the different deletion mutants. Numbers on the right indicate the amino acid regions present in each mutant. (B) The entire SF2/ASF or the indicated deletion mutants shown in (A) were overexpressed as GFP fusions in HeLa cells. After 40 h, the cells were heat shocked (for 1 h at 42°C followed by 1 h at 37°C) and immediately fixed. Cells were stained with a polyclonal antibody to hnRNP HAP and with anti-rabbit rhodamine-conjugated goat antibody. Cells were analyzed by confocal laser microscopy to study the distribution of the GFP protein and of hnRNP HAP. Images of the same cells were taken and merged to reveal co-localization of the two proteins in nSBs.
To understand whether RRM2 was sufficient for the recruitment, we determined the distribution of the GFP-RRM2 fusion in heat-shock HeLa cells. As a control, we also studied the distribution of GFP-RRM1. Upon shock, only RRM2 co-localized with hnRNP HAP in stress bodies (Figure 1) indicating that this domain contains all the elements necessary and sufficient for the association with nSBs.
Mapping the protein determinants for the RRM2 function
The RRM is a highly structured domain composed of a four-strand antiparallel β-sheets packed against two α-helices perpendicularly oriented (27). In this structure, two short motifs, RNP-1 and RNP-2, are located in the central β1 and β3 strands and make direct contacts with the RNA molecule (28) (Figure 2A). The β-sheet constitutes an RNA-binding platform, but the RNA-binding specificity mainly resides in the variable regions of the loops that connect the β-strands and in the terminal portions of the domain. RRM2 is an atypical RRM-like repeat that is present in a subset of SR factors. It lacks the conserved aromatic residues present in RNP-1 and RNP-2 but contains all of the conserved amino acids that form the hydrophobic core of the RRM (29). The role of this domain is still poorly understood. Several experimental data indicate that RRM2 is essential for the specificity of binding of SF2/ASF to RNA (15); moreover, a role of RRM2 in protein–protein interactions has been suggested previously (30,31). In order to gain further insight into the function of the RRM2 in the heat-shock-induced redistribution, we challenged a number of mutants of SF2/ASF bearing amino acid substitution in RRM2 for their ability to associate with nSBs. Amino acid substitutions in these mutants (Figure 2A) were designed not to affect the overall architecture of the domain, as the residues selected for mutations are all surface exposed according to the three-dimensional (3D) RRM structures. In a group of mutants (Y-A, D-R, VV-AA and E-R in Figure 2A), we replaced the amino acids in strands β2 and β3 that are supposed to establish direct contacts with the RNA molecule. In one mutant (R-A), the arginine, immediately upstream of the putative RNP-2, was substituted with an alanine. Instead, the last mutant (WQD-AAA) was generated by replacing three surface residues in the heptapeptide that composes the first α-helix of the domain and is conserved in all the SR proteins containing an atypical RRM. According to the RRM model, this extended α-helix is not involved in RNA binding but has a structural function and could be available for protein–protein interactions. As mentioned earlier, the 3D structure model predicts that the WQD residues are surface exposed, and the mutation should have minimal impact on the overall RRM structure. This is also reflected in our bioinformatic analyses of wild-type and WQD-AAA mutant. A possible local perturbation originating from the substitution of tryptophan, the predicted first residue of the helix, with alanine should not affect our interpretation of the mutational results. All these mutants were expressed as GFP fusions in human HeLa cells and their distribution after the heat shock was assessed by confocal laser microscopy and compared with that of hnRNP HAP. None of the substitutions introduced into the β-strand regions affected the localization of the protein (Figure 2B). Notably, a small fraction of the overexpressed proteins was still detectable in nuclear speckles stained by the monoclonal antibody against splicing factor SC-35 (see Supplementary Figure 1). Although the expression level of the proteins may influence the extent of the recruitment, it is worth noticing that the same phenomenon also occurs with the endogenous protein (data not shown) suggesting that the level or activity of the recruiting factor could be the limiting factor for this event. Contrary to the substitutions in the β-strands, the replacement of WQD stretch, within the first α-helix, with three alanines completely abrogated the recruitment to nSBs (Figure 2B) and the overexpressed protein perfectly co-localized with splicing factor SC-35 in nuclear speckles (Supplementary Figure 1). Thus, the heptapeptide that forms the first part of RRM2 is crucial for the association of SF2/ASF with nSBs.
Figure 2.
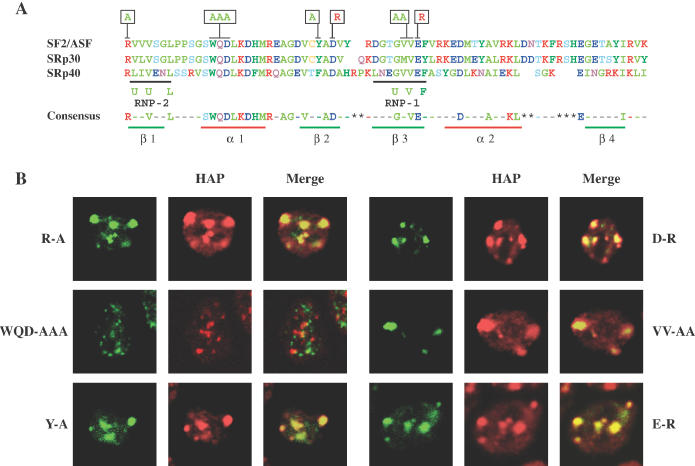
Effect of amino acid substitutions in RRM2 on the recruitment of SF2/ASF to the nSBs. (A) Alignment of the amino acid sequence of RRM2 in three SR proteins recruited to nSBs. The position of RNP-1 and RNP-2 motif is indicated along with their consensus sequence. At the bottom is shown the consensus of the three sequences and the position of the α-helices and β-sheets. Mutated amino acids are indicated in the boxes above the first line. (B) SF2/ASF mutants bearing the indicated amino acid substitutions in RRM2 were overexpressed in HeLa cells as GFP fusions. After 40 h, the cells were heat shocked as in Figure 1B, fixed and stained with a polyclonal antibody to hnRNP HAP and with anti-rabbit rhodamine-conjugate goat antibody. The cells were analyzed by confocal laser microscopy to study the distribution of the GFP proteins and of hnRNP HAP. Images of the same cells were taken and merged to reveal co-localization of the two proteins in nSBs.
Two-hybrid screening
The results in the previous sections are compatible with two alternative scenarios for the redistribution of SF2/ASF upon stress. The association with nSBs, in fact, can be due to a direct binding either to SatIII RNAs or to another protein present in these structures. In both cases, the interaction should be mediated by the wild-type RRM2 and sensitive to the WQD-AAA substitution. In order to identify proteins specifically interacting with RRM2, we screened a human cDNA library by the two-hybrid approach in yeast using RRM2 as a bait (see Materials and Methods). Yeast cells stably expressing a fusion between RRM2 and the Gal4 DNA-binding domain were transformed with a library of HeLa cell cDNAs fused to the activation domain of Gal4. From the screening of 7 × 107 transformants, we selected a few clones in which the expression of URA3 and β-galactosidase marker genes required the presence of both the hybrid proteins. These clones contained the cDNAs for (i) aldolase A, a glycolytic enzyme present also in the cell nucleus (32); (ii) fortilin, a novel antiapoptotic protein (33); and (iii) poly(A)-binding protein PABPN1, which in unstressed cells co-localizes with SF2/ASF in nuclear speckles (34). Interestingly, the interaction of these proteins in the two-hybrid assay was abrogated by the replacement of the WQD motif in RRM2 with three alanines. However, none of them accumulated in nSBs when overexpressed in HeLa cells fused to the GFP (data not shown). In the case of PABPN1, the lack of recruitment was also verified with antibodies directed against the endogenous protein (data not shown). Although it is still possible that an interaction may actually occur in unstressed cells, the lack of co-localization argues against the possibility that these proteins may act as recruiters of SF2/ASF to nSBs.
Interaction with SatIII sequences
The results in the previous section prompted us to investigate whether the recruitment to nSBs could reflect the ability of SF2/ASF to interact with SatIII transcripts through RRM2. This hypothesis was supported by the fact that the transcribed strand of the SatIII element (158 nt; GenBank accession no. X06137) contains four putative high-affinity binding sites for SF2/ASF identified by the ESEfinder program (http://exon.cshl.edu/ESE/) (Figure 3). In particular, one of these (AACACGA at position 88) shows a score higher than the experimentally verified binding site of SF2/ASF in exon 7 of the SMN1 gene (4.08 versus 3.76) (35). Therefore, we decided to challenge in a gel mobility assay, the full-length protein or different portions of it for their ability to bind an oligoribonucleotide comprising the predicted binding site at position 88 (SatIII oligo). As positive and negative controls, we used (i) the well-characterized binding site of SF2/ASF in exon 7 of the SMN1 gene and (ii) the corresponding sequence in the SMN2 gene that differs from SMN1 only for a C to T substitution and is not recognized by SF2/ASF (35). In agreement with the hypothesis, GST-SF2/ASF interacted with SatIII and SMN1 but not with SMN2 (Figure 4A). In this assay, the interaction was completely abrogated upon the deletion of RRM2, indicating a major role of this domain in the ability of SF2/ASF to bind to its target sites. In contrast, deletion of the RS domain (ΔRS mutant) had no detectable effect on binding. Quantification of the retarded bands observed with GST-SF2/ASF and GST-ΔRS indicated that the relative affinity for SatIII and SMN1 sequences (11 and 1% of the total oligonucleotide respectively with GST-ASF/ASF and 8 and 2% with GST-ΔRS) paralleled the score values produced by the ESEfinder program. Finally, no interaction was observed when only the GST moiety was used (data not shown).
Figure 3.

Putative SF2/ASF binding sites in the transcribed strand of the SatIII DNA. The SF2/ASF consensus heptamer motif is shown at the top. The sequence of the putative binding sites identified by the ESEfinder program is shown below. Numbers on the left refer to the Satellite III sequence (GenBank accession no. X06137). Motif scores are indicated on the right.
Figure 4.
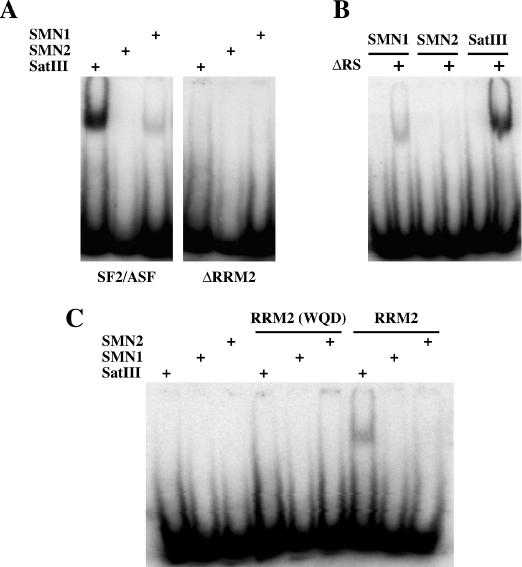
Mobility shift assay. Equal amounts (15 fmol) of the 5′-end labeled oligo-ribonucleotides containing (i) the SF2/ASF site in SMN1 gene, (ii) the corresponding sequence in the SMN2 gene and (iii) the Satellite III sequence with the highest score were incubated with 4 pmol of GST-SF2/ASF or GST-SF2-ΔRRM2 (A) as described in the Materials and Methods. After 20 min, complexes were resolved through a 5% polyacrylamide gel and quantified with a PhosphorImager apparatus. An image of the gel is shown. The same oligos in (A) were tested with GST-ΔRS (B) and with GST-RRM2 and GST-RRM2 (WQD-AAA) (C).
As shown in Figure 1B, the sole RRM2 is sufficient for the recruitment to nSBs. We wondered whether RRM2 was also able to bind to the SatIII oligo by itself. The result in Figure 4C indicates that this is indeed the case. The intensity of the retarded band, however, was lower than that observed with GST-SF2/ASF (1.5% versus 11% of total radioactivity) pointing to a role of RRM1 in stabilizing the interaction. No interaction was instead detected with SMN1 or SMN2 oligos. Finally, the binding of RRM2 to SatIII was completely abrogated by the replacement of the WQD motif in the first α-helix with three alanines (Figure 4C), a substitution that prevents the recruitment to nSBs (Figure 2B).
Mutations in RRM2 affect the activity of SF2/ASF in alternative splicing
The results in the previous section indicate that RRM2 mediates the interaction with a motif in the satellite III sequence that matches the SF2/ASF-binding site consensus. Similar sequences are found in splicing enhancers recognized and bound by SF2/ASF (16). It is conceivable, therefore, that the amino acid substitutions that abrogate the recruitment to nSBs may also affect the activity of SF2/ASF in alternative splicing. To explore this possibility, we challenged the wild-type SF2/ASF and the different mutants described in Figure 2 for their ability to affect the alternative splicing pattern of the adenoviral E1A transcripts in vivo. As schematically shown in Figure 5A, splicing of adenoviral E1A pre-mRNA is characterized by the use of three alternative 5′ splice sites and results in the production of three mature molecules known as 13S, 12S and 9S. Overexpression of SF2/ASF strongly increases the utilization of the 13S 5′ splice site and decreases the production of 12S and 9S molecules (21,36). HeLa cells were co-transfected with the E1A minigene and with vectors driving the expression of SF2/ASF or of different mutants, all N-terminally tagged with the T7 epitope. After 48 h, total RNAs were extracted and subjected to RT–PCR to determine the relative abundance of 13S, 12S and 9S molecules. The result of this analysis is shown in Figure 5B and in Supplementary Figure 2. In agreement with the previous data (21,36), overexpression of the wild-type SF2/ASF promoted the production of 13S mRNAs with the concomitant reduction in 12S and 9S molecules. A similar result was observed with SRp40 (see Supplementary Figure 2), another SR factor that is recruited to nSBs (unpublished data) and contains an atypical RRM. In contrast, overexpression of SRp20, which contains a single RRM and is not recruited to nSBs, promoted the production of 12S RNA (Supplementary Figure 2). As shown in Figure 5B and in agreement with the results obtained in other laboratories (15,37), the ability of SF2/ASF to increase the 13S level depended on RRM2 and a mutant lacking this domain (Δ2) somehow resembled SRp20 in its ability to promote the production of 9S and 12S molecules. Interestingly, the Δ1 and ΔRS mutants, which lacked RRM1 and the RS domain respectively, behaved similarly to the entire protein pointing to RRM2 as the major determinant for the activity of SF2/ASF in alternative splicing (Supplementary Figure 2). Finally, we tested the SF2/ASF mutants bearing amino acid substitutions in RRM2 (Figure 5B). All the substitutions in the β-strands (Y-A, D-R, VV-AA and E-R) had modest effect on the activity of SF2/ASF and, similarly to the wild-type protein, increased the production of 13S molecules. A modest reduction in the SF2/ASF activity was observed with the R-A mutant in which the arginine residue immediately upstream of the putative RNP-2 motif was substituted with an alanine. In contrast, the replacement of three amino acids in the first α-helix (WQD-AAA) had a drastic effect on E1A splicing and the protein behaved as the Δ2 mutant. Thus, the first α-helix of RRM2 has a major role in the recruitment to nSBs, in the interaction with high-affinity binding sites, and in the function of SF2/ASF in the alternative splicing of E1A transcripts.
Figure 5.
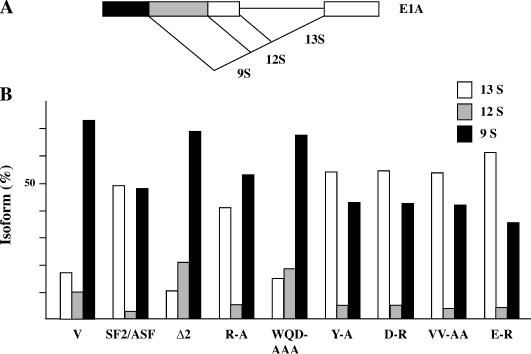
The role of RRM2 in alternative splicing. (A) A diagram of the E1A pre-mRNA. The alternative 5′ splice sites and splicing events that generate 13S, 12S and 9S mRNAs are shown schematically. (B) SF2/ASF or the indicated mutants, all N-terminally tagged with the T7 epitope, were overexpressed in HeLa cells to test the effect on the splicing pattern of transcripts encoded by the E1A minigene. After 48 h, the RNAs were extracted, subjected to RT–PCR with E1A specific oligos in the presence of [α-32P]dCTP. The RT–PCR products were resolved through a 5% polyacrylamide gel and quantified with a PhosphorImager apparatus. The histogram shows the relative amount of 13S, 12S and 9S molecules in cells overexpressing the indicated mutants. The bars represent the average of at least three independent experiments. Δ2, SF2/ASF lacking RRM2. V, E1A minigene co-transfected with the empty pCGTHCFFLT7 expression vector. Amino acid substitutions introduced in SF2/ASF are shown in Figure 2A.
DISCUSSION
In this paper, we have studied the recruitment of splicing factor SF2/ASF to nSBs. The nSBs are intimately connected with RNA metabolism and in fact, their formation and stability is affected by transcription inhibitors and by RNAse treatments (19,38). The link with RNAs is underscored by the recent finding that satellite III (SatIII) transcripts are stable components of nSBs (8,9). This has suggested a model whereby SatIII RNAs are directly involved in the recruitment of RNA-processing factors, among them SF2/ASF. Here, we provide evidence in support of this model, and we show that the atypical RRM2 is necessary and sufficient (i) for the recruitment to nSBs and (ii) for the interaction of SF2/ASF with a high-affinity binding site in the Satellite III sequence.
A central role for RRM2 in the SF2/ASF function
Similar to other splicing factors SF2/ASF, in addition to being diffusely distributed throughout the nucleoplasm, is concentrated in a few specific sub-nuclear compartments known as nuclear speckles. Targeting these structures requires at least two of the three domains (RRM1, RRM2 and the RS domain) of SF2/ASF, implying the existence of additive and redundant signals for this function. In contrast to this, our analysis indicates that the association with nSBs exclusively relies on RRM2, which is both necessary and sufficient for the distribution of SF2/ASF in heat-shocked cells. The physiological role of the atypical RRM2 domain in the SF2/ASF function is still elusive. However, several studies have suggested its importance in both RNA-binding specificity and alternative splicing. In vitro and in vivo studies have led to the isolation of high-affinity binding sites for SF2/ASF (15,18) and to the identification of a binding site consensus (16). Notably, sequences selected by SF2/ASF mutants lacking RRM2 drastically differ from this consensus (18). SF2/ASF recognizes short degenerate sequence motifs (6–8 nt) often located within splicing enhancers. The interaction with the cognate sites is essential for the activity of SF2/ASF in exon recognition and for the ability of this splicing factor to stimulate mRNA translation (39). A program, called ESEfinder, has been recently developed which provides a list of ‘high-score motifs’ for SF2/ASF in sequences of interest (40). By means of this program, we have identified four putative high-affinity binding sites for SF2/ASF within the transcribed strand of the satellite III DNA (158 nt). We have been able to show that at least one of these sites is, indeed, bound by SF2/ASF and that RRM2 is both necessary and sufficient for the interaction. This is the first direct evidence that RRM2 is involved in the binding specificity of SF2/ASF. The RNA-binding property of RRM2 appears to be intimately linked to the recruitment to nSBs. Indeed, the first α-helix of RRM2 is critical for both the aspects as indicated by behavior of the WQD-AAA substitution mutant. In contrast, the RRM2 function in the sub-nuclear distribution of SF2/ASF is not affected by mutations in the β-strand segments that, on the basis of both structural and biochemical analysis of RRM domains, are expected to establish direct contacts with the RNA molecule. It is still possible that, similar to what was observed with other RRMs, the residues between β-strands may be involved in the RNA-binding specificity.
Others have recently investigated the role of RRM2 and of the heptapeptide in the RNA-binding properties and in the alternative splicing activity of SF2/ASF (31,41). In agreement with our results, these authors have shown the central role of RRM2 in alternative splicing and have demonstrated the ability of this atypical RRM to interact with RNA. Moreover, the same authors have observed that RRM2 is both necessary and sufficient to elicit the repressing activity of SF2/ASF on the IIIa distal 3′ splice site of the L1 adenovirus unit, and the heptapeptide has a critical role in this activity (31). Contrary to our observations, however, they have found that the mutation of the heptapeptide does not affect binding of SF2/ASF to a 115 nt RNA spanning the E1A 13S 5′ splice site, suggesting a role of this sequence in protein–protein interactions. We believe that this discrepancy could originate from the different assay that is used. These authors, in fact, applied UV-crosslinking that is suitable to detect even a weak and transient binding, whereas the mobility shift assay can only reveal relatively strong and stable interactions. Whether or not the heptapeptide can mediate protein–protein interactions is still an open question. In effect, our two-hybrid screening has selected a number of proteins able to interact with the wild-type RRM2 but not with the WDQ-AAA mutant. However, none of these proteins appears to co-localize with SF2/ASF in nSBs.
Possible function of SatIII transcripts
Our data indicate that the massive recruitment of SF2/ASF to nSBs is due to the ability of the protein to interact with high-affinity binding sites that are reiterated in the SatIII transcripts. Consistent with this, the WQD-AAA mutant remains confined in nuclear speckles in heat-shocked cells. So far, there are no indications that the SatIII transcripts are substrates for the splicing apparatus, and we have never observed an accumulation in nSBs of other proteins or factors critical for the splicing reaction such as snRNPs (data not shown). We hypothesize that the function of the non-coding satellite III transcripts is to sequester a subset of RNA-processing factors thus preventing their interaction with pre-mRNA molecules.
Our results point to the heptapeptide that forms the first α-helix of RRM2 as a major determinant in the association with nSBs and the interaction with SatIII RNAs. Interestingly, this heptapeptide is perfectly conserved in all the RRM2 domains of SR splicing factors, whereas other residues in the same domain appear more divergent (Figure 2A). Although we have not performed an extensive analysis of the other SR proteins bearing RRM2, the fact that SRp30 (10) and SRp40 (data not shown) are both recruited to nSBs supports the idea that the association with nSBs is a common feature of RNA-binding proteins with RRM2, and is part of the cellular response to heat shock. SR factors have a major role in alternative splicing and, as recently shown, also in the export to the cytoplasm and in controlling translation of mature mRNA (39). Both events are mediated by the interaction of SF2/ASF with high-affinity binding sites in the exonic sequences. In this perspective, the presence of high-affinity binding sites in the Satellite III transcripts would lead to the sequestration of RRM2-containing SR factors and consequently would affect alternative splicing and translation of a large number of genes.
It has recently been reported that the rapid and transient inhibition of splicing triggered by heat shock is due to the dephosphorylation of another SR protein, SRp38, which sequesters the U1 small nuclear ribonucleoprotein particle (42). In HeLa cells, this event is completely reverted after 1 h of recovery. However, SF2/ASF is sequestered in nSBs well beyond this time. Thus, it is conceivable that the restart of splicing is accompanied by the execution of a specific alternative splicing program that relies on the unbalanced distribution of splicing factors with the prevalence of those not recruited to nSBs, such as SC-35 and SRp20 (10). We propose that this could be part of a strategy exploited by human cells to tune the gene expression program at the post-transcriptional level during the recovery from stress.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants of the ‘Associazione Italiana per la Ricerca sul Cancro’ (AIRC) to G.B., of the program MIUR/FIRB ‘Post-genoma’ (contract numbers RBNEOKXC9_001 and RBNE015MPB_003) to S.R. and G.B respectively, and from Progetto CNR-MIUR ‘Genomica Funzionale’ L.449/97 to G.B.
REFERENCES
- 1.Morimoto R.I. (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev., 12, 3788–3796. [DOI] [PubMed] [Google Scholar]
- 2.Pirkkala L., Nykanen,P. and Sistonen,L. (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J., 15, 1118–1131. [DOI] [PubMed] [Google Scholar]
- 3.Holmberg C.I., Tran,S.E., Eriksson,J.E. and Sistonen,L. (2002) Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem. Sci., 27, 619–627. [DOI] [PubMed] [Google Scholar]
- 4.Jolly C., Morimoto,R., Robert-Nicoud,M. and Vourc'h,C. (1997) HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J. Cell. Sci., 110, 2935–2941. [DOI] [PubMed] [Google Scholar]
- 5.Cotto J., Fox,S. and Morimoto,R. (1997) HSF1 granules: a novel stress-induced nuclear compartment of human cells. J. Cell. Sci., 110, 2925–2934. [DOI] [PubMed] [Google Scholar]
- 6.Jolly C., Konecny,L., Grady,D.L., Kutskova,Y.A., Cotto,J.J., Morimoto,R.I. and Vourc'h,C. (2002) In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell. Biol., 156, 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denegri M., Moralli,D., Rocchi,M., Biggiogera,M., Raimondi,E., Cobianchi,F., De Carli,L., Riva,S. and Biamonti,G. (2002) Human chromosomes 9, 12, and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol. Biol. Cell., 13, 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jolly C., Metz,A., Govin,J., Vigneron,M., Turner,B.M., Khochbin,S. and Vourc'h,C. (2004) Stress-induced transcription of satellite III repeats. J. Cell. Biol., 164, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzi N., Denegri,M., Chiodi,I., Corioni,M., Valgardsdottir,R., Cobianchi,F., Riva,S. and Biamonti,G. (2004) Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol. Biol. Cell, 15, 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denegri M., Chiodi,I., Corioni,M., Cobianchi,F., Riva,S. and Biamonti,G. (2001) Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol. Biol. Cell, 12, 3502–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graveley B.R. (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet., 17, 100–107. [DOI] [PubMed] [Google Scholar]
- 12.Smith C.W. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- 13.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 14.Cartegni L., Chew,S.L. and Krainer,A.R. (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet., 3, 285–298. [DOI] [PubMed] [Google Scholar]
- 15.Caceres J.F., Misteli,T., Screaton,G.R., Spector,D.L. and Krainer,A.R. (1997) Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell. Biol., 138, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H.X., Zhang,M. and Krainer,A.R. (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev., 12, 1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tacke R., Chen,Y. and Manley,J.L. (1997) Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc. Natl Acad. Sci. USA, 94, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tacke R. and Manley,J.L. (1995) The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J., 14, 3540–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weighardt F., Cobianchi,F., Cartegni,L., Chiodi,I., Villa,A., Riva,S. and Biamonti,G. (1999) A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J. Cell. Sci., 112, 1465–1476. [DOI] [PubMed] [Google Scholar]
- 20.Mayeda A. and Krainer,A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- 21.Caceres J.F., Stamm,S., Helfman,D.M. and Krainer,A.R. (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science, 265, 1706–1709. [DOI] [PubMed] [Google Scholar]
- 22.Yang X., Bani,M.R., Lu,S.J., Rowan,S., Ben-David,Y. and Chabot,B. (1994) The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl Acad. Sci. USA, 91, 6924–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson A.C., Peterson,M.G. and Herr,W. (1995) The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev., 9, 2445–2458. [DOI] [PubMed] [Google Scholar]
- 24.Montecucco A., Rossi,R., Levin,D.S., Gary,R., Park,M.S., Motycka,T.A., Ciarrocchi,G., Villa,A., Biamonti,G. and Tomkinson,A.E. (1998) DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J., 17, 3786–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghigna C., Moroni,M., Porta,C., Riva,S. and Biamonti,G. (1998) Altered expression of heterogenous nuclear ribonucleoproteins and SR factors in human colon adenocarcinomas. Cancer Res., 58, 5818–5824. [PubMed] [Google Scholar]
- 26.Zuo P. and Manley,J.L. (1993) Functional domains of the human splicing factor ASF/SF2. EMBO J., 12, 4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagai K., Oubridge,C., Jessen,T.H., Li,J. and Evans,P.R. (1990) Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature, 348, 515–520. [DOI] [PubMed] [Google Scholar]
- 28.Gorlach M., Wittekind,M., Beckman,R.A., Mueller,L. and Dreyfuss,G. (1992) Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J., 11, 3289–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birney E., Kumar,S. and Krainer,A.R. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res., 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen-Mahrt S.K., Estmer,C., Ohrmalm,C., Matthews,D.A., Russell,W.C. and Akusjarvi,G. (1999) The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J., 18, 1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dauksaite V. and Akusjarvi,G. (2002) Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J. Biol. Chem., 277, 12579–12586. [DOI] [PubMed] [Google Scholar]
- 32.Satou W., Tanimoto,H., Ukekawa,R., Fujii,M. and Ayusawa,D. (2004) Amplification of nuclear aldolase A in mouse cell mutants resistant to Hoechst 33342. Biochem. Biophys. Res. Commun., 315, 845–849. [DOI] [PubMed] [Google Scholar]
- 33.Li F., Zhang,D. and Fujise,K. (2001) Characterization of fortilin, a novel antiapoptotic protein. J. Biol. Chem., 276, 47542–47549. [DOI] [PubMed] [Google Scholar]
- 34.Calado A. and Carmo-Fonseca,M. (2000) Localization of poly(A)-binding protein 2 (PABP2) in nuclear speckles is independent of import into the nucleus and requires binding to poly(A) RNA. J. Cell. Sci., 113, 2309–2318. [DOI] [PubMed] [Google Scholar]
- 35.Cartegni L. and Krainer,A.R. (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nature Genet., 30, 377–384. [DOI] [PubMed] [Google Scholar]
- 36.Wang J. and Manley,J.L. (1995) Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA, 1, 335–346. [PMC free article] [PubMed] [Google Scholar]
- 37.van Der Houven Van Oordt W., Newton,K., Screaton,G.R. and Caceres,J.F. (2000) Role of SR protein modular domains in alternative splicing specificity in vivo. Nucleic Acids Res., 28, 4822–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiodi I., Biggiogera,M., Denegri,M., Corioni,M., Weighardt,F., Cobianchi,F., Riva,S. and Biamonti,G. (2000) Structure and dynamics of hnRNP-labelled nuclear bodies induced by stress treatments. J. Cell. Sci., 113, 4043–4053. [DOI] [PubMed] [Google Scholar]
- 39.Sanford J.R., Gray,N.K., Beckmann,K. and Caceres,J.F. (2004) A novel role for shuttling SR proteins in mRNA translation. Genes Dev., 18, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cartegni L., Wang,J., Zhu,Z., Zhang,M.Q. and Krainer,A.R. (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res., 31, 3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dauksaite V. and Akusjarvi,G. (2004) The second RNA binding domain of human splicing factor ASF/SF2 is the critical domain controlling adenovirus E1A alternative 5′ splice site selection. Biochem. J., 381, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin C., Feng,Y. and Manley,J.L. (2004) Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature, 427, 553–558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


