Abstract
RNA interference (RNAi) can silence genes at the transcriptional level by targeting locus-specific Lys9H3 methylation or at the post-transcriptional level by targeting mRNA degradation. Here we have cloned and sequenced genomic regions methylated in Lys9H3 in Neurospora crassa to test the requirements for components of the RNAi pathway in this modification. We find that 90% of clones map to repeated sequences and relics of transposons that have undergone repeat-induced point mutations (RIP). We find siRNAs derived from transposon relics indicating that the RNAi machinery targets these regions. This is confirmed by the fact that the presence of these siRNAs depends on components of the RNAi pathway such as the RdRP (QDE-1), the putative RecQ helicase (QDE-3) and the two Dicer enzymes. We show that Lys9H3 methylation of RIP sequences is not affected in mutants of the RNAi pathway indicating that the RNAi machinery is not involved in transcriptional gene silencing in Neurospora. We find that RIP regions are transcribed and that the transcript level increases in the mutants of the RNAi pathway. These data suggest that the biological function of the Neurospora RNAi machinery is to control transposon relics and repeated sequences by targeting degradation of transcripts derived from these regions.
INTRODUCTION
RNA interference (RNAi) is the process by which double-stranded RNA (dsRNA) leads to post-transcriptional silencing (PTGS) of cognate genes (1) and is related to transgene-induced PTGS in plants and in Neurospora (2). The cellular machinery responsible for RNAi has been shown to include the RNAse III enzyme Dicer, an RNA-dependent RNA polymerase (RdRP), and proteins of the PPD (Piwi Paz Domain) family. PTGS is triggered when Dicer cleaves dsRNAs into 21–23 bp siRNA (3) which guide an RNA-induced silencing complex (RISC) to degrade homologous mRNA (4).
Components of the RNAi machinery have also been shown to be involved in transcriptional or chromatin-based silencing by promoting locus-specific methylation of lysine 9 on histone H3 (lys9H3) (5–9). In Schizosaccharomyces pombe, for example, mutants in the PPD protein AGO1 (ago1), Dicer (dcl1) or the RdRP (rdp1) are defective in Lys9H3 methylation in the centromeric region (5) and in the re-establishment of Lys9H3 methylation of the mating type locus (10). Likewise, mutants in genes encoding proteins of the PPD family are defective in locus-specific Lys9H3 methylation in Arabidopsis (ago4) (7), Drosophila (piwi) (8) and Tetrahymena (twi1) (9). It is thought that locus-specific Lys9H3 methylation is guided by a complex called RITS (for RNA-induced initiation of transcriptional gene silencing) (11). Like the RISC complex that targets RNA degradation, the RITS complex contains Dicer-derived siRNAs and a protein of the PPD family (4,11).
Lys9H3 methylation has been associated with silent genes in mammals (12) plants (13,14), fungi (15) and yeast (16) and is required for transposon silencing in plants (17) and heterochromatin formation in yeast (10). Recent work indicates that Lys9H3 methylation triggers gene silencing by serving as a mark for the recruitment of Heterochromatin Protein 1 (HP1) or its yeast homolog SWI6 (10,12,18–20) and for DNA methylation (15).
In addition to targeting Lys9H3 methylation, the biological function of RNAi is to silence transposons in Caenorhabditis elegans (21) and viruses in plants (22,23). Although resistance to some viruses in plants has been shown to be a post-transcriptional process (24), there are indications that in plants the RNAi machinery could also be involved in chromatin-based silencing. Arabidopsis mutants defective in transgene-induced PTGS such as in RdRP (SDE1) or in the PPD protein (AGO1) showed a reduction of transgene methylation (23,25). Furthermore, the Arabidopsis mutant (ago4) encoding a protein of the PPD family is defective in loci-specific Lys9H3 methylation (7). Transposons in C.elegans have been shown to be silenced at the post-transcriptional level (21), but it is not known if transposon silencing also involves RNAi-dependent chromatin-based silencing. The fact that transposons are known targets of Lys9H3 methylation make this an attractive possibility.
In Neurospora crassa, the RNAi machinery has been shown to be required for transgene-induced post-transcriptional gene silencing [(26) and references within]. Here we have cloned and sequenced genomic regions methylated in Lys9H3 in N.crassa to test the requirements for components of the RNAi pathway in this modification. We find that 90% of clones map to repeated sequences and relics of transposons that have undergone repeat-induced point mutation (RIP). We find siRNAs derived from transposon relics, indicating that the RNAi machinery targets these regions. This is confirmed by the fact that the presence of these siRNAs depends on components of the RNAi pathway such as the RdRP (QDE-1), the putative RecQ helicase (QDE-3) and the Dicers. We show that Lys9H3 methylation of these regions is not affected in mutants of the RNAi pathway indicating that RNAi is not involved in transcriptional gene silencing in Neurospora. We find that these RIP regions are transcribed and that the transcript levels increase in the mutants of the RNAi pathway. These data suggest that the biological function of the Neurospora RNAi machinery is to control transposon relics and repeated sequences by targeting degradation of transcripts derived from these regions.
MATERIALS AND METHODS
Chromatin immunoprecipitation
Chromatin immunoprecipitations (ChIPs) were carried out as described previously (27) with some modifications. Conidia of 1 × 108 were inoculated in Neurospora minimal media and grown for 24 h. The mycelia were fixed in 2.5% formaldehyde for 10 min, filtered and washed with cold phosphate-buffered saline (PBS). One milligram of dry mycelia was sonicated in 5 ml of TEE and 1 ml of glass beads for 10 pulses of 30 s with 30 s resting. The insoluble debris was pellet by centrifugation and the chromatin in the supernatant was obtained by CsCl ultracentrifugation as described in (27). Fractions with a density between 1.35 and 1.45 g/cm3 were pooled, dialyzed and stored at −20°C. A fraction of chromatin was de-crosslinked to determine the concentration. The equivalent of 15 μg of chromatin were used for immunoprecipitation (IP) with three different anti-trimethyl Lys9H3 antibodies that were provided as gifts from Dr Thomas Jenuwein or Dr Prim Singh or purchased from AbCam. DNA was resuspended in 100 μl of H2O and 2 μl was used for the PCR.
Cloning of Lys9H3 methylated regions
Cloning was done as described previously (28). Briefly, DNA from five immunoprecipitations carried out with an anti-trimethyl Lys9H3 antibody (A gift from Dr Prim Sing) was combined and treated with Mung bean nuclease to make blunt ends. The treated DNA was phenol extracted, ethanol precipitated and cloned into the pCR4Blunt-TOPO (Invitrogen). Randomly selected clones were sequenced.
Quantification of immunoprecipitated DNA
The quantification was done on the Roche's LightCycler using Roche's FastStart DNA Master SYBR green 1 Kit. Only IPs that showed a more than 5-fold enrichment over mock IPs of the same chromatin were used for quantification.
The following primers were used.
HMC3 forward ATCGGCTGCTCCGACTATCC
HMC3 reverse ACGTCCTCTCCGGTCATCATC
HMC4 forward GCGGGTTCAAAACAGGCTTG
HMC4 reverse CCCCCTCCAATTAAAGGCTCA
HMC6 forward TTTTCTATTTACCCTACTACG
HMC6 reverse TTATTAAAGGAAAAGGTAGAGT
HMC8 forward GCCTGATGCTGGGAGACATTC
HMC8 reverse GCCACGAGATACGGTTTCAAGG
HMC10 forward GCCCTACGGTCCGAAGTTATCG
HMC10 reverse GTAATCGCCGGGGTGAATCTG
HMC15 forward GGTGGTGCGGCTGGTTTTC
HMC15 reverse AGCCGCGGCGTTTGTACC
HMC17 forward ACCCTACCCAACTTAGCCCTCT
HMC17 reverse TAAGCGAACCCCCTTCTTTGT
HMC34 forward CGTTTAAAAGTACATCTAAGC
HMC34 reverse CGTACTTACTTAATTGGAATAG
HMC37 forward CTTGCCGGACAAACATCTGTG
HMC37 reverse GGTGGCCTTTTGGAAATCCTC
COPIA forward AATAGCAGCAACGCCAAAGAC
COPIA reverse CTGTCCACCTCTTGAATCAACC
Wc-1 forward TCAACATCTTCCGCCTCATCTC
Wc-1 reverse ATGCTGCTGATGCTGCTTATGC
Pund forward CCCCACCCTCGGTCATATAAC
Pund reverse GCCCTCCAGCCAATCCTTTAT
The relative values were obtained by dividing the values obtained for the cloned gene by the values obtained for the wc-1 gene, which were extrapolated from individual external standard curves made with serial dilutions of input DNA. The error bars represent the standard deviation of two different immunoprecipitations carried out with two different antibodies and analyzed in duplicates.
Small RNA purification and northern analysis
Small RNA purification was performed as described previously (29) with modifications (30). Single-stranded RNA probes were prepared by T3 RNA polymerase transcription of Not1-digested pCR4Blunt vector containing the desired sequence using 32P-labeled uridine triphosphate (50 μCi per 20 μL reaction volume; specific activity 3000 Ci/mmol; New England Nuclear). To remove plasmid template, the reaction was incubated at 37°C for 15 min with RNase-free DNase I (Roche). To break labeled transcripts to an average size of 50 nt, 300 μL of 80 mM sodium bicarbonate and 120 mM sodium carbonate were added to the transcriptional reaction and incubated at 60°C for 3 h. To stop the hydrolysis reaction of the transcript, 20 μL of 3 M sodium acetate (pH 5.0) was added.
Reverse transcription and quantitative PCR
Reverse transcription (RT) was carried out with SuperScript II H–Reverse transcriptase (Invitrogen) according to the manufacturer's instructions except for the following: the amount of total RNA was 2.5 μg and the amount of gene specific primer was 2 pmol. One-tenth of the RT reaction volume was used for quantitative PCR. The quantification was done using an external standard curve made with a serial dilution of one of the RT reaction. The efficiency of reverse transcription among different samples was normalized by including an actin-specific primer in all RT reactions and quantifying the amount of actin RNA.
N.crassa strains, media and growth conditions
Strains were grown in Vogel's minimal medium for Neurospora in the presence of appropriate nutritional supplements and/or selectable markers (30). The stably silenced strain (6xw) was described previously (31) as well as the qde mutant strains (32), Δdim5 strain (Chicas et al., submitted) and Δdcl1/dcl2 strain (26). Strains 74-OR23A (FGSC N° 987) and 74-OR8a (FGSC N° 988) were obtained from the Fungal Genetics Stock Center, University of Kansas, Kansas City, KS.
RESULTS
Identification of genomic regions methylated in Lys9 of histone H3
Lys9H3 methylation has been shown to be associated with promoters of silenced genes (12), with repeated DNA sequences (28) and transposons (17) and is required for the formation of heterochromatin in S.pombe (5,6) and for DNA elimination in Tetrahymena (33). In Neurospora, Lys9H3 methylation is carried out by DIM-5 and is required for DNA methylation (15). A recent study showed that DNA methylation in Neurospora is associated exclusively with sequences that have undergone RIP (34). The dim-5 mutant, however, shows growth and fertility defects that are not observed in the DNA methyltransferase mutant dim-2, suggesting that Lys9H3 methylation has functions in addition to directing DNA methylation (15). To examine the role of the RNAi machinery in locus-specific Lys9H3 methylation and to uncover possible new roles for this modification, we set out to clone and sequence Lys9H3 methylated regions in N.crassa. This was carried out by immunoprecipitating formaldehyde crosslinked chromatin with an antibody that recognizes trimethylated Lys9H3. The chromatin was obtained from a strain that was transformed with a plasmid containing the qa-2 gene and a fragment of the al-1 gene (31). We have shown previously that this transgene is hypermethylated on Lys9H3 (Chicas et al., manuscript in preparation) and it was used as an internal control for our experiment. The co-immunoprecipitated DNA was cloned as described in (28) and randomly selected clones were sequenced. As would be expected, one of the histone H3 methylated clones (HMCs) contained the qa-2 transgene (Table 1, HMC7). To map the remaining HMCs, we used BLAST to search for our sequences in the N.crassa databank at www.broad.mit.edu/annotation/fungi/neurospora. We found that 37 of 50 (74%) HMCs mapped to repeated DNA regions and transposon relics that have undergone RIP (Table 1 and Supplementary Material). Seventy-four percent is a substantial enrichment for these regions considering that RIP sequences represent 10% of the genome of Neurospora (35). Five of the sequences (10%) mapped to single genes, and no match was found for eight sequences (16%) (Table 1 and Supplementary Material). RIP causes G:C to A:T point mutations of repeated sequences in Neurospora that create sequences with particular dinucleotide frequencies and high A + T content. We therefore determined if the eight sequences that did not find a match in the Neurospora databank were RIP sequences by calculating dinucleotide frequencies or RIP indices (36). All eight HMCs showed indices typical of RIP sequences (see Supplementary Material). Thus RIP sequences are 90% enriched in our pool of clones. These results are consistent with the previous observations that DNA methylation in Neurospora is found associated exclusively with RIP sequences (34) and that Lys9H3 methylation is the mark for DNA methylation (37).
Table 1. Neurospora crassa Lys9H3 methylated clonesa.
| Clone | Contig | Location | Identifier |
|---|---|---|---|
| HMC1 | No match | — | — |
| HMC2 | 3.303 | 134767–135311 | Gypsy relic |
| HMC4 | 3.690 | 700–300 | dTad2 relic |
| HMC5 | 3.523 | 4745–4000 | Repeat 14×;dTad2 relic |
| HMC6 | 3.697 | 300–100 | Repeat 6× |
| HMC7 | 3.350 | 13904–13063 | Qa-2 transgenic DNA |
| HMC9 | No match | — | — |
| HMC16 | No match | — | — |
| HMC17 | 3.690 | 11805–11490 | dTad2 relic |
| HMC18 | No match | — | — |
| HMC19 | No match | — | — |
| HMC20 | 3.275 | 14692–14625 | Yoyo relic |
| HMC22 | 3.690 | 727–616 | dTad2 relic |
| HMC23 | 3.285 | 1544–2167 | Pogo relic |
| HMC23 | 3.488 | 4–557 | Repeat 2×; |
| HMC25 | 3.631 | 1057–1232 | Repeat 2× |
| HMC26 | 3.746 | 9036–8657 | Repeat 2× |
| HMC27 | 3.503 | 48791–48205 | Repeat 3× |
| HMC29 | 3.661 | 853–1326 | Mariner class 2 relic |
| HMC30 | No match | — | — |
| HMC31 | No match | — | — |
| HMC32 | 3.12 | 492230–491648 | Mariner class 2 relic |
| HMC33 | 3.781 | 857–657 | Repeat 2× |
| HMC34 | 3.537 | 1468–1232 | Repeat 2× |
| HMC36 | 3.68 | 12832–12963 | Repeat 2× |
| HMC38 | 3.321 | 2000–2300 | Maggy relic |
| HMC39 | 3.384 | 2100–2600 | Repeat 4× |
| HMC40 | 3.695 | 1620–1507 | Repeat 13× |
| HMC42 | No match | ||
| HMC43 | 3.766 | 2431–2059 | Repeat 2× |
| HMC45 | 3.553 | 23996–24282 | Repeat 269× Pot relic |
| HMC46 | 3.787 | 604–5 | Repeat 14× |
| HMC47 | 3.289 | 95730–96166 | Repeat 2× |
| HMC48 | 3.250 | 3166–31627 | Gypsy relic |
| HMC49 | 3.711 | 4945–4364 | Repeat 17× |
| HMC50 | 3.318 | 987–696 | Repeat 27× |
| HMC51 | 3.253 | 18647–18771 | Repeat 3× Pot2 relic |
| HMC52 | 3.423 | 1394–1750 | Repeat 3× |
| HMC53 | 3.97 | 663–180 | Repeat 18× |
| HMC54 | 3.553 | 26122–26000 | Repeat 29× |
| HMC55 | 3.122 | 6870–6627 | Repeat 2× near Tto |
| HMC56 | 3.553 | 23995–24282 | Repeat 269× |
| HMC57 | 3.208 | 696–492 | Repeat of 5× |
| HMC58 | 3.228 | 68569–68422 | Repeat 11× |
| HMC60 | 3.637 | 2301–2613 | Repeat 34 small rDNA |
aThe discontinuity in the numbering is due to clones that were not sequenced due to technical problems or clones from single genes that were found not to be hypermethylated in Lys9H3 by ChIP and quantitative PCR.
To confirm that the genomic HMCs were effectively hypermethylated in Lys9H3, we determined the enrichment in Lys9H3 methylation at the HMCs using ChIP and quantitative PCR. Chromatin was immunoprecipitated with the anti-trimethylated Lys9H3 antibody and the co-precipitated DNA was analyzed by quantitative PCR using SYBR green 1 and region-specific primers. As shown in Figure 2, four HMCs that mapped to RIP sequences showed enrichment in Lys9H3 methylation (Figure 2A–D; wt lanes). In contrast, no enrichment in Lys9H3 methylation was found in the five HMCs that mapped to single copy genes (data not shown). Some of the single copy genes were found to be enriched in trimethylated Lys4H3 (data not shown). Since the antibody used for the immunoprecipitation for the cloning has been reported to cross-react with trimethyl Lys4H3 (38), this would explain why these single copy genes were also cloned. Analysis of RIP regions revealed that they are hypomethylated in Lys4H3 relative to our reference gene (data not shown). These data indicate that Lys9H3 methylation, like DNA methylation, is mainly associated with RIP sequences in Neurospora.
Figure 2.
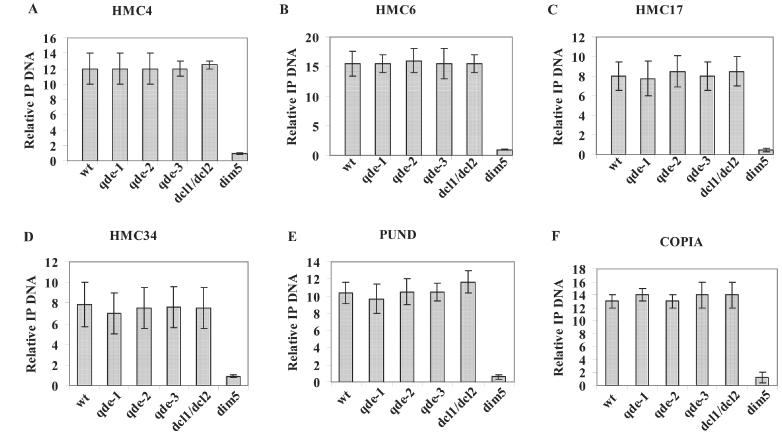
The Neurospora RNAi machinery is not required for loci-specific Lys9H3 methylation. DNA co-immunoprecipitated with anti-trimethyl antibodies was quantified by real-time PCR. The relative values were obtained by dividing the values obtained for the HMCs by the values obtained for the wc-1 gene, which were extrapolated from individual external standard curves made with serial dilutions of input DNA. The error bars represent the standard deviation of two different immunoprecipitation carried out with two different antibodies and analyzed in duplicates.
siRNA derived from transposons relics
siRNAs are thought to be the determinants of specificity in both RNAi-dependent transcriptional and post-transcriptional gene silencing. In fact, siRNAs have been associated with both transcriptional and post-transcriptional gene silencing (39) and are part of the RITS and RISC complexes that target locus-specific Lys9H3 methylation and RNA degradation, respectively (11,4). To test whether RNAi is required for Lys9H3 methylation of our cloned regions, we first looked for siRNAs derived from the HMCs. siRNAs corresponding to three transposon relics were detected in a wild-type strain by northern blot analysis (Figure 1A–C). To determine the requirements for components of the Neurospora RNAi machinery in the accumulation of the HMCs-derived siRNAs, we also looked for these siRNAs in the qde-1, qde-2 and qde-3 mutants corresponding to mutations in the RdRP, the PPD protein and the putative RecQ helicase, respectively, and in a strain deleted in the two Neurospora Dicer genes (dcl-1/dcl-2). We have shown previously that these mutants are defective in transgene-induced PTGS in Neurospora [(26) and references within]. Similar to what we have observed for transgene-induced PTGS, the presence of these siRNAs depend on a functional QDE-1, QDE-3 and the Dicers as they are not detected in the qde-1, qde-3 and dcl-1/dcl-2 mutants (Figure 1A–C). Similar to the case of transgene-induced PTGS, there appears to be an accumulation of the siRNAs in the qde-2 mutant (Figure 1) (30). These data indicate that the RNAi machinery controls transposon relics in Neurospora.
Figure 1.
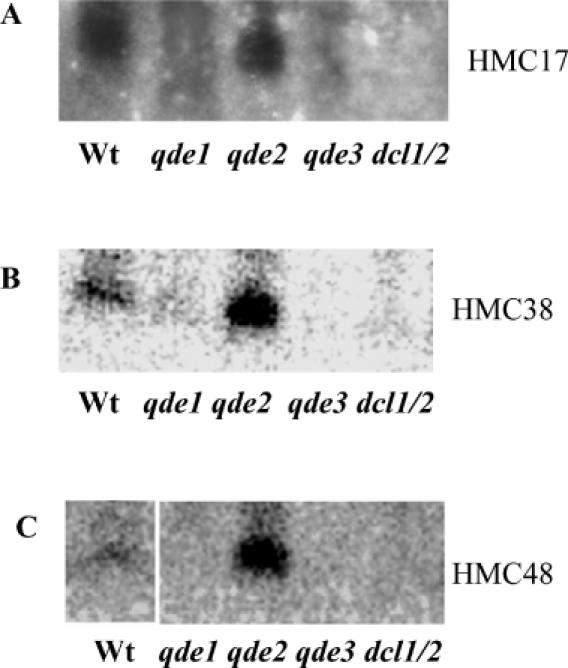
Transposon relics derived siRNAs. RNA blots were hybridized with probes for the indicated regions. HMC17 is a relic of Tad, HMC38 is a maggy relic and HMC48 is a gypsy relic. The low-molecular-weight RNAs were extracted from a wild-type strain or from the indicated mutants. qde-1 is a mutant in the RdRP, qde-2 is a mutant in the PPD protein, qde-3 is a mutant in the putative RecQ helicase and dcl-1/dcl-2 is a double mutant in the two Neurospora Dicer genes.
Locus-specific Lys9H3 methylation is independent of the Neurospora RNAi machinery
RNAi can silence genes at the transcriptional level by targeting locus-specific Lys9H3 methylation or at the post-transcriptional level by targeting mRNA degradation. Components of the RNAi pathway are known to be required for transgene-induced post-transcriptional gene silencing in Neurospora [(26) and references within]. To examine whether components of the Neurospora RNAi pathway are also involved in chromatin-based silencing, we tested the requirement for components of the Neurospora RNAi machinery for Lys9H3 methylation of the HMCs. Chromatin from the different strains was immunoprecipitated with an anti-trimethyl Lys9H3 antibody and the DNA co-precipitated was analyzed by quantitative PCR. Figure 2 shows that Lys9H3 methylation was not affected in any of the qde mutants or the Δdcl-1/dcl-2 strain in four regions analyzed (HMC4, HMC6, HMC17, HMC34). In contrast, and as expected, a clear loss of Lys9H3 methylation was detected in a strain in which we have deleted DIM-5 (Chicas et al., unpublished data) (Figure 2). These data indicate that the Neurospora RNAi machinery is not required for locus-specific LysH3 methylation. To confirm our results, we examined whether the qde mutants affected Lys9H3 methylation in other regions. We examined the pund locus, which has been shown to be hypermethylated at Lys9H3 (37) and the Tcen region of centromere VII (40) because it has been shown that Lys9H3 methylation of the S.pombe centromeric regions requires the components of the RNAi machinery (5). Quantitative PCR analysis with primers specific for the Tcen region, a centromere-specific, copia-like element indicates that this region is enriched in Lys9 methylated H3 (Figure 2F). No significant changes in Lys9H3 methylation were detected in the qde mutants or in Δdcl-1/dcl-2 strain at the Tcen or pund regions (Figure 2E and F). Again as expected, a clear loss of this mark is detected in the Δdim-5 strain. These data demonstrate that components of the RNAi pathway are not required for locus-specific Lys9H3 methylation in Neurospora.
Transcripts derived from repeated DNA sequences are targets of the Neurospora RNAi machinery
The presence of siRNAs derived from the HMCs suggests that these regions are transcribed despite the hypermethylation of Lys9H3, the DNA methylation and the mutations caused by RIP. To search for transcripts derived from these regions, we carried out RT–PCR. We detected bidirectional transcripts derived from four repeated regions tested although at low levels (Figure 3A), confirming that these regions are effectively transcribed. To determine if the RNAi machinery targeted these transcripts for degradation, we quantified transcripts derived from the pund region in the wild type and qde mutant strains. Figure 3B shows that the levels of the transcripts increased in the qde mutants, suggesting that the RNAi machinery targets these transcripts for degradation. These data suggest that the biological function of the Neurospora RNAi machinery is to control RIP sequences by targeting degradation of RNAs derived from these regions.
Figure 3.
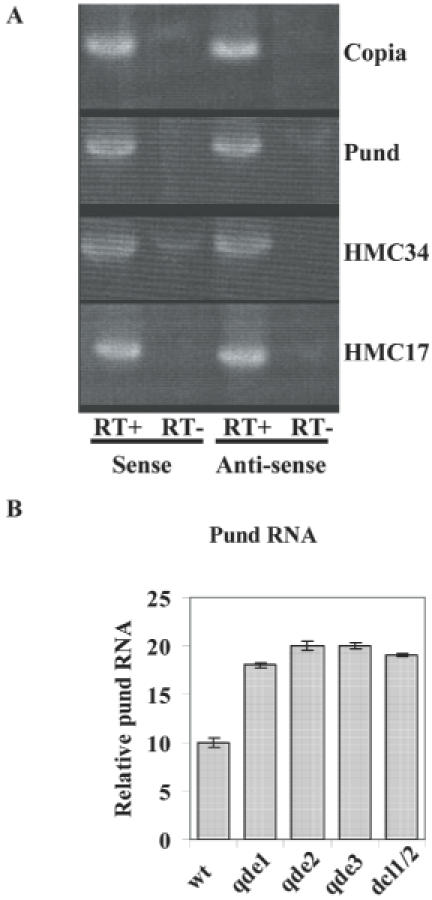
Transcripts derived from repeated sequences are targets of the Neurospora RNAi machinery. (A) RT–PCR analysis showing the presence of bidirectional transcripts derived from repeated sequences. (B) Quantification of pund transcripts in wild type and mutants in genes of the Neurospora RNAi pathway. The error bars represent the standard deviation of an RT reaction analyzed in triplicates.
DISCUSSION
Lys9H3 methylation has been associated mainly with silenced repeated sequences (5,17,28) but also with silenced single genes (12,13,16). In Neurospora, Lys9H3 methylation is required for DNA methylation (15) and DNA methylation is exclusively associated with sequences that have undergone RIP (34). Our results that the HMCs mapped predominantly to RIP sequences, such as repeated sequences and relics of transposons, is consistent with the previous observation that Lys9H3 methylation is the mark that triggers DNA methylation (15).
N.crassa contains many relics of transposons (representing 46% of the Neurospora RIP-mutated repetitive sequences) but no active transposon elements, which is a testament to the efficiency of RIP in inactivating transposons (35). RIP takes place during the sexual cycle of Neurospora; therefore cells have to go through meiosis in order to inactivate transposons by RIP. During vegetative growth, active transposons experimentally introduced into Neurospora have been shown to be silenced by DNA methylation (41). Even though RIP by itself can inactivate transposons, the products of RIP are also the only known targets for DNA methylation in Neurospora (34). The presence of siRNA derived from transposon relics indicates surprisingly that these relics are also under the control of the RNAi machinery. siRNAs are thought to silence genes at the transcriptional level by targeting locus-specific Lys9H3 methylation. Consistent with this idea, siRNAs derived from heterochromatic regions have been found in Drosophila (42) and in S.pombe (43), and components of the RNAi machinery are required for locus-specific Lys9H3 methylation in S.pombe (5,6), Arabidopsis (7), Drosophila (8) and Tetrahymena (9). Our analyses of diverse loci indicate that siRNAs and other components of the Neurospora RNAi machinery are not required for locus-specific Lys9H3 methylation. In contrast, we found that transcripts derived from RIP sequences increase in RNAi mutants. Consistent with previous results (30), in a qde-2 mutant background siRNAs are accumulated normally since the QDE-2 protein was found acting downstream of the siRNA production. Together, these results suggest that the Neurospora RNAi machinery is mainly dedicated to post-transcriptional gene silencing and is not involved in chromatin-based silencing, although in Neurospora a RITS-like complex may exist as suggested by the presence of predicted homologs of S.pombe Chp1 and Tas3.
Interestingly, it appears that RIPed sequences are targeted contemporarily by two different and independent silencing pathways. First, these sequences are hypermethylated at Lys9H3 and are presumably transcriptionally silenced. Second, the transcripts are also degraded by post-transcriptional silencing. This redundancy may lead to an increased silencing efficiency. It is worth noting that although hypermethylated the RIPed sequences are transcribed, suggesting that the transcriptional silencing is not able to completely block transcription. Moreover, this seems to be true not only in Neurospora but also in S.pombe where the transcriptionally silent centromeric regions produce transcripts that are the target of the RNAi machinery.
The divergence in the requirement for the RNAi machinery for loci-specific Lys9H3 methylation between Neurospora and other organisms could be due to the known structural differences between the Neurospora Lys9H3 methyltransferase and that of other organisms. DIM-5, the Lys9H3 methyltransferase in Neurospora, unlike its counterparts in S.pombe, Drosophila and mammals lacks a chromodomain (15). This domain has been shown to bind RNA (44) in addition to recognizing the Lys9H3 methylation mark (18). It could be that since DIM-5 lacks this chromodomain, it uses a different mechanism for recognizing sequences to be Lys9H3 methylated. As we have demonstrated here, Lys9H3 methylation mainly targets RIP sequences. RIP causes G:C to A:T point mutations of repeated sequences in Neurospora with a strict sequence preference that creates sequences with high A + T content (36). DIM-5 could, therefore, directly or indirectly recognize RIP sequences due to their characteristic high A:T content. This is consistent with the observation that RIP sequences or oligonucleotides with a rich A:T content are sufficient to induce de novo DNA methylation in Neurospora (45) and with the observation that both the DNA methylated regions and the HMCs map exclusively to RIP sequences. It is worth noting that the cenH region of S.pombe, like the RIP sequences, has a high A:T content and that Lys9H3 methylation of the A:T-rich regions of cenH is independent of the RNAi machinery (6), suggesting that CLR-4 like DIM-5 can also methylate A:T-rich regions independent of the RNAi machinery.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Prim Singh and Dr Thomas Jenuwein for providing us with their antibodies against trimethyl Lys9H3, Dr Valerio Orlando for valuable suggestions, Dr Baroni and the Dipartimento di Scienze Cliniche for use of their LightCycler instrument, and the Whitehead Institute for providing access to the Neurospora genome database. This work is supported by grants of The European Community (#QLK3-CT-2000-00078), the Instituto Pasteur Fondazione Cenci Bolognetti, FIRB-MIUR 2001 (RBNEO15MPB_001/RBNE01KXC9_006) and CNR 2003 (Progetto Strategico MIUR—legge 449/97)
REFERENCES
- 1.Denli A.M. and Hannon,G.J. (2003) RNAi: an ever-growing puzzle. Trends Biochem. Sci., 28, 196–201. [DOI] [PubMed] [Google Scholar]
- 2.Chicas A. and Macino,G. (2001) Characteristics of post-transcriptional gene silencing. EMBO Rep., 2, 992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 4.Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 5.Volpe T.A., Kidner,C., Hall,I.M., Teng,G., Grewal,S.I. and Martienssen,R.A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science, 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- 6.Hall I.M., Shankaranarayana,G.D., Noma,K., Ayoub,N., Cohen,A. and Grewal,S.I. (2002) Establishment and maintenance of a heterochromatin domain. Science, 297, 2232–2237. [DOI] [PubMed] [Google Scholar]
- 7.Zilberman D., Cao,X. and Jacobsen,S.E. (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science, 299, 716–719. [DOI] [PubMed] [Google Scholar]
- 8.Pal-Bhadra M., Leibovitch,B.A., Gandhi,S.G., Rao,M., Bhadra,U., Birchler,J.A. and Elgin,S.C. (2004) Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science, 303, 669–672. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Mochizuki,K. and Gorovsky,M.A. (2004) Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc. Natl Acad. Sci. USA, 101, 1679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakayama J., Rice,J.C., Strahl,B.D., Allis,C.D. and Grewal,S.I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. [DOI] [PubMed] [Google Scholar]
- 11.Verdel A., Jia,S., Gerber,S., Sugiyama,T., Gygi,S., Grewal,S.I. and Moazed,D. (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science, 303, 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen S.J., Schneider,R., Bauer,U.M., Bannister,A.J., Morrison,A., O'Carroll,D., Firestein,R., Cleary,M., Jenuwein,T., Herrera,R.E. and Kouzarides,T. (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature, 412, 561–565. [DOI] [PubMed] [Google Scholar]
- 13.Bastow R., Mylne,J.S., Lister,C., Lippman,Z., Martienssen,R.A. and Dean,C. (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature, 427, 164–167. [DOI] [PubMed] [Google Scholar]
- 14.Sung S. and Amasino,R.M. (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature, 427, 159–164. [DOI] [PubMed] [Google Scholar]
- 15.Tamaru H. and Selker,E.U. (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature, 414, 277–283. [DOI] [PubMed] [Google Scholar]
- 16.Schramke V. and Allshire,R. (2003) Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science, 301, 1069–1074. [DOI] [PubMed] [Google Scholar]
- 17.Gendrel A.V., Lippman,Z., Yordan,C., Colot,V. and Martienssen,R.A. (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science, 297, 1871–1873. [DOI] [PubMed] [Google Scholar]
- 18.Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- 19.Lachner M., O'Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- 20.Freitag M., Hickey,P.C., Khlafallah,T.K., Read,N.D. and Selker,E.U. (2004) HP1 is essential for DNA methylation in Neurospora. Mol. Cell, 13, 427–434. [DOI] [PubMed] [Google Scholar]
- 21.Sijen T. and Plasterk,R.H. (2003) Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature, 426, 310–314. [DOI] [PubMed] [Google Scholar]
- 22.Mourrain P., Beclin,C., Elmayan,T., Feuerbach,F., Godon,C., Morel,J.B., Jouette,D., Lacombe,A.M., Nikic,S., Picault,N., Remoue,K., Sanial,M., Vo,T.A. and Vaucheret,H. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- 23.Dalmay T., Hamilton,A., Rudd,S., Angell,S. and Baulcombe,D.C. (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- 24.Ratcliff F.G., MacFarlane,S.A. and Baulcombe,D.C. (1999) Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell, 11, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fagard M., Boutet,S., Morel,J.B., Bellini,C. and Vaucheret,H. (2000) AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl Acad. Sci. USA, 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catalanotto C., Pallotta,M., ReFalo,P., Sachs,M.S., Vayssie,L., Macino,G. and Cogoni,C. (2004) Redundancy of the two Dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol., 24, 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orlando V., Strutt,H. and Paro,R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- 28.Kondo Y. and Issa,J.P. (2003) Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J. Biol. Chem., 278, 27658–27662. [DOI] [PubMed] [Google Scholar]
- 29.Hamilton A.J. and Baulcombe,D.C. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- 30.Catalanotto C., Azzalin,G., Macino,G. and Cogoni,C. (2002) Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev., 16, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cogoni C., Irelan,J.T., Schumacher,M., Schmidhauser,T.J., Selker,E.U. and Macino,G. (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA–DNA interactions or DNA methylation. EMBO J., 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- 32.Cogoni C. and Macino,G. (1997) Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl Acad. Sci. USA, 94, 10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taverna S.D., Coyne,R.S. and Allis,C.D. (2002) Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell, 110, 701–711. [DOI] [PubMed] [Google Scholar]
- 34.Selker E.U., Tountas,N.A., Cross,S.H., Margolin,B.S., Murphy,J.G., Bird,A.P. and Freitag,M. (2003) The methylated component of the Neurospora crassa genome. Nature, 422, 893–897. [DOI] [PubMed] [Google Scholar]
- 35.Galagan J.E., Calvo,S.E., Borkovich,K.A., Selker,E.U., Read,N.D., Jaffe,D., FitzHugh,W., Ma,L.J., Smirnov,S., Purcell,S. et al. (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature, 422, 859–868. [DOI] [PubMed] [Google Scholar]
- 36.Margolin B.S., Garrett-Engele,P.W., Stevens,J.N., Fritz,D.Y., Garrett-Engele,C., Metzenberg,R.L. and Selker,E.U. (1998) A methylated Neurospora 5S rRNA pseudogene contains a transposable element inactivated by repeat-induced point mutation. Genetics, 149, 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamaru H., Zhang,X., McMillen,D., Singh,P.B., Nakayama,J., Grewal,S.I., Allis,C.D., Cheng,X. and Selker,E.U. (2003) Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nature Genet., 34, 75–79. [DOI] [PubMed] [Google Scholar]
- 38.Peters A.H., Kubicek,S., Mechtler,K., O'Sullivan,R.J., Derijck,A.A., Perez-Burgos,L., Kohlmaier,A., Opravil,S., Tachibana,M., Shinkai,Y. et al. (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell, 12, 1577–1589. [DOI] [PubMed] [Google Scholar]
- 39.Bailis J.M. and Forsburg,S.L. (2002) RNAi hushes heterochromatin. Genome Biol., 3, REVIEWS1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cambareri E.B., Aisner,R. and Carbon,J. (1998) Structure of the chromosome VII centromere region in Neurospora crassa: degenerate transposons and simple repeats. Mol. Cell. Biol., 18, 5465–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y., Cambareri,E.B. and Kinsey,J.A. (2001) DNA methylation inhibits expression and transposition of the Neurospora Tad retrotransposon. Mol. Genet. Genomics, 265, 748–754. [DOI] [PubMed] [Google Scholar]
- 42.Aravin A.A., Lagos-Quintana,M., Yalcin,A., Zavolan,M., Marks,D., Snyder,B., Gaasterland,T., Meyer,J. and Tuschl,T. (2003) The small RNA profile during Drosophila melanogaster development. Dev. Cell, 5, 337–350. [DOI] [PubMed] [Google Scholar]
- 43.Reinhart B.J. and Bartel,D.P. (2002) Small RNAs correspond to centromere heterochromatic repeats. Science, 297, 1831. [DOI] [PubMed] [Google Scholar]
- 44.Akhtar A., Zink,D. and Becker,P.B. (2000) Chromodomains are protein–RNA interaction modules. Nature, 407, 405–409. [DOI] [PubMed] [Google Scholar]
- 45.Tamaru H. and Selker,E.U. (2003) Synthesis of signals for de novo DNA methylation in Neurospora crassa. Mol. Cell. Biol., 23, 2379–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


