Abstract
Phenotype-based functional genomic methods are useful for the identification of genes that are related to a particular biological function or disease. Essential to this approach is the ability to regulate the expression of selected genes. Artificial transcription factors (ATFs) are key molecular tools that selectively regulate gene expression in vivo. Here, we use an ATF library to identify genes that participate in rendering a cell resistant to the drug Taxol, a potent anti-cancer drug that binds to tubulin and inhibits cell division. The library, which encodes ATFs that activate (rather than inhibit) transcription, was introduced into a HeLa cell line, and Taxol-resistant cells were selected. After eight rounds of selection, we identified two ATFs that significantly increased the level of Taxol resistance (TR) in HeLa cells. Gene expression microarray experiments using these ATFs identified 37 co-regulated genes, including genes already known to participate in TR. This study demonstrates that ATF libraries can be used to induce phenotypic alterations in eukaryotic cells and then identify specific genes that are associated with the phenotype of choice.
INTRODUCTION
Now that the human genome and those of many other organisms have been sequenced, the next goal is to annotate the products of the thousands, to tens of thousands of newly identified genes. Various phenotype-based functional genomic approaches have been developed and are currently being used to identify the functions of previously uncharacterized genes (1–6). Such methods employ libraries that encode molecular tools that alter gene expression, which can give rise to phenotypic changes. After transfection into cells, the components of the libraries are expressed randomly in individual cells, and each perturbs the expression of one or a small subset of genes. One then selects for a phenotype of interest, such as reporter gene expression (7), cellular morphology (8), cell invasion (9) or a change in cellular growth rate (1). Finally, the identity of the molecular tool that induced the phenotype of interest is revealed, and this information is used to determine the genes that are responsible for the selected phenotype.
Various kinds of molecular libraries have been used to screen for phenotypes of interest in mammalian cells. Examples include libraries of small molecules (4), cDNAs (5), antisense RNAs (3), ribozymes (2,10), small inhibitory RNAs (siRNAs) (1), peptides (6) and artificial transcription factors (ATFs) (8,11). Although most of these molecular libraries contain or encode molecules that either up- or down-regulate the activity of target gene products, ATFs can both activate and inhibit target gene expression, which generates a broader range of phenotypic patterns.
Zinc-finger-containing proteins represent a large and diverse class of transcriptional regulatory proteins. One common type of zinc finger is the Cys2His2 finger. Each zinc finger consists of ∼30 amino acids, which usually binds to 3 bp of DNA in a sequence-specific manner. Because of the modular nature of zinc fingers, ATFs can be constructed by fusing three or four zinc fingers together to form multi-finger proteins that recognize specific 9 or 12 bp DNA sequences, respectively (12–14). The fused zinc finger domains (ZFDs) can serve as gene-specific transcriptional repressors on their own, or can be joined with a transcriptional activation or repression domain to form a gene-specific transcriptional activator or repressor protein (ATFs). These ATFs enable researchers to modulate, at will, the transcription of a gene of interest (15–19). Several studies have shown that the artificial zinc-finger-containing transcription factors can regulate the expression of a variety of genes, such as VEGF (15,16) and PPARγ (17), in mammalian cells and other selected genes in plant (18) and bacterial (19) cells. Recently, we and others have shown that transfection of cells with randomized ATF libraries is an effective method for inducing phenotypes of interest, such as resistance to heat shock, osmotic shock or a toxic chemical in yeast, and cell differentiation or proliferation in mammalian cells (8,11). After screening for cells that display the phenotype of interest, the specific ATF that induced the phenotype can be determined. Then, if the genome of the organism of interest has been sequenced, the selected ATFs can be used to identify the genes that give rise to a specific phenotype (8).
Taxol, a brand name of paclitaxel, is one of the most prescribed chemotherapeutic agents (20). Although it is widely used to treat breast, non-small cell lung and ovarian cancers, the development of drug resistance is a major problem in Taxol chemotherapy. Over-expression or mutation of β-tubulin (21) and over-expression of P-glycoprotein [P-gp, ABCB1 (22)], which is encoded by the multiple-drug-resistance 1 (MDR1) gene, have been linked to the acquisition of Taxol resistance (TR). Lamendola et al. (23) identified genes associated with TR by comparing the genome-wide gene expression profiles of Taxol-resistant and control cell lines.
Another recently described approach used a peptide library-based functional genomic screen to generate Taxol-resistant cells. They then carried out a yeast two-hybrid screen to identify protein targets whose inhibition leads to TR (6). The drawback of the peptide library-based approach and potentially of other molecular tool-based screens is that in most cases, the molecular tool acts as an inhibitor of gene expression. It is plausible that the optimal targets of drugs that battle TR will be genes that are up-regulated when cells become resistant to Taxol.
Thus, to identify genes that are up-regulated by TR, we used a library that encodes zinc-finger-based artificial transcriptional activator proteins to generate Taxol-resistant cells. A recent report that involved transient transfection of a randomized ribozyme library into HeLa cells has been used successfully to isolate ribozymes that make cells resistant to FAS-mediated apoptosis (10). Therefore, we also transiently transfected HeLa cells with the zinc finger library and screened for ATFs inducing TR. We then used the selected ATFs and genome-scale gene expression profiles to identify the genes involved in inducing the Taxol-resistant phenotype. Drugs that inhibit the expression of these genes may be useful for blocking the development of TR.
MATERIALS AND METHODS
Construction of plasmids and an ATF library
We used 25 distinct ZFDs to construct a plasmid library that encodes ATFs each of which contain four ZFDs fused to the p65 transcriptional activation domain (NCBI accession number NP_068810; amino acids 275–535) as reported previously (24).
To confirm that the TR induced by transfection of the TR17-p65 ATF was caused by the binding of TR17-p65 to DNA, the ZFDs of TR17-p65 were mutated to change critical arginines in the two RDNQ ZFDs to alanines. A sequential PCR protocol that introduced a point mutation in TR17-p65 was carried out (25) with a pair of oligomers that contained the specified mutation (ZFP17mut-F: 5′-TAAGCGCTTCATGGCATCCGACAACCTGAC-3′ and ZFP17mut-R: 5′-GTCAGGTTGTCGGATGCCATGAAGCGCTTA-3′).
Screening for ATFs that induce TR in HeLa cells
The procedure for screening HeLa cells for TR is shown in Figure 1. HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum and antibiotics, at 37°C, in a humidified atmosphere containing 95% air and 5% CO2. HeLa cells (1 × 106) were seeded onto a 100 mm plate, incubated for 1 day under the above conditions, and then transfected with ATF library DNA (4 μg) using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's protocol. After 1 day of incubation under the above conditions, one-third of the transfected HeLa cells were transferred onto a new 100 mm plate and incubated for one more day. After treatment with Taxol (Sigma, Co.) (100 nM) for 48 h, the cells that survived the treatment were harvested, and their DNA was isolated using a G-spin kit (Intron, Co., Korea). The DNA from these cells was used to transform Escherichia coli for amplification of the ATF-encoding plasmids. The amplified ATF-encoding plasmids were used to re-transfect HeLa cells. The same selection procedure was repeated eight times under increasingly stringent conditions (Figure 1b). After the eighth selection cycle, the ATF-encoding plasmids were harvested from HeLa cells that had survived the selection and amplified in and isolated from E.coli transformants, and the ATFs were identified by nucleic acid sequencing. These ATFs were then tested for their ability to induce TR in HeLa cells.
Figure 1.
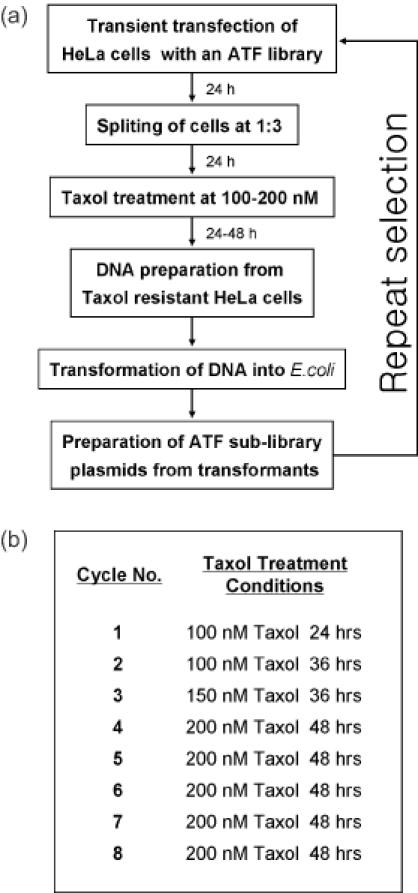
Procedure for screening an ATF library for ATFs that induce TR in HeLa cells. (a) An ATF library was transiently transfected into HeLa cells, and the transfected cells were incubated in the presence of Taxol. Colonies of cells that survived the Taxol treatment were pooled, and genomic DNA was isolated and used to transform E.coli for plasmid amplification. A selected ATF library was prepared from a pool of E.coli transformants and used again for HeLa cell transfection. The whole procedure was repeated eight times, and two ATFs were finally isolated from individual E.coli transformants. (b) Conditions of Taxol treatment for each selection cycle. To maximize the effect of the screening, Taxol concentrations and exposure times were increased as the cycles proceeded.
Confirmation of the ability of the selected ATFs to induce TR
Using the protocol described above, HeLa cells (2 × 105) were seeded into each well of a 12-well plate and then were transfected with either 500 ng of one of the purified ATF-encoding plasmids or an empty plasmid vector and 200 ng of the lacZ reporter plasmid pcDNA3.1/His/LacZ (Invitrogen). The transfected HeLa cells were then treated with Taxol at 100 nM for 48 h. Following the Taxol treatment, the cells were washed once with phosphate-buffered saline, stained to detect lacZ activity (to differentiate transfected cells from untransfected ones) and photographed. The β-galactosidase (lacZ) positive cells were then counted, and the survival rate was calculated using the following formula:
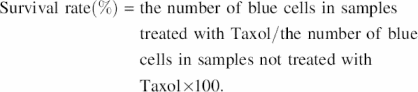
Gene expression microarray experiments using cells rendered Taxol-resistant by transfected ATFs
Microarray chips containing ∼17 000 human expressed sequence tag (EST) cDNAs were purchased from Genomic Tree, Inc. (Daejeon, South Korea). HeLa cells were transfected with one of two TR-inducing ATFs (TR 17-p65 or TR 216-p65) or an empty control vector (pLFD-p65) and incubated at 37°C for 24 h in the absence of Taxol. Total RNA was then prepared from each cell population using Trizol reagent (Invitrogen) according to the manufacturer's protocol. RNA isolated from the empty vector sample was used as the reference sample (labeled with Cy3, a fluorescent dye), and RNAs from cells with TR17-p65- or TR216-p65-containing vectors constituted the experimental (labeled with Cy5, a fluorescent dye) samples. Gene expression microarray experiments were performed and analyzed according to the protocol of the chip manufacturer (Genomic Tree, Inc., South Korea).
RESULTS AND DISCUSSION
Screening and selection of ATFs that induce TR in HeLa cells
Previously, we performed well-by-well transfection of cells with ATF libraries in 96-well plates to screen for desired phenotypes, such as cellular differentiation or change of cell growth rate (8). To develop a more efficient and large-scale screening process, we attempted to perform a batch screen. In a recent report, Kawasaki et al. (10), transiently transfected a batch of cells with ribozyme libraries and then selected for surviving clones that were resistant to Fas-mediated apoptosis. Because a transient transfection-based batch screen is more efficient than stable or well-by-well transfection or retrovirus-mediated colony formation, we decided to deliver our ATF library into HeLa cells via transient batch transfection.
Figure 1a shows a scheme of this screening procedure. HeLa cells were transiently transfected with an ATF library, and cells that survived Taxol treatment were pooled and subjected to subsequent longer treatments with higher concentrations of Taxol (Figure 1b). When HeLa cells were treated with 200 nM Taxol, >99% died within 48 h of incubation. After completion of five selection cycles, DNA from a pool of Taxol-resistant cells was used to transform E.coli, and ATF-encoding plasmids were isolated, both individually and as a pool, from the transformed cells. The DNA pool was tested for its ability to induce TR in fresh HeLa cells, but little difference was observed when these cells were compared with cells that had been transformed with an empty vector. In addition, we analyzed the plasmids for enrichment of target sequences deduced from individual zinc finger proteins, but no enrichment was observed. The reason why five cycles were not enough to yield an enrichment of plasmids of interest might be because a single transfected cell can contain tens of thousands of plasmids (26). Thus, the enrichment achieved in a single cycle is small compared to other selection screening methods, such as SELEX. Therefore, we carried out three more selection cycles and repeated the procedures mentioned above. This time, we observed positive results with a pool of plasmids both in the TR assays and in analysis for enrichment of target sequences. We then tested plasmids that were selected twice from randomly picked E.coli colonies after completion of the eighth cycle. From more than 100 individual plasmids sequenced, six identical pairs of ZFP plasmids were obtained. Among them, one pair was disregarded because the ZFP gene had undergone recombination. We tested the five other pairs for TR and observed positive results after individual transfection. We also tested the TR-promoting ability of several ZFP plasmids that were selected only once from randomly picked E.coli colonies after completion of the eighth cycle and found one ZFP positive vector.
Two ATFs, TR17-p65 and TR216-p65, were finally selected because they exhibited the highest level of resistance to Taxol when compared to the vector control (Figure 2), and the identities of these two ATFs were determined by nucleic acid sequencing of the plasmid inserts (Table 1). To assess the ability of the TR17-p65 ATF to induce TR in HeLa cells, the TR17-p65-encoding DNA was mutated in such a way that the TR17 ATF was unable to bind to its target DNA sequence (the arginines in the two RNDQ fingers, which are critical for DNA binding, were changed to alanine, yielding mTR17). In addition, the p65 transcriptional activation domain was removed from TR17-p65, resulting in the TR17-no FD protein. Cells transfected with either mTR17-p65 or TR17-no FD showed a level of Taxol sensitivity similar to that of cells transfected with the vector control (Figure 3). These experiments confirmed that the induction of TR by TR17-p65 resulted from the protein's ability to activate transcription of a target gene(s) that contained the appropriate DNA-binding sequences.
Figure 2.
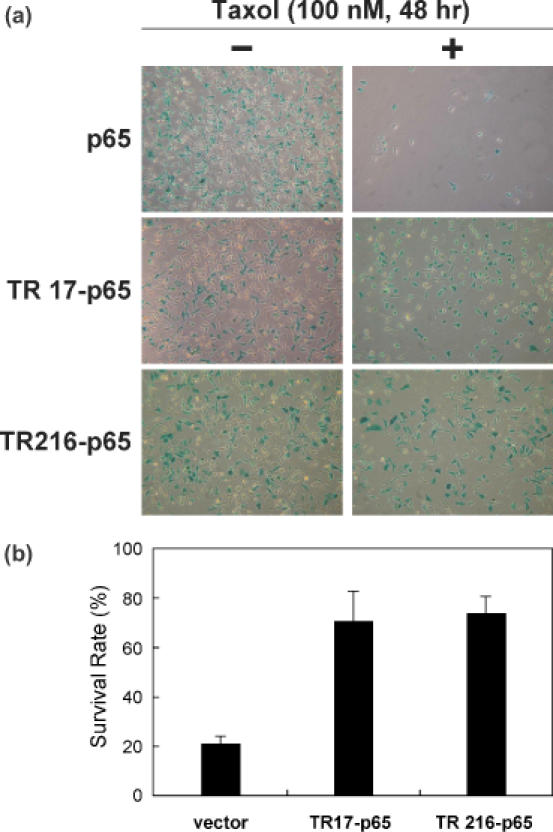
Expression of two ATFs, TR17-p65 and TR216-p65, increased TR in HeLa cells. (a) HeLa cells grown in a 12-well plate were transfected with 500 ng of a plasmid encoding one of the ATFs and 200 ng of a lacZ reporter plasmid and were treated with Taxol at 100 nM for 48 h. Cells were stained for lacZ activity to differentiate transfected cells from untransfected ones. (b) The survival rate of HeLa cells transfected with each of the ATF-encoding plasmids was calculated as follows: Survival rate (%) = the number of blue cells in samples treated with Taxol/the number of blue cells in samples not treated with Taxol × 100.
Table 1. Identities and binding sequences of TR17 and TR216.
| ATFs | ZFDs (N- to C-termini) | Target DNA-binding sites (5′ to 3′) | |||
|---|---|---|---|---|---|
| TR17 | RDNQ | RDNQ | ISNR | VSTR | GCWGAWAAGAAG |
| TR216 | RDKR | ISNR | ISNR | QSNK | DAAGAWGAWRGG |
W: A or T; D: A, G or T; and R: A or G.
Figure 3.
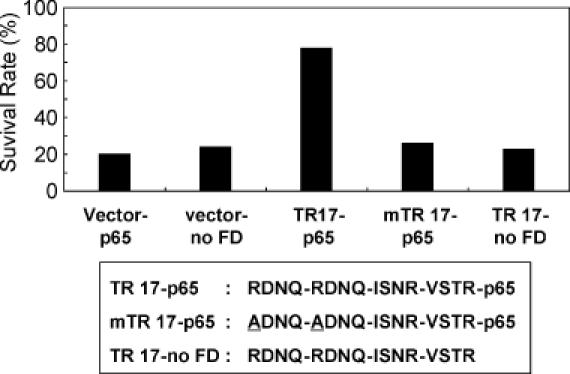
The survival rates of HeLa cells transfected with TR17-p65 intact and mutated plasmids. The TR17-p65 sequence was mutated such that either the RDNQ-encoding sequences in the first two ZFDs were mutated to yield ADNQ (mTR17-p65) or the p65 functional domain was deleted (TR17-no FD). Two control plasmids, empty vector (Vector-no FD) and vector expressing only p65 without any ZFDs (Vector-p65), were also tested. The transfections and Taxol treatments were carried out as shown in Figure 2.
One must keep in mind that the target gene(s) that were activated by TR17-p65 and TR216-p65 might not be the direct regulators of the TR phenotype; it is possible that these ATFs induced the expression of gene products that, in turn, modulated the expression of other genes that conferred TR, as shown in the case of ketoconazole resistance in yeast cells (8).
Genome-scale gene expression profile of Taxol-resistant cells
To identify genes whose expression is associated with TR, genome-scale gene expression experiments using cDNA microarrays were performed with RNA prepared from HeLa cells transfected with each of two TR-inducing ATFs (TR17-p65 or TR216-p65) or an empty control vector (pLFD-p65). We looked for genes whose expression was co-activated by both ATFs. Cells that expressed TR17-p65 and TR216-p65 showed a >2-fold increase in the expression of 187 and 152 of the 17 000 EST clones, respectively, compared to control cells. When we compared the expression profiles of the two ATFs, we found that the expression of 37 genes were co-activated >2-fold by both TR17-p65 and TR216-p65 (Table 2).
Table 2. Genes co-activated >2-fold by both TR17-P65 and TR216-P65.
| Function | Protein name | Gene ID |
|---|---|---|
| Enzymes | Cytochrome P450 3A5 | R92425 |
| Phospholipase C, delta 4 | AI203283 | |
| Aurora/Ipl1-related kinase 3 | AA421265 | |
| Aldehyde dehydrogenase 6 | AI859300 | |
| G-protein-coupled receptor kinase 5 | AA862435 | |
| Glucosamine-6-sulfatase | AI358910 | |
| RNA helicase | AA126958 | |
| RNase 4 | T60163 | |
| TNF related | TNF receptor-associated factor 5 | AA102634 |
| TNF superfamily member 6 (FASL receptor) | AA293571 | |
| p53 related | p53-induced protein SIP27 | AA459364 |
| Sestrin 1 (p53-regulated PA26 nuclear protein) | AA447661 | |
| Breast cancer related | DEME-6 protein | AA886199 |
| Breast tumor novel factor 1 (bnf-1) | AI343625 | |
| IFIT family | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) | AA489640 |
| Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) | N63988 | |
| Transcriptional regulation | Neural crest transcription factor SLUG | N64741 |
| Basal transcriptional activator ABT1 | AA480876 | |
| Ion channel/transporter | Na+/K+/Ca2+ exchange protein 1 | W02204 |
| Na+/K+ ATPase 2 | R73570 | |
| Sodium channel alpha subunit | AI091722 | |
| Brain-specific Na-dependent inorganic phosphate cotransporter | AA702627 | |
| Monocarboxylate transporter 8 (MCT 8) | AA425395 | |
| Others | Crystallin, alpha B | AA504891 |
| Allograft inflammatory factor 1 | W69954 | |
| Sphingolipid G-protein-coupled receptor (EDG3) | AI141395 | |
| Insulin-like growth factor binding protein 6 (IGFBP-6) | AA478724 | |
| Apolipoprotein L6 | AI391658 | |
| Ca2+-promoted Ras inactivator (CAPRI) | AA989217 | |
| Histidine-rich calcium-binding protein (HRC) | AI769340 | |
| Matrilin 2 | AA071473 | |
| Retinoic acid receptor responder protein 3 | W47350 | |
| Signal sequence receptor, alpha (SSR1) | AA910877 | |
| SRY (sex determining region Y)-box 3 | AI359981 | |
| Neurexin 1 homolog | AI201652 | |
| Unknown gene | AA505067 |
Some of the up-regulated genes were easily recognized as encoding gene products that are directly involved in the TR mechanism. One of them was the cytochrome P450 3A5 gene (R92425, Table 2), which encodes an enzyme previously shown to metabolize and detoxify Taxol (27,28). It has been reported that expression of the cytochrome P450 3A family members are induced by Taxol (29). Another gene we identified that appears to play a role in the TR mechanism was the Aurora/Ipl1-related kinase 3 gene (AA421265, also called Aurora-C kinase). The Aurora kinase family contains three highly conserved members, A, B and C (30). A recent report has shown that the over-expression of Aurora-A kinase results in TR by overriding the mitotic spindle assembly checkpoint (31). Therefore, the Aurora-C kinase might also be involved in TR by a similar mechanism.
The functions of the remaining expression-enhanced genes in the TR mechanism are at present unclear. However, the fact that our screen identified genes known to be related to TR suggests that at least some of these remaining genes might also contribute to TR, either directly or indirectly.
Previous studies reported that the β-tubulin and P-gp genes are over-expressed in some Taxol-resistant cells, and forced over-expression of these genes in cells results in TR (21,22). Xu et al. (6) have screened a peptide library in HeLa cells to isolate a peptide(s) that induce TR and found peptide RGP8.5.1, which was suggested to induce the over-expression of P-gp (ABCB-1) by inhibiting proteasome-mediated protein degradation. However, in both TR17-p65- and TR216-p65-expressing HeLa cells, the expression levels of both β-tubulin and P-gp were maintained at a level similar to that of control cells. It is likely that the two ATFs selected in our screen achieve TR by a novel mechanism or via genetic pathways that do not involve in β-tubulin or P-gp.
It is intriguing that two seemingly independent TR mechanisms, Taxol detoxification mediated by cytochrome P450 and overriding of the microtubule assembly checkpoint by an Aurora-related kinase, were both induced by the two ATFs selected in our screen. An interesting interpretation of this observation is that different cellular mechanisms or pathways that confer TR are co-regulated rather than individually regulated. However, at this point, we cannot discard the possibility that the two ATFs just happen to target multiple, unrelated TR pathways. Future studies will aim at resolving this issue.
A recent study by Kang et al. (32) using oligonucleotide microarrays identified genes that are differentially expressed in acquired cisplatin-resistant gastric cancer cells. While direct comparison between our data and their data may not be feasible, because different anticancer drugs and cell lines were used, we found that the gene encoding insulin-like growth factor binding protein-6 (IGFBP-6) was up-regulated in both the cisplatin-resistant gastric cancer cells in their study (32) and the Taxol-resistant HeLa cells in our study. Thus, comparison studies using resistant cell lines induced by drug treatment and those induced by ATFs will be beneficial in pinpointing direct mechanisms of drug resistance.
Library-based approaches with ribozymes or random peptides have widely been used to discover genes or biological pathways responsible for a variety of cellular phenotypes, many of which are relevant to disease progression. However, most of these approaches uncover only genes whose loss-of-function results in the desired phenotype. In many cases, however, genes or pathways give rise to particular phenotypes when their expression or activity is up-regulated. To identify genes or pathways in this class, a phenotype-based approach that can randomly up-regulate a subset of genes is needed. cDNA library screening is one approach (5). However, this method requires the transfection of individual (preferably full-length) cDNA clones, which is a time-consuming and labor-intensive process.
Compared to the methods described above, a random ATF library has several advantages. First, an ATF library can be constructed with transcriptional activators as well as repressors. Second, ATFs can change the expression level of target genes subtly depending on the binding affinities of the ATFs for target DNAs. Third, the same ATF library can be used in a wide range of organisms, such as yeast, plants and mammalian cells.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mr Jin Woo Park (ToolGen) for his help in analyzing the microarray data. This work was partially supported by a Molecular and Cellular BioDiscovery Research Program grant (M1-0311-00-0076) from the Ministry of Science and Technology in Korea.
REFERENCES
- 1.Berns K., Hijmans,E.M., Mullenders,J., Brummelkamp,T.R., Velds,A., Heimerikx,M., Kerkhoven,R.M., Madiredjo,M., Nijkamp,W., Weigelt,B. et al. (2004) A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature, 428, 431–437. [DOI] [PubMed] [Google Scholar]
- 2.Suyama E., Kawasaki,H. and Taira,K. (2003) Use of a ribozyme library for validation of gene functions and cellular pathways. Nucleic Acids Res. (Suppl.), 3, 253–254. [DOI] [PubMed] [Google Scholar]
- 3.Yin D., Fox,B., Lonetto,M.L., Etherton,M.R., Payne,D.J., Holmes,D.J., Rosenberg,M. and Ji,Y. (2004) Identification of antimicrobial targets using a comprehensive genomic approach. Pharmacogenomics, 5, 101–113. [DOI] [PubMed] [Google Scholar]
- 4.Kuruvilla F.G., Shamji,A.F., Sternson,S.M., Hergenrother,P.J.and Schreiber,S.L. (2002) Dissecting glucose signalling with diversity-oriented synthesis and small-molecule microarrays. Nature, 416, 653–657. [DOI] [PubMed] [Google Scholar]
- 5.Michiels F., van Es,H., van Rompaey,L., Merchiers,P., Francken,B., Pittois,K., van der Schueren,J., Brys,R., Vandersmissen,J., Beirinckx,F. et al. (2002) Arrayed adenoviral expression libraries for functional screening. Nat. Biotechnol., 20, 1154–1157. [DOI] [PubMed] [Google Scholar]
- 6.Xu X., Leo,C., Jang,Y., Chan,E., Padilla,D., Huang,B.C., Lin,T., Gururaja,T., Hitoshi,Y., Lorens,J.B. et al. (2001) Dominant effector genetics in mammalian cells. Nature Genet., 27, 23–29. [DOI] [PubMed] [Google Scholar]
- 7.Beger C., Pierce,L.N., Kruger,M., Marcusson,E.G., Robbins,J.M., Welcsh,P., Welch,P.J., Welte,K., King,M.C., Barber,J.R. et al. (2001) Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc. Natl Acad. Sci. USA, 98, 130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park K.S., Lee,D.K., Lee,H., Lee,Y., Jang,Y.S., Kim,Y.H., Yang,H.Y., Lee,S.I., Seol,W. and Kim,J.S. (2003) Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat. Biotechnol., 21, 1208–1214. [DOI] [PubMed] [Google Scholar]
- 9.Suyama E., Kawasaki,H., Nakajima,M. and Taira,K. (2003) Identification of genes involved in cell invasion by using a library of randomized hybrid ribozymes. Proc. Natl Acad. Sci. USA, 100, 5616–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawasaki H. and Taira,K. (2002) A functional gene discovery in the Fas-mediated pathway to apoptosis by analysis of transiently expressed randomized hybrid-ribozyme libraries. Nucleic Acids Res., 30, 3609–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blancafort P., Magnenat,L. and Barbas,C.F.,III (2003) Scanning the human genome with combinatorial transcription factor libraries. Nat. Biotechnol., 21, 269–274. [DOI] [PubMed] [Google Scholar]
- 12.Wolfe S.A., Nekludova,L. and Pabo,C.O. (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct., 29, 183–212. [DOI] [PubMed] [Google Scholar]
- 13.Lee D.K., Seol,W. and Kim,J.S. (2003) Custom DNA-binding proteins and artificial transcription factors. Curr. Top. Med. Chem., 3, 645–657. [DOI] [PubMed] [Google Scholar]
- 14.Beerli R.R. and Barbas,C.F., III (2002) Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol., 20, 135–141. [DOI] [PubMed] [Google Scholar]
- 15.Snowden A.W., Zhang,L., Urnov,F., Dent,C., Jouvenot,Y., Zhong,X., Rebar,E.J., Jamieson,A.C., Zhang,H.S., Tan,S. et al. (2003) Repression of vascular endothelial growth factor A in glioblastoma cells using engineered zinc finger transcription factors. Cancer Res., 63, 8968–8976. [PubMed] [Google Scholar]
- 16.Rebar E.J., Huang,Y., Hickey,R., Nath,A.K., Meoli,D., Nath,S., Chen,B., Xu,L., Liang,Y., Jamieson,A.C. et al. (2002) Induction of angiogenesis in a mouse model using engineered transcription factors. Nature Med., 8, 1427–1432. [DOI] [PubMed] [Google Scholar]
- 17.Ren D., Collingwood,T.N., Rebar,E.J., Wolffe,A.P. and Camp,H.S. (2002) PPARgamma knockdown by engineered transcription factors: exogenous PPARgamma2 but not PPARgamma1 reactivates adipogenesis. Genes Dev., 16, 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez J.P., Ullman,C., Moore,M., Choo,Y. and Chua,N.H. (2002) Regulation of gene expression in Arabidopsis thaliana by artificial zinc finger chimeras. Plant Cell Physiol., 43, 1465–1472. [DOI] [PubMed] [Google Scholar]
- 19.Joung J.K., Ramm,E.I. and Pabo,C.O. (2000) A bacterial two-hybrid selection system for studying protein–DNA and protein–protein interactions. Proc. Natl Acad. Sci. USA, 97, 7382–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowinsky E.K. and Donehower,R.C. (1995) Paclitaxel (taxol). N. Engl. J. Med., 332, 1004–1014. [DOI] [PubMed] [Google Scholar]
- 21.Orr G.A., Verdier-Pinard,P., McDaid,H. and Horwitz,S.B. (2003) Mechanisms of Taxol-resistance related to microtubules. Oncogene, 22, 7280–7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yusuf R.Z., Duan,Z., Lamendola,D.E., Penson,R.T. and Seiden,M.V. (2003) Paclitaxel resistance: molecular mechanisms and pharmacologic manipulation. Curr. Cancer Drug Targets, 3, 1–19. [DOI] [PubMed] [Google Scholar]
- 23.Lamendola D.E., Duan,Z., Yusuf,R.Z. and Seiden,M.V. (2003) Molecular description of evolving paclitaxel resistance in the SKOV-3 human ovarian carcinoma cell line. Cancer Res., 63, 2200–2205. [PubMed] [Google Scholar]
- 24.Bae K.H., Kwon,Y.D., Shin,H.C., Hwang,M.S., Ryu,E.H., Park,K.S., Yang,H.Y., Lee,D.K., Lee,Y., Park,J. et al. (2003) Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat. Biotechnol., 21, 275–280. [DOI] [PubMed] [Google Scholar]
- 25.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (2001) Current Protocols in Molecular Biology. John Wiley Sons, Inc., New York, NY. [Google Scholar]
- 26.Tseng W.-C., Haselton,F.R. and Giorgio,T.D. (1997) Transfection by cationic liposomes using simultaneous single cell measurements of plasmid delivery and transgene expression. J. Biol. Chem., 272, 25641–25647. [DOI] [PubMed] [Google Scholar]
- 27.Martinez C., Garcia-Martin,E., Pizarro,R.M., Garcia-Gamito,F.J. and Agundez,J.A. (2002) Expression of paclitaxel-inactivating CYP3A activity in human colorectal cancer: implications for drug therapy. Br. J. Cancer, 87, 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kivisto K.T., Kroemer,H.K. and Eichelbaum,M. (1995) The role of human cytochrome P450 enzymes in the metabolism of anticancer agents: implications for drug interactions. Br. J. Clin. Pharmacol, 40, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kostrubsky V.E., Lewis,L.D., Strom,S.C., Wood,S.G., Schuetz,E.G., Schuetz,J.D., Sinclair,P.R., Steven,A., Wrighton,S.A. and Sinclair,J.F. (1998) Induction of cytochrome P4503A by taxol in primary cultures of human hepatocytes. Arch. Biochem. Biophys., 355, 131–136. [DOI] [PubMed] [Google Scholar]
- 30.Katayama H., Brinkley,W.R. and Sen,S. (2003) The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev., 22, 451–464. [DOI] [PubMed] [Google Scholar]
- 31.Anand S., Penrhyn-Lowe,S. and Venkitaraman,A.R. (2003) AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell, 3, 51–62. [DOI] [PubMed] [Google Scholar]
- 32.Kang H.C., Kim,I.-J., Park,J.-H., Shin,Y., Ku,J.-L., Jung,M.S., Yoo,B.C., Kim,H.K. and Park,J.-G. (2004) Identification of genes with differential expression in acquired drug-resistant gastric cancer cells using high-density oligonucleotide microarrays. Clin. Cancer Res., 10, 272–284. [DOI] [PubMed] [Google Scholar]


