Abstract
The expression of Rad51p, a DNA repair protein that mediates homologous recombination, is induced by DNA damage and during both meiosis and exconjugant development in the ciliate Tetrahymena thermophila. To completely investigate the transcriptional regulation of Tetrahymena RAD51 expression, reporter genes consisting of the RAD51 5′ non-translated sequence (5′ NTS) positioned upstream of either the firefly luciferase or green fluorescent protein coding sequences have been targeted for recombination at the macronuclear btu1-1 (K350M) locus of T. thermophila strain CU522. Expression from RAD51-luciferase reporter constructs has been directly quantified from transformant whole cell lysates. Luciferase is induced to maximum levels in transformants harboring the full-length RAD51-luciferase reporter gene following exposure to DNA damaging UV irradiation. A series of truncations, deletions, insertions, substitutions and inversions of the RAD51 5′ NTS have led to the identification of three distinct transcriptional promoter elements. The first of these sequence elements is required for basal levels of transcription. The second modulates expression in the absence of DNA damage, whereas the third ensures increased RAD51 transcription in response to DNA damage and during meiosis. Tetrahymena RAD51 is tightly regulated through these transcriptional elements to produce the appropriate expression during conjugation, and in response to DNA damage.
INTRODUCTION
Homologous recombination fulfills two seemingly conflicting roles. Recombination between sister chromatids during meiosis generates genetic diversity, while the same mechanism helps to maintain genetic stability when cells repair DNA damage incurred as a result of normal cellular processes and environmental agents. At the core of homologous recombination is the identification of homologous DNA by a ‘recombinase’ protein, represented in eukaryotes by Rad51p and its various homologs (1). The Rad51p binds to single-stranded DNA and helps to scan double-stranded DNA until a homologous sequence is found, where it forms a nucleofilament on the single-stranded DNA and catalyzes homologous strand exchange [reviewed in (2,3)].
The expression of RAD51 from the ciliate Tetrahymena thermophila varies in response to both environmental conditions and developmental signals. The Rad51 mRNA levels increase following exposure to DNA damaging agents (4), a property shared by homologs from other eukaryotes. There is a distinct pattern of RAD51 expression through the cell cycle, with minimum levels coincident with micronuclear M phase and maximum expression corresponding to a period of macronuclear DNA replication that immediately follows cytokinesis (5). There is also a bimodal increase in the mRNA levels during conjugation, with periods of peak expression coinciding with meiotic prophase and exconjugant macronuclear development (5). Tetrahymena differs from most eukaryotes in its dependence on a single RAD51 paralog for homologous recombination in both vegetatively dividing cells and cells undergoing meiosis (5). Other eukaryotes express multiple RAD51 paralogs, one of which (DMC1) is only expressed during meiosis, where it is essential for sister chromatid exchange in yeast and humans (6,7).
Variations detected for Tetrahymena Rad51 mRNA levels are similar to those seen for RAD51 in other eukaryotes. Changes in yeast and human Rad51 mRNA levels are largely mediated by either induction or repression of transcription (8–12), although post-translational inactivation of human RAD51p by caspase-mediated cleavage during apoptosis (13) or phosphorylation by c-Abl after ionizing radiation has also been documented (14,15).
It was unclear whether the increases in Tetrahymena RAD51 expression in response to DNA damaging agents (4) and during conjugation (5) are due to transcriptional or post-transcriptional regulation. Although the major mechanism for regulating mRNA abundance in Tetrahymena is differential transcription (16), mRNA degradation can play a role in gene products such as the SerH3 surface antigen (17). In an effort to determine whether the Tetrahymena RAD51 expression is under transcriptional control, a series of reporter genes under control of the putative RAD51 transcriptional promoter have been constructed and introduced into Tetrahymena transformants. We have used this methodology to define the inducible RAD51 transcriptional promoter and to identify cis elements critical to its differential expression in response to DNA damage and the developmental stages of conjugation.
MATERIALS AND METHODS
General methods
Tetrahymena genomic DNA was isolated by detergent lysis as described previously (18). RT–PCR protocols and molecular techniques were as described in (19). Total RNA was isolated from vegetatively dividing and conjugating cells using Qiagen RNeasy Total RNA kits (Valencia, CA). The specific 32P-radiolabeled DNA probes were generated by a PCR strategy as described previously (20), and Southern-blot analysis was performed as described previously (5). Treatment of total cellular RNA from Tetrahymena with glyoxal prior to agarose gel electrophoresis and northern-blot analysis was as described previously (19).
Tetrahymena thermophila strains and growth conditions
The T.thermophila strains CU522, CU725 and CU727 (Jacek Gaertig, University of Georgia, Athens, GA) express the mutant β-tubulin btu1-1 (K350M) allele that confers hypersensitivity to the microtubule-stabilizing drug paclitaxel (Table 1). These strains were transformed with various reporter constructs by the targeted disruption of the btu1-1 allele throughout this study (21). The cells were grown in 2% PPYS (2% proteose peptone, 0.2% yeast extract and 0.1% sequestrene) at 30°C on a platform shaker (100 r.p.m.) as described previously (18). All Tetrahymena cultures were maintained in 1× PSF (Penicillin, Streptomyocin and Fungizone; Life Technologies, Rockville, MD) to prevent bacterial and fungal growth.
Table 1. Tetrahymena thermophila strains.
| Strain | Micronuclear genotype | Macronuclear genotype | Macronuclear phenotype |
|---|---|---|---|
| CU522 | mpr1/mpr1, btu1-1/btu1-1 | mpr1, btu1-1 | mp-R, ory-R, pac-S, VI |
| CU725 | chx1/chx1, btu1-1/btu1-1 | chx1, btu1-1 | cy-R, ory-R, pac-S, VII |
| TC202 | mpr1/mpr1, btu1-1/btu1-1 | mpr1, btu1-1::Luc | mp-R, ory-S, pac-R, VI BtLuc |
| TC232 | mpr1/mpr1, btu1-1/btu1-1 | mpr1, btu1-1::RdLuc | mp-R, ory-S, pac-R, VI RdLuc |
| TC296 | chx1/chx1, btu1-1/btu1-1 | chx1, btu1-1::RdLuc | cy-R, ory-S, pac-R, VII RdLuc |
| TC368 | mpr1/mpr1, btu1-1/btu1-1 | mpr1, btu1-1::RdGFP | mp-R, ory-S, pac-R, VI RdGFP |
| TC370 | chx1/chx1, btu1-1/btu1-1 | chx1, btu1-1::RdGFP | cy-R, ory-S, pac-R, VII RdGFP |
chx1, cycloheximide resistance (cy-R); mpr1, 6-methylpurine resistance (mp-R); btu1-1, paclitaxel sensitivity (pac-S) and oryzalin resistance (ory-R). BtLuc, luciferase expressed from the BTU1 promoter; RdLuc, luciferase expressed from the RAD51 promoter; RdGFP, GFP expressed from the RAD51 promoter. The genotypes and phenotypes of representative transgenic strains are listed in this table. Transgenic strains not listed include those expressing reporter genes from variously mutated promoters described in the text.
Radiolabeled probes
The PCR products were radiolabeled by the incorporation of [α−32P]dATP (sp. act. 3000 Ci/mmol) as described in (20), to be used as probes in Southern- and northern-blot analyses. The PCR primer pairs P1, P2, P3, P4 and P5 were used to synthesize probes specific for the 3′ non-translated sequence (3′ NTS) of the Tetrahymena β-tubulin 1 gene (accession no. L01415), and coding regions of Tetrahymena Rad51 (accession no. AF064516) Tetrahymena α-tubulin (accession no. M86723), luciferase and green fluorescent protein (GFP), respectively.
P1(+) TCGGTCAGCTAAACCAAC
P1(−) ATGCGGGTGAGTGCAGAA
P2(+) GACGAATTCGGTATTGC
P2(−) TCACTCGTTGAAGTC
P3(+) GTTATTTCAATTCACG
P3(−) AAGAAGACAGCTCTGG
P4(+) AAGAAAGGCCCGGCGC
P4(−) GAGGCAGAGCGACACC
P5(+) GCCAATTGGAGTATTT
P5(−) GTTGTCCCAATTCTTG
Construction of reporter genes for expression in Tetrahymena
The Rad51-luciferase reporter construct was initiated by introducing a BamHI site at −1.3 kb of the RAD51 5′ NTS, and the HindIII and EcoRV sites at the third codon and the stop codon of the Rad51p coding sequence, respectively (4). The luciferase coding sequence present in the plasmid pGL3-Basic (Promega, Madison, WI) was amplified by PCR, using primers that introduce HindIII and EcoRV sites at the third codon and the stop codon, respectively, to accommodate replacement of the Rad51p coding sequence with that of luciferase. The resultant BamHI–EcoRV fragment was subcloned into the Litmus 28 vector (New England Biolabs). The BTU2 3′ NTS, includes a polyadenylation site, was amplified by PCR from pHAB2 (J. Gaertig, University of Georgia) and cloned into the pLit28::RdLuc construct at unique EcoRV and SpeI sites. Site-directed mutagenesis by the method of Kunkel (22) was used to introduce unique BamHI and NsiI sites flanking the H4-Neo cassette in pHAB1, a plasmid designed for the targeted disruption of the Tetrahymena BTU1 locus (23). The introduction of BamHI and NsiI sites in pHAB1 facilitated the replacement of the H4-Neo cassette with the Rad51-luciferase-BTU2 reporter construct described above. The resultant reporter construct, targeted for the disruption of the btu1-1 locus in strain CU522 and consisting of the RAD51 promoter, luciferase coding sequence and the BTU2 polyadenlyation site, is referred to throughout the text as RdLuc (Figure 1A). Similar methodologies were used to construct other reporter genes targeted for btu1-1 disruption, including luciferase expressed from the BTU1 promoter (BtLuc; Figure 1A), and GFP expressed from the RAD51 promoter (RdGFP; Figure 8A).
Figure 1.
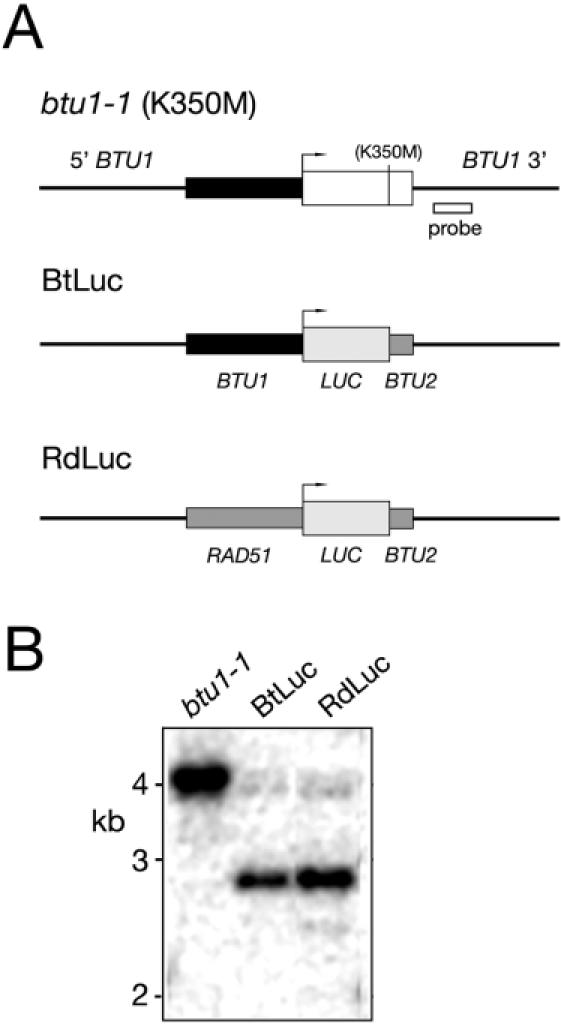
Reporter gene constructs. (A) Schematic representation of the btu1-1 mutant allele that expresses β-tubulin (K350M) (white box), which is responsible for hypersensitivity of strain CU522 to the microtubule-stabilizing drug paclitaxel. Targeted disruption of the btu1-1 allele with BtLuc [a reporter construct consisting of luciferase (light grey box) transcribed from the BTU1 transcriptional promoter (black box), with a polyadenylation site from the BTU2 gene (short dark grey box)] or RdLuc [luciferase expressed from the RAD51 transcriptional promoter (long dark grey box)] is facilitated by the 5′ and 3′ BTU1 flanking sequence (thin black lines). The location of the probe sequence used for Southern-blot analysis is also indicated. Not drawn to scale. (B) Southern-blot analysis of total DNA from paclitaxel resistant transformants. A restriction digest polymorphism (BglII and SphI digest) makes it possible to distinguish between intact and disrupted btu1-1 alleles (4.0 and 2.8 kb, respectively).
Figure 8.
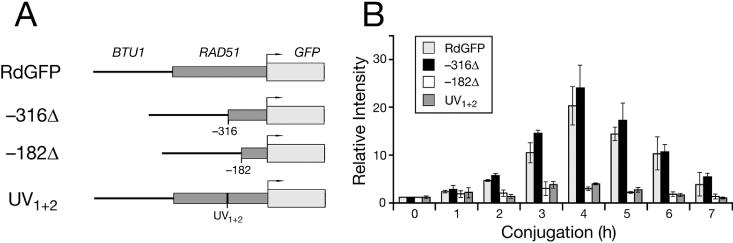
UV1 and UV2 repeats in the RAD51 promoter are responsible for proper expression during conjugation. (A) Reporter constructs in transformants expressing GFP from the full-length, wild-type promoter (RdGFP), two promoter truncations [RdGFP (−316Δ) and RdGFP(−182Δ)], and the full-length promoter with mutated UV1 and UV2 repeats [RdGFP(UV1+2)] are shown schematically as in Figures 1 and 4 for the RdLuc reporter. (B) GFP mRNA was monitored during the first 7 h of conjugation between transformant clonal lines. Each conjugation was repeated in triplicate. A northern blot of total RNA (10 μg per lane), isolated from each time point and hybridized with a GFP-specific radiolabeled probe, was quantified by PhosphorImager analysis. GFP mRNA levels are indicated by light grey bars for RdGFP, black bars for RdGFP(−316Δ), white bars for RdGFP(−182Δ) and dark grey bars for RdGFP(UV1+2). Error bars represent the standard deviation.
Modification of the RAD51-luciferase reporter constructs
Manipulation of the RAD51 promoter, including various mutations, truncations, deletions and inversions, were accomplished by introducing unique restriction sites by the site-directed mutagenesis method of Kunkel (22) and/or PCR amplification of selected regions of the promoter. For example, the introduction of unique BglII sites within the promoter, followed by a double digest with BglII and BamHI and ligation of the compatible termini, resulted in a variety of reporter constructs with truncated promoters. Insertions and internal deletions were constructed using similar techniques. The sequence of constructs was confirmed by restriction digest analysis and DNA sequencing.
Biolistic transformation of Tetrahymena
Tetrahymena strain CU522 (also CU725 and CU727 for conjugation experiments) was grown in 100 ml cultures (2% PPYS) to a density between 1.0 and 3.0 × 105 cells/ml, and starved in 100 ml of 10 mM Tris–HCl (pH 7.5) for 14–21 h. The cells were pelleted by centrifugation and resuspended in 1–3 ml of 10 mM HEPES (pH 7.5) to a density of ∼1 × 107 cells/ml. The concentrated cells were then transferred onto a Petri dish (100 mm diameter) that contained a sterile Whatman 114 filter paper that was presoaked using 2 ml of 10 mM HEPES (pH 7.5). The Au particles (1.0 μm diameter), coated with ∼1.0 μg of various reporter constructs digested previously with restriction enzymes KpnI and SacI, were introduced to the cell biolistically with the BioRad Gene Gun™, using 900 psi and a vacuum of 27 mm Hg as per the manufacturer's instructions. After biolistic transformation, the cells on the filter paper were incubated at 30°C for 2 h in 50 ml of pre-warmed 2% PPYS and then distributed to 96-well plates in 100 μl aliquots. Selection of transformants resistant to 20 μM paclitaxel at 30°C was monitored for 4–7 days. Southern-blot analysis of total DNA from resistant clonal cell lines confirmed the presence or absence of the reporter constructs in the btu1-1 locus. Positive clonal lines were grown at low cell densities in 2% PPYS under continuous selective pressure (20–40 μM paclitaxel) until 100% phenotypic assortment to the reporter construct at the targeted btu1-1 locus had been achieved.
UV irradiation
Tetrahymena transformants and wild-type cells were irradiated with UV by the method described previously (4). Cells were allowed to recover at 30°C for 2 h before the preparation of cell extracts for luciferase activity assays.
Tetrahymena cell extract preparation for luciferase activity assays
The transformants were grown in 2% PPYS to densities of 1–3 × 105 cells/ml and pelleted by centrifugation (4 min at 1100 g). The cells were lysed in 300–800 μl of 1× CCLR (Cell Culture Lysis Reagent; Promega), and lysates clarified by centrifugation at 16 250 g. The protein concentration of cell extracts was determined using BioRad Bradford Reagent (BioRad, Hercules, CA). The extracts were normalized for total protein (1.0 mg/ml), and the luciferase activity assay was performed using the procedure outlined in the Luciferase Assay Kit (Promega). A Turner Designs Luminometer (Model TD-20/20) was used to measure the luciferase activity. The sensitivity and Relative Light Units (RLUs) were normalized using luciferase levels present in BtLuc transformants, which constitutively express high levels of luciferase from the BTU1 promoter (Figure 1A).
RESULTS
Reporter gene expression in Tetrahymena
The mutant β-tubulin allele btu1-1 (K350M) confers hypersensitivity to the microtubule-stabilizing drug paclitaxel (23). Targeted disruption of this non-essential gene (a second β-tubulin gene, BTU2, is expressed in Tetrahymena) confers a selective advantage to transformants (paclitaxel resistance), making it possible to introduce and study foreign gene expression in T.thermophila (21). We have used this methodology to create stably transformed clonal lines that express foreign genes from either the BTU1 or the RAD51 transcriptional promoters. Chimeric reporter genes were constructed with either the BTU1 or the RAD51 5′ NTS positioned upstream of either the firefly Photinus pyralis luciferase coding sequence (Promega) or the GFP coding sequence (D. L. Chalker, Washington University, St Louis, MO). These reporter genes were subsequently cloned in a vector suitable for targeted disruption of the btu1-1 (K350M) locus (Figure 1A). Following biolistic transformation of strain CU522, paclitaxel-resistant clonal lines were maintained under continuous selective pressure for 20–30 fissions to ensure 100% phenotypic assortment to the reporter allele (24,25), which was subsequently confirmed by Southern-blot analysis of the total DNA from transformants (Figure 1B; data not shown).
Whole cell lysates from clonal lines transformed with the luciferase reporter constructs were routinely assayed for luciferase activity, making it possible to quantify its expression from a variety of different promoter constructs, and from cells subjected to a variety of environmental conditions. Luciferase expression from the BTU1-luciferase reporter (BtLuc) was ∼100 times greater than that from the RAD51-luciferase construct (data not shown). All subsequent luciferase activity assays were normalized with respect to the constitutively high levels of luciferase detected in lysates from BtLuc transformants.
Induction of the RAD51-luciferase reporter by DNA damage
Transformants expressing the RAD51-luciferase reporter (RdLuc, Figure 1A) were irradiated with UV(c) (240 J/m2) and assayed for luciferase activity over the next 6 h. An increase in luciferase was detected ∼1 h after irradiation, with peak levels attained within 2 h (Figure 2). A similar pattern of induction was observed for transformants expressing the RAD51-GFP reporter (RdGFP, data not shown). The induction kinetics for the reporter genes reiterates those previously seen for Rad51 mRNA following UV irradiation (4).
Figure 2.
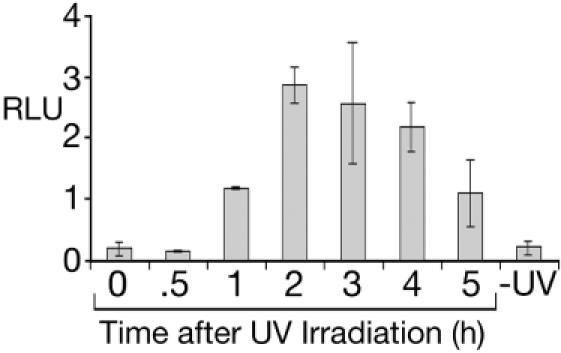
Luciferase expression from the RAD51 promoter following UV irradiation of transformants. RdLuc transformants were irradiated with UV as described in the Materials and Methods, and allowed to recover for the time intervals indicated prior to the preparation of whole cell lysates. The cell extracts (20 μg total protein) were assayed for luciferase activity in triplicate and normalized to the luciferase activity from BtLuc transformants, which constitutively express luciferase independent of UV irradiation (data not shown). The relative light units (RLUs) for non-irradiated RdLuc transformants are also shown. The bars represent the standard deviation for luciferase assays from three independently isolated RdLuc transformants.
In order to determine whether the de novo protein synthesis is required for the increased transcription of RAD51 in response to DNA damage, transformants were UV-irradiated in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX). The northern-blot analysis indicates that comparable levels of both Rad51 and luciferase mRNA from RdLuc transformants are attained with or without CHX, although maximal levels are delayed in the presence of the drug (Figure 3). In contrast, ATU1 (α-tubulin) mRNA transcription decreases over time in the presence of CHX. The increase in Rad51 expression in response to UV irradiation is at the level of transcription and not due to changes in mRNA stability, since both luciferase mRNA and the endogenous Rad51 mRNA follow the same kinetics in this experiment (Figure 3).
Figure 3.
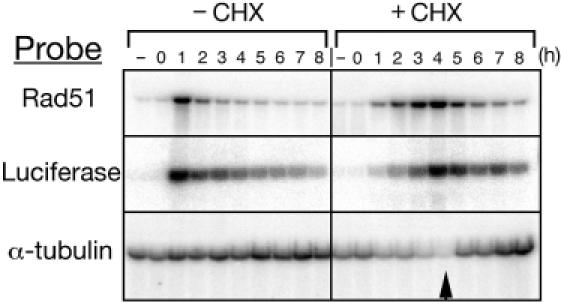
Transcription from the RAD51 promoter in response to DNA damage is independent of de novo protein synthesis. RdLuc transformants were UV irradiated as described in the Materials and Methods in the presence (+CHX) or absence (−CHX) of the protein synthesis inhibitor CHX. Total RNA was prepared from the cells at the times indicated following irradiation, as well as from non-irradiated cells (−). CHX-treated cultures were washed extensively after 4 h and transferred to fresh media without CHX for an additional 4 h (arrow). Equal amounts (10 μg) of total RNA from each sample were separated in three identical electrophoretic gels. The RNA was transferred onto Nytran membranes, which were subsequently hybridized with probes specific for Rad51, luciferase and α-tubulin coding sequences.
Identification of RAD51 promoter elements
A series of truncations of the 1.3 kb 5′ NTS in the RdLuc reporter construct have revealed three separable cis-sequence elements within the RAD51 transcriptional promoter. The results from these experiments are summarized in Figures 4 and 5. Truncation of the Rad51 5′ NTS to nucleotide positions −7 or −82 (relative to the translational start codon) eliminates all measurable luciferase from transformant lysates, with or without UV irradiation. In contrast, luciferase is still expressed in transformants with a reporter that includes nucleotides from −182 to −1 of the 5′ NTS [RdLuc(−182Δ)], although expression from this construct is unaffected by exposure to UV(c). Only when the proximal 316 bp (or more) of the Rad51 5′ NTS is retained in reporter constructs do luciferase levels increase in response to UV(c) irradiation [RdLuc(−316Δ); Figure 4B]. A similar response was observed during deletion analysis of the RdGFP reporter (data not shown). Taken together, these data indicate that at least two cis-sequence elements within the Rad51 5′ NTS are required for (i) basal transcription (located between nucleotides −182 and −82) and (ii) inducible expression in response to DNA damage (located between nucleotides −316 and −182).
Figure 4.
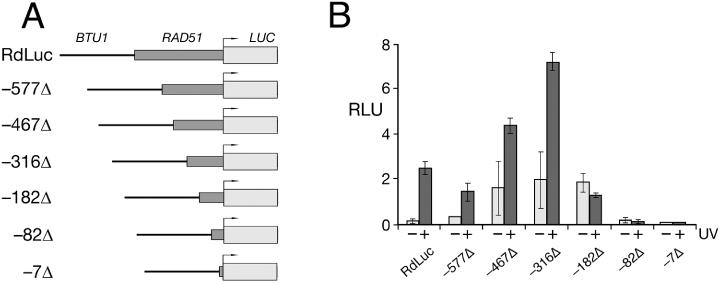
Luciferase expression from the truncated RAD51 promoter. (A) The full-length RAD51 promoter (1.3 kb) in the RdLuc construct as well as six successively larger truncations are shown schematically. The length of each truncated promoter (from its 5′ terminus to the luciferase initiator codon) is indicated. (B) Luciferase activity from cells transformed with the various truncated RdLuc reporter constructs, with (+) and without (−) UV irradiation. Whole cell extracts were prepared 2 h following irradiation and assayed (20 μg total protein) in triplicate for luciferase activity. Whole cell extracts from transformants that constitutively express luciferase (BtLuc) were used to establish the RLUs. Error bars represent the standard deviation.
Figure 5.
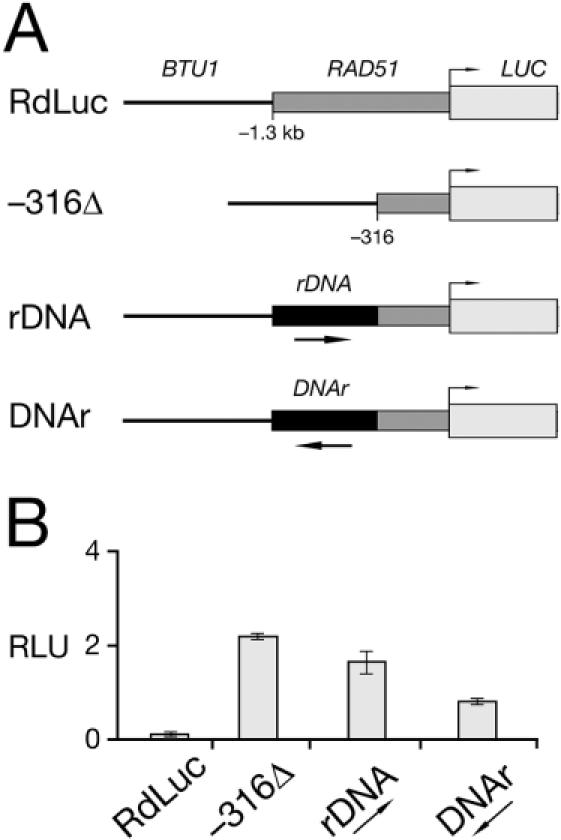
An upstream cis element contributes to low steady-state levels of expression from the RAD51 promoter. (A) Schematic diagrams of the full-length RdLuc reporter construct, the −316 truncation [RdLuc(−316Δ)], and the substitution of 1.0 kb of the Rad51 promoter (−1.3 kb to −0.3 kb) with 0.9 kb of Tetrahymena rDNA 3′ NTS (accession no. X54512; 9363–10262 nt) in both sense [RdLuc(rDNA)] and anti-sense [RdLuc(DNAr)] orientations. Not drawn to scale. (B) Basal luciferase activities of lysates from transformants expressing the four reporter constructs (Figure 5A) was as described for non-irradiated cells in Figure 4. Error bars represent the standard deviation. See text for discussion.
Steady-state luciferase activity was relatively low for transformants expressing the full-length RdLuc reporter without UV irradiation. This basal activity increased significantly for the various truncation constructs, with the increase somewhat proportional to the length of the 5′ NTS truncation, being the most pronounced for the −467Δ, −316Δ and −186Δ reporter constructs (Figure 4B). It was unclear whether the basal level increase was due to the loss of a putative control element between −1.3 kb and −316 of the Rad51 5′ NTS or the proximity of the BTU1 5′ targeting sequence in these constructs. In order to distinguish between these two possibilities, reporter constructs were tested that included 0.9 kb of a non-specific sequence derived from the rDNA 3′ non-transcribed sequence (26), which was inserted between the BTU1 5′ targeting sequence and −316 of the Rad51 5′ NTS (Figure 5A). Insertion of the non-specific ‘spacer’ rDNA in either orientation reduced the basal luciferase levels, but not to those observed for full-length RdLuc transformants (Figure 5B). These data reveal the presence of a cis-element within the Rad51 5′ NTS that contributes to the maintenance of low, steady-state levels of Rad51 observed in the absence of DNA damage.
Characterization of the cis-element required for RAD51 induction
Luciferase induction by UV(c) from variously truncated reporter constructs is dependent on a relatively short cis-element within the Rad51 5′ NTS (Figure 4). A more complete investigation of this putative DNA damage response element (DRE) is outlined below. Both inversion and deletion of the 134 bp segment between nt −316 and −182 completely eliminates any increase in the luciferase expression in response to UV(c) irradiation (Figure 6). Although luciferase is induced when full-length RdLuc transformants were treated with other DNA damaging agents, including UV(a), ionizing gamma irradiation (137Cs source, 4 Gy) and methylmethane sulfonate, there is no induction in RdLuc(DEL) transformants (data not shown).
Figure 6.
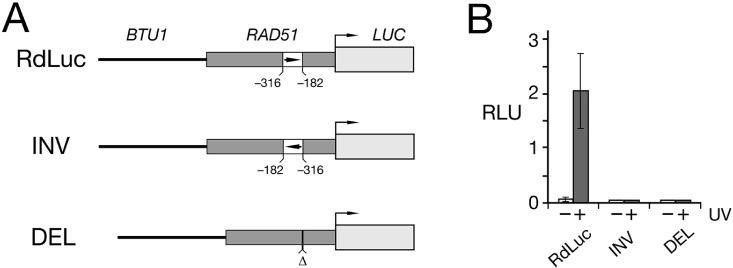
The induction region of the RAD51 promoter. (A) A schematic diagram of luciferase reporter constructs expressed from the RAD51 promoter with the 134 bp from positions −316 to −182 either inverted [RdLuc(INV)] or deleted [RdLuc(DEL)]. The reporter construct with the full length, wild-type RAD51 promoter is also shown (RdLuc). Not drawn to scale. (B) Luciferase activity from trasformants expressing the three reporter constructs in Figure 6A, with (+) and without (−) UV irradiation. Luciferase assays of whole cell lysates were as described for Figure 4.
A search of the GenBank database failed to reveal any significant similarity between this sequence and any other known transcriptional promoter elements (data not shown). However, close examination of the 134 bp sequence revealed a 7 bp repeat (TTTCAAT) separated by a 14 bp. The direct repeat, identified as UV1 and UV2 (Figure 7A), is not found in either orientation elsewhere within the 1.3 kb Rad51 5′ NTS.
Figure 7.
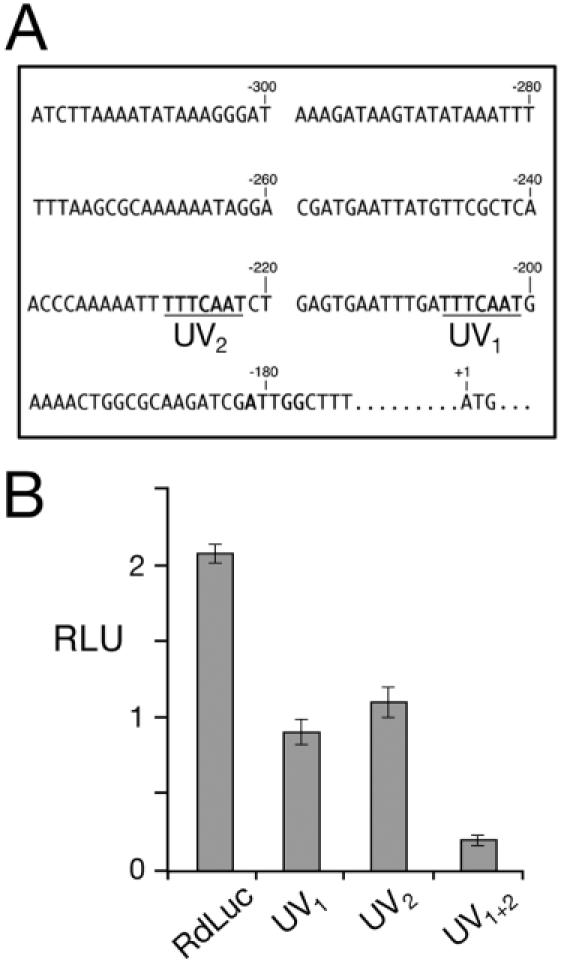
Analysis of a 7 bp tandem repeat within the RAD51 promoter induction region. (A) Sequence of the RAD51 promoter between nucleotide positions −320 and −180 is shown. Two 7 bp repeats (TTTCAAT) unique to the promoter region are underlined (UV1 and UV2). (B) Transformants expressing RdLuc reporter constructs that include UV1 and UV2 mutated singly [RdLuc(UV1), RdLuc(UV2)], and together [RdLuc(UV1+2)] were assayed for luciferase activity 2 h after UV irradiation. Extracts from BtLuc transformants that constitutively express luciferase were used to establish the RLUs. Error bars represent the standard deviation.
The possibility that the UV1 and/or UV2 sequences are involved in Rad51 inducible expression was tested in a series of luciferase reporter constructs. Mutation of 5 of the 7 bp for each TTTCAAT repeat (UV1 changed to TCCTAGG and UV2 changed to GCTAGCT) had nearly identical effects, reducing luciferase induction in transformants by ∼50% (Figure 7B). The effect of mutating both repeats in the same construct was additive, with the response to UV(c) reduced to ∼10% that of the unaltered RdLuc construct. In contrast, the response to UV irradiation of reporter constructs with nucleotide substitutions at two randomly chosen sites relatively close to the UV1/UV2 repeats (‘ATTGG’ at positions −177 to −181 and ‘AATATC’ at positions −321 to −316) were indistinguishable from the full-length RdLuc reporter (data not shown).
The control of RAD51 expression during meiosis
Maximal Rad51 expression in mating Tetrahymena is coincident with meiotic prophase, which normally occurs ∼4 h after conjugation has been initiated (5). In order to determine whether the Rad51 expression is under transcriptional control during meiosis, the kinetics of RdGFP expression during conjugation of transformants was investigated (Figure 8). Total RNA from cell lines expressing various RdGFP reporter constructs were monitored throughout the early stages of conjugation by northern-blot analysis. As an internal control, Rad51 mRNA levels (expressed from the endogenous RAD51 locus) were also monitored in duplicate northern blots (data not shown). The results from these experiments are summarized below.
The GFP mRNA from transformants expressing the full-length RdGFP reporter, as well as from the −316 truncated version [RdGFP(−316Δ)], exhibited the same kinetics during conjugation as that of Rad51 mRNA expressed from the endogenous gene (5). In contrast, there was no marked increase in GFP mRNA during conjugation when expressed from RdGFP(−182Δ). Furthermore, mutation of the TTTCAAT direct repeat (UV1 and UV2; Figure 7) eliminated the dramatic increase in GFP mRNA levels during meiotic prophase. The expression pattern for these transformants closely resembled that from cells expressing RdGFP(−182Δ), as opposed to the pattern seen for RdGFP(−316Δ) transformants at the same stage of conjugation (Figure 8).
DISCUSSION
Tetrahymena transcriptional promoters
Although nuclear run-on experiments have shown that the vast majority of Tetrahymena genes are under transcriptional control (16), the mechanism(s) that mediate(s) this control are poorly understood. It is generally assumed that the cis-sequence elements and trans-acting factors involved in this process for ciliates are similar to those described for other eukaryotes (27–30). One of the difficulties encountered in identifying Tetrahymena cis-sequence elements by homology to well-characterized promoters from other model organisms is the 75% A + T content for the Tetrahymena genome (31). As yet, no conclusive ‘TATA’ or ‘GC’ boxes have been identified for Tetrahymena genes, although a handful of cis-elements have been assigned that status. Characterization of the well-conserved histone genes has revealed a loose ‘CCAAT’ box consensus in the 5′ non-transcribed sequence for Tetrahymena species (32–34). The T.thermophila telomerase RNA gene (TER1), a RNA polymerase III transcript (35), was found to contain a proximal sequence element at −55 (relative to the transcriptional start site) and an important A/T-rich element at −25 that are essential for the expression of a ‘reporter’ telomerase RNA (36).
In previous studies, the detection of foreign reporter gene expression in Tetrahymena has been limited to northern-blot analysis (36,37) and fluorescence microscopy (38). We have exploited the powerful technique of stable transformation by targeted disruption of the non-essential Tetrahymena btu1-1 gene (23) to introduce a variety of luciferase reporter constructs. Our ability to quantify luciferase activity levels from transformant whole cell lysates has made it possible to characterize the Tetrahymena RAD51 transcriptional promoter.
Once transformant cell lines under the selective pressure of 20–40 μM paclitaxel have undergone 100% phenotypic assortment, transgenes are present at approximately the same copy number as endogenous macronuclear genes (Figure 1). The integrated reporters are positioned within macronuclear chromatin, placing it in the proper context for normal transcriptional control. This is borne out by the similar kinetics of endogenous Rad51 mRNA with those of luciferase or GFP mRNA from transformants in response to DNA damage (Figure 2) and conjugation (Figure 8). The Tetrahymena RAD51 expression patterns (5) and transcriptional regulation appear to be similar to that from other eukaryotes. For example, the induction of RAD51 expression in response to DNA damage, despite the presence of protein synthesis inhibitors (Figure 3), is similar to that seen for RAD51 homologs in other eukaryotes (8,10,39).
It is unclear why there is a delay in peak levels of Rad51 mRNA when induced by UV in the presence of CHX. A possible explanation for this phenomenon is that the detection of DNA damage, and the subsequent induction of RAD51 expression, may be coincident with a specific stage of the cell cycle. It is conceivable that a delay in the majority of cells reaching this particular stage in the cell cycle by treatment with CHX results in the observed delay (Figure 3). The UV induction of Rad51 in a culture treated with or without CHX that has been synchronized by starvation may show the same kinetics of Rad51 mRNA accumulation.
RAD51 transcriptional promoter cis-elements
Sequence analysis of transcriptional promoters for 10 DNA repair genes (including RAD51) from Saccharomyces cerevisiae revealed a high occurrence of MCB, HAP and UASH regulatory boxes, all of which are speculated to be involved in transcriptional regulation following irradiation (12). Increased expression of the human paralog RAD51B after exposure to DNA damaging agents is assumed to be mediated by ‘consensus’ promoter binding sites for both the AP2 and p53 proteins (40). However, due to the rapid divergence of sequences upstream of the relatively well-conserved Rad51p coding sequences, the identification of promoter elements by comparative analyses of RAD51 homologs from yeast to ciliates to humans has not been possible.
We have identified three separate cis-sequence elements within the RAD51 transcriptional promoter, not by comparative analysis, but by expression of various reporter constructs in vivo. The first element, positioned between −182 and −82 bp (relative to the translational start site), ensures basal levels of expression, since truncation of the RAD51 promoter to −82 in RdLuc(−82Δ) transformants eliminates significant expression of the luciferase reporter (Figure 4). As the RAD51 5′ terminus has been mapped to position −100 (4), this basal transcriptional promoter element is most likely contained within the 62 bp between −182 and −120.
A second sequence element, located within the 1000 bp between −1.3 kb and −316 bp, specifically limits Rad51 expression to relatively low levels in the absence of DNA damage or developmental signals. Progressively, larger truncations of this sequence from the 5′ terminus of reporter constructs results in significantly higher basal luciferase activity, with the most pronounced increase detected at the transition between −577 and −467 (Figure 5B). These effects are not due simply to changes in the proximity of BTU1 5′ targeting sequences in the truncation constructs, since insertion of a non-specific rDNA 3′ NTS ‘spacer’ in either orientation does not restore luciferase activity to the low level seen in full-length RdLuc transformants (Figure 5). In lieu of a more extensive deletion analysis, this regulatory element can be broadly defined as contained within the 200 bp between −600 and −400 bp.
A third promoter element (located between −316 and −182) is required for the induction of RAD51 expression in response to both DNA damage and meiosis (Figures 7 and 8). Within this inducible element are 28 bp (from −228 to −201) that include 7 bp direct repeats, TTTCAAT, which are separated by 14 bp. Mutagenesis of the UV1 + UV2 repeats drastically reduces the induction of the luciferase reporter to ∼10% of wild type (Figure 7). A very similar DRE has been mapped within the promoter for RHP51, the Schizzosaccharomyces pombe RAD51 homolog (9). Although DRE1 and DRE2 are not direct repeats, they are similar in length (11 bp), distance from each other (10 bp) and overall position (from −233 to −204) to those of UV1 and UV2 in Tetrahymena. The EMSA experiments revealed specific binding of two S.pombe proteins (59 and 45 kDa) to DRE1 and DRE2, although an increase in the presumed ‘activator’ proteins was not detected following DNA damage (9). A similar response is likely to exist for UV1 and UV2 in Tetrahymena cells that have incurred DNA damage or that have initiated meiosis.
In summary, Tetrahymena RAD51 expression is controlled at the level of transcription, with levels varying in response to DNA damage and conjugation. The stable transformation of Tetrahymena with reporter constructs has made it possible to define three RAD51 promoter elements, one of which is critical to increased expression in response to these conditions. The identification of this DRE will make it possible to more completely characterize trans-acting factors involved in the transcriptional induction of RAD51, and possibly that of other genes, expressed during the early stages of conjugation in Tetrahymena.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by NSF grant MCB 0220085 (D.P.R.) and NSF RU1 grant MCB 0131175 (E.S.C.).
REFERENCES
- 1.Gasior S.L., Olivares,H., Ear,U., Hari,D.M., Weichselbaum,R. and Bishop,D.K. (2001) Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl Acad. Sci. USA, 98, 8411–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West S.C. (2003) Molecular views of recombination proteins and their control. Nature Rev. Mol. Cell Biol., 4, 435–445. [DOI] [PubMed] [Google Scholar]
- 3.Sung P., Krejci,L., Van Komen,S. and Sehorn,M.G. (2003) Rad51 recombinase and recombination mediators. J. Biol. Chem., 278, 42729–42732. [DOI] [PubMed] [Google Scholar]
- 4.Campbell C. and Romero,D.P. (1998) Identification and characterization of the RAD51 gene from the ciliate Tetrahymena thermophila. Nucleic Acids Res., 26, 3165–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsh T.C., Cole,E.S., Stuart,K.R., Campbell,C. and Romero,D.P. (2000) RAD51 is required for propagation of the germinal nucleus in Tetrahymena thermophila. Genetics, 154, 1587–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) DMC1: a meiosis-specific yeast homolog of E.coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- 7.Habu T., Taki,T., West,A., Nishimune,Y. and Morita,T. (1996) The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res., 24, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flygare J., Benson,F. and Hellgren,D. (1996) Expression of the human RAD51 gene during the cell cycle in primary human peripheral blood lymphocytes. Biochim. Biophys. Acta, 1312, 231–236. [DOI] [PubMed] [Google Scholar]
- 9.Jang Y.K., Jin,Y.H., Shim,Y.S., Kim,M.J., Yoo,E.J., Choi,I.S., Lee,J.S., Seong,R.H., Hong,S.H. and Park,S.D. (1996) Identification of the DNA damage-responsive elements of the rhp51+ gene, a recA and RAD51 homolog from the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 251, 167–175. [DOI] [PubMed] [Google Scholar]
- 10.Jang Y.K., Jin,Y.H., Myung,K., Seong,R.H., Hong,S.H. and Park,S.D. (1996) Differential expression of the rhp51+ gene, a recA and RAD51 homolog from the fission yeast Schizosaccharomyces pombe. Gene, 169, 125–130. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto A., Taki,T., Yagi,H., Habu,T., Yoshida,K., Yoshimura,Y., Yamamoto,K., Matsushiro,A., Nishimune,Y. and Morita,T. (1996) Cell cycle-dependent expression of the mouse Rad51 gene in proliferating cells. Mol. Gen. Genet., 251, 1–12. [DOI] [PubMed] [Google Scholar]
- 12.Mercier G., Denis,Y., Marcb,P., Picard,L. and Dutreix,M. (2001) Transcriptional induction of repair genes during slowing of replication in irradiated Saccharomyces cerevisiae. Mutat. Res., 487, 157–172. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y., Nakada,S., Ishiko,T., Utsugisawa,T., Datta,R., Kharbanda,S., Yoshida,K., Talanian,R.V., Weichselbaum,R., Kufe,D. et al. (1999) Role for caspase-mediated cleavage of Rad51 in induction of apoptosis by DNA damage. Mol. Cell. Biol., 19, 2986–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Z.M., Huang,Y., Ishiko,T., Kharbanda,S., Weichselbaum,R. and Kufe,D. (1997) Regulation of DNA damage-induced apoptosis by the c-Abl tyrosine kinase. Proc. Natl Acad. Sci. USA, 94, 1437–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan Z.M., Huang,Y., Ishiko,T., Nakada,S., Utsugisawa,T., Kharbanda,S., Wang,R., Sung,P., Shinohara,A., Weichselbaum,R. et al. (1998) Regulation of Rad51 function by c-Abl in response to DNA damage. J. Biol. Chem., 273, 3799–3802. [DOI] [PubMed] [Google Scholar]
- 16.Stargell L.A., Karrer,K.M. and Gorovsky,M.A. (1990) Transcriptional regulation of gene expression in Tetrahymena thermophila. Nucleic Acids Res., 18, 6637–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love H.D., Allen-Nash,A., Zhao,Q.A. and Bannon,G.A. (1988) mRNA stability plays a major role in regulating the temperature-specific expression of a Tetrahymena thermophila surface protein. Mol. Cell. Biol., 8, 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G.L. and Blackburn,E.H. (1990) Amplification of tandemly repeated origin control sequences confers a replication advantage on rDNA replicons in Tetrahymena thermophila. Mol. Cell. Biol., 10, 2070–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 20.McCormick-Graham M. and Romero,D.P. (1996) A single telomerase RNA is sufficient for the synthesis of variable telomeric DNA repeats in ciliates of the Genus Paramecium. Mol. Cell. Biol., 16, 1871–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaertig J., Gao,Y., Tishgarten,T., Clark,T.G. and Dickerson,H.W. (1999) Surface display of a parasite antigen in the ciliate Tetrahymena thermophila. Nat. Biotechnol., 17, 462–465. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel T.A., Roberts,J.D. and Zakour,R.A. (1987) Rapid and efficient site-directed mutagenesis without phenotypic selection. Methods Enzymol., 155, 166–178. [DOI] [PubMed] [Google Scholar]
- 23.Gaertig J., Thatcher,T.H., Gu,L. and Gorovsky,M.A. (1994) Electroporation-mediated replacement of a positively and negatively selectable β-tubulin gene in Tetrahymena thermophila. Proc. Natl Acad. Sci. USA, 91, 4549–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doerder F.P., Deak,J.C. and Lief,J.H. (1992) Rate of phenotypic assortment in Tetrahymena thermophila. Dev. Genet., 13, 126–132. [DOI] [PubMed] [Google Scholar]
- 25.Merriam E.V. and Bruns,P.J. (1988) Phenotypic assortment in Tetrahymena thermophila: assortment kinetics of antibiotic-resistance markers, tsA, death, and the highly amplified rDNA locus. Genetics, 120, 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu G.L., Hasson,M. and Blackburn,E.H. (1988) Circular ribosomal DNA plasmids transform Tetrahymena thermophila by homologous recombination with endogenous macronuclear ribosomal DNA. Proc. Natl Acad. Sci. USA, 85, 5151–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeder R.H. (1999) Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acid Res. Mol. Biol., 62, 293–327. [DOI] [PubMed] [Google Scholar]
- 28.Chedin S., Ferri,M.L., Peyroche,G., Andrau,J.C., Jourdain,S., Lefebvre,O., Werner,M., Carles,C. and Sentenac,A. (1998) The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harb. Symp. Quant. Biol., 63, 381–389. [DOI] [PubMed] [Google Scholar]
- 29.Kornberg R.D. (1998) Mechanism and regulation of yeast RNA polymerase II transcription. Cold Spring Harb. Symp. Quant. Biol., 63, 229–232. [DOI] [PubMed] [Google Scholar]
- 30.Struhl K. (1993) Yeast transcription factors. Curr. Opin. Cell Biol., 5, 513–520. [DOI] [PubMed] [Google Scholar]
- 31.Bannon G.A., Bowen,J.K., Yao,M.C. and Gorovsky,M.A. (1984) Tetrahymena H4 genes: structure, evolution and organization in macro- and micronuclei. Nucleic Acids Res., 12, 1961–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen L.K. and Kristiansen,K. (1995) Transcription in vitro of Tetrahymena class II and class III genes. J. Biol. Chem., 270, 7601–7608. [DOI] [PubMed] [Google Scholar]
- 33.Brunk C.F. and Sadler,L.A. (1990) Characterization of the promoter region of Tetrahymena genes. Nucleic Acids Res., 18, 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiuwen L. and Gorovsky,M.A. (1993) Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNA using RNA ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res., 21, 4954–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu G.L., Bradley,J.D., Attardi,L.D. and Blackburn,E.H. (1990) In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature, 344, 126–132. [DOI] [PubMed] [Google Scholar]
- 36.Hargrove B.W., Bhattacharyya,A., Domitrovich,A.M., Kapler,G.M., Kirk,K., Shippen,D.E. and Kunkel,G.R. (1999) Identification of an essential proximal sequence element in the promoter of the telomerase RNA gene of Tetrahymena thermophila. Nucleic Acids Res., 27, 4269–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang Y., Song,X., Bowen,J., Corstanje,R., Gao,Y., Gaertig,J. and Gorovsky,M.A. (2002) A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl Acad. Sci. USA, 99, 3734–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haddad A. and Turkewitz,A.P. (1997) Analysis of exocytosis mutants indicates close coupling between regulated secretion and transcription activation in Tetrahymena. Proc. Natl Acad. Sci. USA, 94, 10675–10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan R., Fan,S., Wang,J.A., Meng,Q., Ma,Y., Schreiber,D., Goldberg,I.D. and Rosen,E.M. (1999) Coordinate alterations in the expression of BRCA1, BRCA2, p300, and RAD51 in response to genotoxic and other stresses in human prostate cancer cells. Prostate, 40, 37–49. [DOI] [PubMed] [Google Scholar]
- 40.Peng L., Rice,M. and Kmiec,E. (1998) Analysis of the human RAD51L1 promoter region and its activation by UV light. Genomics, 54, 529–541. [DOI] [PubMed] [Google Scholar]


