Abstract
Neuregulin (NRG) (also known as ARIA, GGF, and other names) is a heparin sulfate proteoglycan secreted into the neuromuscular junction by innervating motor and sensory neurons. An integral part of synapse formation, we have analyzed NRG-induced changes in gene expression over 48 h in primary human myotubes. We show that in addition to increasing the expression of acetylcholine receptors on the myotube surface, NRG treatment results in a transient increase of several members of the early growth response (Egr) family of transcription factors. Three Egrs, Egr1, -2, and -3, are induced within the first hour of NRG treatment, with Egr1 and -3 RNA levels showing the most significant increases of ≈9- and 16-fold, respectively. Also noted was a corresponding increase in protein levels for both of these transcription factors. Previous literature indicates that Egr3 expression is required for the formation of muscle spindle fibers, sensory organs that are distinct from skeletal muscle contractile fibers. At the molecular level, muscle spindle fibers express a unique subset of myosin heavy chains. Two isoforms of the myosin heavy chain, the slow development and neonatal, were found to be increased in our myotube cultures after 48 h of treatment with NRG. Taken together, these results indicate that not only can NRG induce the expression of a transcription factor key to spindle fiber development (Egr3), but that a portion of this developmental process can be replicated in vitro.
The neuregulins (NRGs) are a family of glycoproteins that activate ErbB2–4 receptor tyrosine kinases to mediate the proliferation, differentiation, migration, and survival of a number of cell types (1–5). In the motor neuron–muscle system, NRG has been implicated in the maturation of the postsynaptic membrane at the neuromuscular junction (6–8), the survival of Schwann cell precursors (9), the maturation of Schwann cells in peripheral nerve (10), glucose metabolism (11), and myogenesis (12). The NRG/ErbB system has also been implicated in muscle spindle formation (13–15). Here we extend previous results regarding spindles by demonstrating that the NRG β1 epidermal growth factor (EGF)-like domain is sufficient to induce the expression of immediate early genes and proteins that characterize myogenesis within muscle spindles.
In skeletal myotubes maintained in vitro, NRG regulates the number of acetylcholine receptors (AChRs) (16, 17), the subunit compositions of AChRs (18), and the expression of other critical proteins (11, 19, 20). These findings suggest that NRG acts at the neuromuscular junction (NMJ) to help maintain the AChR density essential for effective transmission at this synapse. It is unlikely, however, that the effects of NRG in skeletal muscle are restricted to the NMJ. We therefore sought to identify further roles by surveying NRG-induced gene expression changes in cultured human skeletal muscle.
Muscle spindles appear as encapsulated bundles containing three specialized myofiber types. The differentiation of these fibers depends on trophic factors presumably released by sensory neurons (21–27). Neurons originating in the dorsal root ganglion (DRG) (28), proprioceptive sensory neurons in particular, express NRG early in development (14, 29, 30). While these experiments were ongoing, reports appeared implicating NRG in the development of muscle spindles. Hippenmeyer et al. (14) showed that NRG induces the expression of early growth response 3 (Egr3), a transcription factor that is critical to the differentiation of muscle spindle fibers (31). Evidence for NRG's role in spindle formation is re-enforced by the phenotypic similarities between conditional Erb2 knockout animals and Egr3 null mice (13, 15, 24).
Several isoforms of NRG protein encoded by the Nrg1 gene have been described. The biological effects of all products of the Nrg1 gene appear to depend on an EGF-like domain in the extracellular half of the protein. Here we describe the effects of a NRG, EGF β1 domain (human amino acid residues 176–246) on Egr3 transcription factor and other members of this family in cultured primary human myotubes. We also find that a significant number of other muscle genes are regulated by NRG.
Materials and Methods
Cell Culture. H. Blau (Stanford University, Stanford, CA) provided human primary myoblasts. These cells were cultured on collagen (0.1%)-coated tissue culture plastic in F-10 media supplemented with 15% FBS (Life Technologies, Rockville, MD), 100 units/ml penicillin/streptomycin (Life Technologies), and 0.5% Chicken embryo extract (Sera Labs, Salisbury, U.K.). At confluence, myoblasts were switched and maintained in fusion media comprised of DMEM with high glucose (DMEM-HI), 2% horse serum, 1% insulin-transferrin-selenium supplement, 100 units/ml penicillin/streptomycin, and 2.5 × 10-6 M dexamethasone (Sigma). Under these conditions, the majority of the cultured human myoblasts fused to form multinucleate myotubes within 3 days. Mouse C2 muscle cells were cultured as described (32).
Reagents. Egr transcription factor polyclonal antibodies (Egr1 and Egr3) were purchased from Santa Cruz Biotechnology. The anti-slow developmental myosin heavy chain antibody (MyHC) was a gift from F. Stockdale (Stanford University). The anti-neonatal-MyHC monoclonal antibody was obtained from Novacastra. S. Tzaros (Hellenic Pasteur Institute, Athens, Greece) provided the antibodies specific to the AChR α-subunit. Recombinant NRG (NRG-β1 EGF domain) was obtained from R & D Systems. The agrin used in these experiments has been described (33).
Visualization of AChR Clusters. AChR clusters were labeled as described by Jacobson et al. (34) and visualized with a Nikon fluorescence microscope at a final magnification of ×400.
Radiodetection of AChRs and Quantification. The number of surface AChRs was estimated by [125I]α-bungarotoxin binding. Cultures of primary human myotubes were treated with either 1 nM of the NRG EGF β1 domain for 18 h or left untreated. At the conclusion of this incubation 1 nM of [125I]α-bungarotoxin was added to these cultures for 1 h. Labeled cells were then repeatedly washed with PBS, scraped from the culture dishes and the level of radioincorporation determined. Nonspecific binding was determined by preincubation with 1 μM of unlabeled α-bungarotoxin for 30 min before the addition of 1 nM of [125I]α-bungarotoxin. AChR extraction, immunoprecipitation, gel electrophoresis, and densitometry were performed as described in Jacobson et al. (35).
Expression Analysis. RNA was isolated from cultured human myotubes for hybridization onto cDNA microarrays. Control myotubes were compared with myotubes treated with NRG (1 nM) or with 500 pM neural agrin. Deposition microarrays containing 6,758 features were generated from image clones (ResGen, Huntsville, AL) as described (36, 37). Myotubes were washed twice with PBS and extracted directly with Trizol (5 ml per 10-cm dish, Invitrogen). After chloroform extraction, samples were loaded onto RNeasy columns (Qiagen, Valencia, CA) and processed as per Qiagen's instructions. The resulting RNA was then labeled with either Cy3-dUTP or Cy5-dUTP by using the Amersham Pharmacia first-strand labeling kit. Hybridizations were at 65°C overnight in an aqueous-based hybridization solution. Detailed RNA isolation, labeling, and hybridization protocols are available at http://research.nhgri.nih.gov/microarray. Slide images were acquired by using an Agilent scanner (Agilent Technologies, Palo Alto, CA), gene assignments and intensity data were extracted by using the DeArray Suite (38) for IPLab Spectrum, and the resulting data analyzed by using filemaker pro (FileMaker, Santa Clara, CA) and genespring (Silicon Genetics, Redwood City, CA) software.
Western Blot Analysis. Immunoblotting methods are as shown in Jacobson et al. (35), with the exception of those blots probed with anti-MyHC specific antibodies. Blots probed with anti-MyHC specific antibodies were incubated in 10 mM Tris·HCl (pH 7.4), 0.15 M NaCl and 0.1% Tween-20 supplemented with 2.5% milk/2.5% BSA. Expression levels were quantified via densitometric analysis using the gel analysis macro included in the nih image software package available at ftp://rsbweb.nih.gov/pub/nih-image. To average several independent experiments, all values were expressed as a percentage of the control value (zero time point). Quantification was always performed on film with subsaturating exposure levels.
Immunocytochemistry. Cultures were washed twice with PBS and fixed with a solution of ice-cold 70% ethanol for 15 min at -20°C. After two additional washes with PBS, cells were further permeabilized with 0.1% Triton X-100 in 10% goat serum in PBS for 10 min at room temperature. After permeabilzation, cultures were incubated for 1 h in blocking buffer, 10% goat serum in PBS. Primary antibodies were diluted in blocking buffer and incubated overnight at 4°C. After several washes with PBS, bound primary antibody was detected with Fluorescein-conjugated secondary antiserum. Cultures were washed several times before the addition of ProLong antifade (Molecular Probes) mounting medium and coverslips. Fluorescence was viewed with a Nikon fluorescence microscope at a final magnification of ×200. To document observed differences in the intensity of antibody labeling, experimental and control samples were photographed and reproduced under identical conditions.
Results and Discussion
Human Myotubes Respond to NRG and Agrin. The human myotubes used in this study have not previously been characterized in regard to NRG (ARIA) or agrin effects; therefore, our initial experiments were designed to determine the effects of these factors on AChR number and distribution. Increases in the surface expression of AChRs and the induction of AChR clustering on human primary myotubes were compared to the more extensively studied mouse C2C12 cell line.
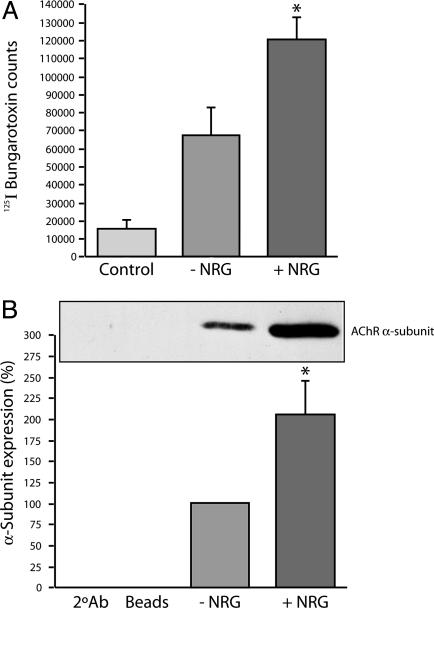
The number of surface AChRs was assayed by the binding of [125I]α-bungarotoxin and by immunoblot of precipitated AChRs (Fig. 1). At a concentration of 1 nM, NRG produced a nearly 2-fold increase in specific [125I]α-bungarotoxin binding in four separate experiments, 120,381 ± 11,896 (SEM) counts vs. 66,697 ± 15,271 (SEM) counts (P < 0.05, t test; Fig. 1 A). An ≈2-fold increase was also noted when AChR levels were compared by immunoblot (n = 5, P < 0.05, t test; Fig. 1B). By either method, NRG induced a significant increase in AChR level in human myotubes.
Fig. 1.
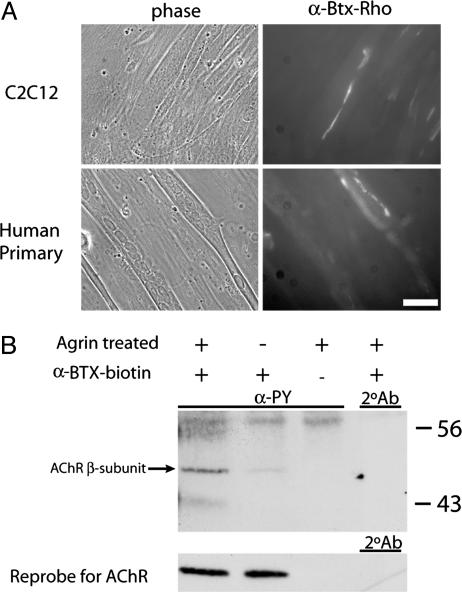
NRG induces the expression of AChRs on the surface of myotubes. The number of AChRs on the surface of myotubes was determined by two methods. The first method quantified the number of AChRs directly by using 125I-labeled α-bungarotoxin (A); in the second method, AChRs were quantified indirectly by using densitometry (B) by combining toxin precipitation with Western blotting (Lower). By either method, we saw a significant ≈2-fold increase in the level of surface AChRs as a result of NRG treatment. In both experiments, n ≥ 4 with P ≤ 0.05. (Scale bar, 10 μM.)
The formation of AChR aggregates was determined after an overnight treatment of myotubes with 500 pM agrin. Agrin-induced AChR clusters were evident in both human and mouse myotubes as shown in Fig. 2A. The phosphotyrosylation of the AChR β-subunit was used as an independent assessment of AChR clustering (39–42). Fig. 2B shows agrin induced phosphorylation of the toxin precipitated β-subunit of the AChR in human myotubes (arrow). Fig. 2B also shows the reprobe for the α-subunit of the AChR, indicating, as suspected, that similar levels of AChR were precipitated in both the agrin-treated and untreated lanes and that, as expected, there was no increase in AChR number accompanying agrin-induced aggregation.
Fig. 2.
Agrin induces the phosphorylation and aggregation of AChRs on primary human myotubes. (A) Primary human or C2C12 myotubes are shown (left column) with fluorescent photomicrographs of the same fields (right column). Agrin-induced AChR aggregates are clearly visible on the surface of both human and mouse myotubes. (B) The increase in the phosphotyrosylation of the AChR β-subunit resulting from similar agrin treatments. In the left lane, an increase in the ≈50 kDa corresponding to the β-subunit of the AChR in the agrin-treated sample is clearly visible but is not present in the untreated sample in the lane at right. This increase is clearly attributable to agrin, as the amount of precipitated AChRs is equivalent in both lanes as shown in the reprobe at the bottom.
Expression Profiling of NRG-Treated Myotubes. We have used deposition microarrays to examine the effects of NRG on cultured muscle gene expression over a period of 48 h. The RNA used in these experiments was isolated from human primary myotubes that had been maintained in fusion media for 3 days before treatment. RNA was isolated pair wise from both NRG-treated and untreated myotube cultures at 13 distinct time points. The RNA isolated from NRG treated myotubes was then hybridized to the cDNA microarray with its time-matched untreated RNA partner.
Expression data were obtained for 4,605 of the 6,758 genes printed on our human array. These 4,605 clones passed a quality metric in each of the 13 hybridizations designed to measure the size, intensity, and morphology of each individual gene spot (43). Changes in gene expression were considered significant if two criteria were met: first, if the fold change was outside a calculated 99% confidence interval (38); second, if a significant change in gene expression was evident in two consecutive time points. With a 99% confidence interval, there is a 1 in 100 chance that a gene falling outside the interval does so at random. A 99% confidence interval over two hybridizations therefore corresponds to a probability of 1 in 10,000 that the variation is due to chance, provided that the experiments in two time points are independent. In fact, many genes exceeded these criteria and were significant over three continuous time points, a 1 in 1,000,000 chance of random occurrence.
Overall, 28% met the first criterion but not the second. Two hundred and fifteen genes (5% of the genes represented on the chip) met both criteria. By current estimates, our microarray analysis of ≈6,800 genes may have included perhaps 20% of the total genome (44). Our experiments may thus have highlighted only a fraction of NRG effects in skeletal muscle. Nonetheless, many of the genes identified as significantly changed in our analysis had no previously known association with NRG. Of the 215 genes meeting our criteria for significance, the gene with largest fold increase (22×) was thrombospondin-1, an extracellular matrix glycoprotein that regulates proliferation, migration, and apoptosis (45). Robust increases in expression were also noted for Egr3, RAB6, sarcosin, and kinesin family member 5B. Each increased in excess of 10-fold. Several genes regulated by NRG could be related by function or homology. In particular, we noted significant changes in transcripts involved in cell growth, mitochondrial function, extracellular matrix composition, axonal guidance, and protein stability. Finally, the expression of genes that may be associated with the diseases amyotrophic lateral sclerosis (IGF binding protein 5) (46), schizophrenia (discs large homolog 1) (47, 48), and fragile X syndrome (fragile X mental retardation autosomal homolog 1) (49) were significantly affected in muscle by NRG treatment. A complete list of genes, accession numbers, and the NRG induced fold change in expression is available in Table 1, which is published as supporting information on the PNAS web site.
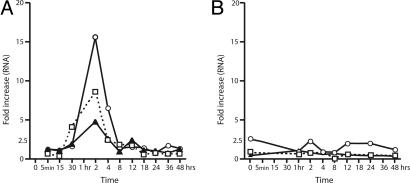
NRG Induces the Expression of Several Members of the Egr Family of Transcription Factors. Three Egr genes cosegregated when the expression profiles of our significant gene list was subjected to clustering using the K-means clustering algorithm within the genespring program. K-means divides genes into groups based on their expression patterns. Applied to time course data, it will identify classes of genes that are similarly up or down regulated in a time-dependent manner. Changes in the Egrs were among the largest observed. As shown in Fig. 3A, Egr1, -2, and -3 all increased significantly and peak by 2 h, remain elevated at 4 h, and return to control levels by 10 h despite the continuous presence of the NRG EGF β1 domain. The increase in RNA expression is particularly dramatic for Egr1 and -3, which were up-regulated 9- and 16-fold, respectively.
Fig. 3.
Time course of NRG-induced Egr gene expression. (A) The Egr transcription factors all show similar expression patterns in response to NRG treatment. The expression levels of the three Egr genes, Egr1 (W), Egr 2 (H), and Egr 3 (E), are all significantly increased within the first hour (as determined by extrapolation) and peak by the second hour of exposure to NRG (measured). Furthermore, in all three cases, the increases induced by NRG are transient and return to normal by the eighth hour of NRG treatment. This pattern is not seen in myotubes treated with agrin over a similar time period (B), indicating that the induction of Egr expression in human myotubes is specific to our NRG treatments.
A similar time course experiment in which human myotubes were treated with agrin is shown in Fig. 3B. A small increase was noted for Egr3 at 2 h, but this change was within the 99% confidence interval for that particular hybridization and it was not sustained. Therefore, the effect of NRG on the Egrs is not replicated by agrin.
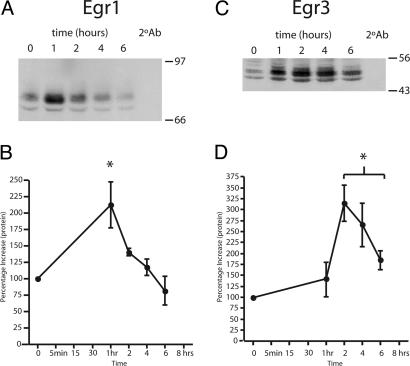
We determined that the increase in Egr mRNA was accompanied by an increase in Egr protein in cultured human myotubes. Representative immunoblots for Egr1 and -3 are shown in Fig. 4 A and C, respectively. Egr2 was not examined. The Egr1 protein level measured at 1 h was increased >2-fold (n = 3, P < 0.05, t test; Fig. 4B). This is not inconsistent with the mRNA measurements if one assumes (by extrapolation between the 30-min and 2-h time points) that Egr1 mRNA is elevated at 1 h. Likewise, Egr3 protein expression increased 3.2-fold. However, Egr3 protein levels peaked later, at 2 h, and remained elevated for a longer period (6 h, n = 4, P < 0.01, t test; Fig. 4D). Overall, the protein and RNA data support an early and significant effect of the NRG EGF domain on Egr gene and protein expression in human skeletal muscle.
Fig. 4.
The expression patterns of the Egr1 and Egr3 proteins are similar to those found for their corresponding RNAs. Extracts from NRG-treated human myotubes were isolated and immunoblotted with antibodies specific to Egr1 and Egr3, representative blots of which are shown in A and C, respectively. The level of expression was subsequently determined by densitometry, and images in B and D represent the averages (±SEM) relative to time 0 (n = 3, P < 0.05). The expression of Egr1 protein was transient and significant (asterisk) at only the 1-h time point in our assay (B). This is contrasted by the expression of Egr3, which showed increases later, at 2 h, and remained significantly increased at all of the later time points assayed (D).
Tourtellotte and Milbrandt (31) examined the role of Egr3 in muscle development by using Egr3 null animals. Mice null for Egr3 have profound gait ataxia and the complete loss of muscle spindles (31), a defect that is apparently a direct result of the loss of Egr3 expression in muscle, not in the innervating sensory afferents (24). Hippenmeyer et al. (14) provided strong evidence that NRG is involved in the regulation of muscle Egr. They found, by using mice with two different Nrg1 mutations, that the Ig containing NRG isoforms were crucial for spindle fiber formation. The ablation of all NRG isoforms resulted in the absence of Egr3 expression and muscle spindles. In contrast, mice lacking only the cysteine-rich domain (CRD)-containing isoforms of NRG developed muscle spindles normally. These results implied that the Ig-NRG isoforms are sufficient for spindle fiber formation (14). However, this implication seems at odds with the efficacy of the EGF-like domain in vitro, demonstrated here. It may be that, in vivo, the absence of the Ig-containing NRG isoforms reduces or ablates the pool of NRG that is proximal to the developing muscle when bound to heparin sulfate proteoglycans in the basal lamina (50). In vitro, relatively high concentrations of the NRG EGF fragment may make up for the lack of binding and concentration of NRG in the basal lamina.
NRG Induces Muscle Spindle Fiber-Specific Isoforms of the MyHC. Intrafusal myofibers are morphologically and biochemically distinct from contractile (extrafusal) muscle fibers (25, 27). Molecularly, these fibers can be differentiated based on their expression of specific MyHC isoforms (51, 52). With this in mind, we investigated the potential of NRG to induce the expression of MyHC isoforms that are specific to muscle spindle fibers (52–56).
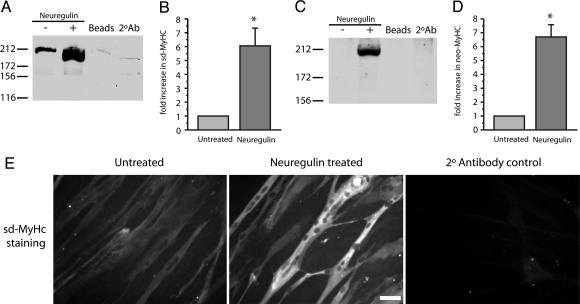
The level of expression was determined by quantifying MyHC-specific fluorescence on immunoblots. The results shown in Fig. 5 demonstrate that NRG induces a significant increase in slow developmental and neonatal isoforms of the MyHC. The expression levels of the slow developmental (Fig. 5B) and neonatal (Fig. 5D) MyHC both increase just over 6-fold, 6.07 ± 1.23 (SEM) and 6.61 ± 0.78 (SEM) fold, respectively, with NRG treatment (n = 4, P < 0.01, t test). Human myotubes stained with anti-slow developmental MyHC monoclonal antibody are shown in Fig. 5E. These photomicrographs show an increase in slow developmental MyHC expression in NRG-treated cultures and that this increase does not appear to be the result of gross morphological changes or increased myotube numbers in treated vs. untreated cultures.
Fig. 5.
NRG induces the expression of muscle-spindle-specific MyHCs. Representative Western blots showing slow developmental MyHC (sd-MyHC) and neonatal MyHC (neo-MyHC) staining are shown in A and C, respectively. Increases in MyHC expression levels were quantified and are shown in B and D. NRG induces a ≈6-fold increase in sd-MyHC (B) and neo-MyHC (D) protein expression in primary human myotubes when compared to untreated cells (n = 4, P < 0.01). Finally, the increase in sd-MyHC by immunocytochemistry is shown in E. Untreated, NRG-treated, and a control for the fluorescently labeled secondary antibody are shown from Left to Right. (Scale bar, 5 μM.)
Potential Spindle Fiber-Related Gene Expression Changes. Beyond the Egr transcription factors and the MyHC isoforms, other NRG induced changes in gene expression may be indicative of muscle spindle formation. The down-regulation of mitochondrial genes involved in ion and proton transport as well as the down-regulation of several components of the cytochrome c complex may reflect a reduction in the energy required by the cultured myofibers consistent with their differentiation into spindle fibers. Prolonged disuse, limb immobilization, and muscle denervation have been shown to have similar effects on mitochondria function in extrafusal muscle (57–59). Furthermore, measurements of contractile strength in frog indicate that spindle fibers generate significantly less force, one-third to one-half the force for a given cross-sectional area, than extrafusal fibers (60). Perhaps the decrease in these transcripts seen here reflects reduced contractile activity in our cultures. An increase in the transcription of several ubiquitin protein ligase transcripts gives further weight to this idea. The E3A and several E2 isoforms of ubiquitin protein ligase have been shown to play a role in muscle atrophy (61). Here, their induction may signal the metabolism of specific contractile proteins that are no longer required. Finally, the induction of collagen IV is noteworthy because it is a significant component of the basal lamina surrounding muscle spindles (62–64).
In synapse formation, the need for reciprocal signaling between innervating sensory neurons and myofibers is obvious. Trophic support from muscle spindle fibers is required to maintain sensory neurons in vivo (22, 65, 66). Our results indicate that the transcripts of two neuronal guidance molecules are up-regulated in response to NRG. In our cultures, exogenous NRG induced the expression of neuropilin 1, a guidance and stop signal for sensory neurons (67), and semaphorin 3C, a molecule that can function as either an attractive or repulsive signal in nerve growth (68) (Table 1). The interplay of these two molecules may effect both sensory and motor neuron innervation of intrafusal and extrafusal muscle.
Conclusion
We have used cDNA microarrays to identify a significant number of NRG induced gene expression changes that together imply a significant role for NRG in the formation of muscle spindle fibers. Egr3, a key transcription factor in spindle fiber development, is regulated by the EGF β1 domain of NRG, as are a significant number of genes that may be involved in myofiber differentiation and contractile function. These results provide additional insight into the development of muscle spindle fibers and an in vitro system that may be useful for studies of muscle spindle development.
Supplementary Material
Acknowledgments
We thank Drs. Daniel Kastner, Story Landis, and Jeffery Trent for supporting this work at the National Institutes of Health. Further thanks go to Mary Anne Mann and Yidong Chen for comments and suggestions provided during the preparation of this manuscript.
Abbreviations: NRG, neuregulin; EGF, epidermal growth factor; AChR, acetylcholine receptor; Egr, early growth response; MyHC, myosin heavy chain.
References
- 1.Burden, S. & Yarden, Y. (1997) Neuron 18, 847-855. [DOI] [PubMed] [Google Scholar]
- 2.Lemke, G. (1996) Mol. Cell. Neurosci. 7, 247-262. [DOI] [PubMed] [Google Scholar]
- 3.Buonanno, A. & Fischbach, G. D. (2001) Curr. Opin. Neurobiol. 11, 287-296. [DOI] [PubMed] [Google Scholar]
- 4.Marchionni, M. A., Grinspan, J. B., Canoll, P. D., Mahanthappa, N. K., Salzer, J. L. & Scherer, S. S. (1997) Ann. N.Y. Acad. Sci. 825, 348-365. [DOI] [PubMed] [Google Scholar]
- 5.Meyer, D. & Birchmeier, C. (1995) Nature 378, 386-390. [DOI] [PubMed] [Google Scholar]
- 6.Falls, D. L., Rosen, K. M., Corfas, G., Lane, W. S. & Fischbach, G. D. (1993) Cell 72, 801-815. [DOI] [PubMed] [Google Scholar]
- 7.Chu, G. C., Moscoso, L. M., Sliwkowski, M. X. & Merlie, J. P. (1995) Neuron 14, 329-339. [DOI] [PubMed] [Google Scholar]
- 8.Jo, S. A., Zhu, X., Marchionni, M. A. & Burden, S. J. (1995) Nature 373, 158-161. [DOI] [PubMed] [Google Scholar]
- 9.Trachtenberg, J. T. & Thompson, W. J. (1996) Nature 379, 174-177. [DOI] [PubMed] [Google Scholar]
- 10.Kopp, D. M., Trachtenberg, J. T. & Thompson, W. J. (1997) J. Neurosci. 17, 6697-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suarez, E., Bach, D., Cadefau, J., Palacin, M., Zorzano, A. & Guma, A. (2001) J. Biol. Chem. 276, 18257-18264. [DOI] [PubMed] [Google Scholar]
- 12.Kim, D., Chi, S., Lee, K. H., Rhee, S., Kwon, Y. K., Chung, C. H., Kwon, H. & Kang, M. S. (1999) J. Biol. Chem. 274, 15395-15400. [DOI] [PubMed] [Google Scholar]
- 13.Andrechek, E. R., Hardy, W. R., Girgis-Gabardo, A. A., Perry, R. L., Butler, R., Graham, F. L., Kahn, R. C., Rudnicki, M. A. & Muller, W. J. (2002) Mol. Cell. Biol. 22, 4714-4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hippenmeyer, S., Shneider, N. A., Birchmeier, C., Burden, S. J., Jessell, T. M. & Arber, S. (2002) Neuron 36, 1035-1049. [DOI] [PubMed] [Google Scholar]
- 15.Leu, M., Bellmunt, E., Schwander, M., Farinas, I., Brenner, H. R. & Muller, U. (2003) Development (Cambridge, U.K.) 130, 2291-2301. [DOI] [PubMed] [Google Scholar]
- 16.Usdin, T. B. & Fischbach, G. D. (1986) J. Cell Biol. 103, 493-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falls, D. L., Harris, D. A., Johnson, F. A., Morgan, M. M., Corfas, G. & Fischbach, G. D. (1990) Cold Spring Harbor Symp. Quant. Biol. 55, 397-406. [DOI] [PubMed] [Google Scholar]
- 18.Martinou, J. C., Falls, D. L., Fischbach, G. D. & Merlie, J. P. (1991) Proc. Natl. Acad. Sci. USA 88, 7669-7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corfas, G. & Fischbach, G. D. (1993) J. Neurosci. 13, 2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gramolini, A. O., Angus, L. M., Schaeffer, L., Burton, E. A., Tinsley, J. M., Davies, K. E., Changeux, J. P. & Jasmin, B. J. (1999) Proc. Natl. Acad. Sci. USA 96, 3223-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milburn, A. (1973) J. Cell Sci. 12, 175-195. [DOI] [PubMed] [Google Scholar]
- 22.Ernfors, P., Lee, K. F., Kucera, J. & Jaenisch, R. (1994) Cell 77, 503-512. [DOI] [PubMed] [Google Scholar]
- 23.Liebl, D. J., Tessarollo, L., Palko, M. E. & Parada, L. F. (1997) J. Neurosci. 17, 9113-9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tourtellotte, W. G., Keller-Peck, C., Milbrandt, J. & Kucera, J. (2001) Dev. Biol. 232, 388-399. [DOI] [PubMed] [Google Scholar]
- 25.Boyd, I. A. (1968) Electroencephalogr. Clin. Neurophysiol. 25, 406-407. [PubMed] [Google Scholar]
- 26.Werner, J. K. (1974) Am. J. Phys. Med. 53, 127-142. [PubMed] [Google Scholar]
- 27.Maier, A. (1997) Int. J. Dev. Biol. 41, 1-17. [PubMed] [Google Scholar]
- 28.Meyer, D., Yamaai, T., Garratt, A., Riethmacher-Sonnenberg, E., Kane, D., Theill, L. E. & Birchmeier, C. (1997) Development (Cambridge, U.K.) 124, 3575-3586. [DOI] [PubMed] [Google Scholar]
- 29.Marchionni, M. A., Goodearl, A. D., Chen, M. S., Bermingham-McDonogh, O., Kirk, C., Hendricks, M., Danehy, F., Misumi, D., Sudhalter, J., Kobayashi, K., et al. (1993) Nature 362, 312-318. [DOI] [PubMed] [Google Scholar]
- 30.Loeb, J. A., Khurana, T. S., Robbins, J. T., Yee, A. G. & Fischbach, G. D. (1999) Development (Cambridge, U.K.) 126, 781-791. [DOI] [PubMed] [Google Scholar]
- 31.Tourtellotte, W. G. & Milbrandt, J. (1998) Nat. Genet. 20, 87-91. [DOI] [PubMed] [Google Scholar]
- 32.Ferns, M., Hoch, W., Campanelli, J. T., Rupp, F., Hall, Z. W. & Scheller, R. H. (1992) Neuron 8, 1079-1086. [DOI] [PubMed] [Google Scholar]
- 33.Ferns, M. J., Campanelli, J. T., Hoch, W., Scheller, R. H. & Hall, Z. (1993) Neuron 11, 491-502. [DOI] [PubMed] [Google Scholar]
- 34.Jacobson, C., Cote, P. D., Rossi, S. G., Rotundo, R. L. & Carbonetto, S. (2001) J. Cell Biol. 152, 435-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobson, C., Montanaro, F., Lindenbaum, M., Carbonetto, S. & Ferns, M. (1998) J. Neurosci. 18, 6340-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan, J., Bittner, M. L., Chen, Y., Meltzer, P. S. & Trent, J. M. (1999) Biochim. Biophys. Acta 1423, M17-M28. [DOI] [PubMed] [Google Scholar]
- 37.Khan, J., Simon, R., Bittner, M., Chen, Y., Leighton, S. B., Pohida, T., Smith, P. D., Jiang, Y., Gooden, G. C., Trent, J. M. & Meltzer, P. S. (1998) Cancer Res. 58, 5009-5013. [PubMed] [Google Scholar]
- 38.Chen, Y., Dougherty, E. & Bittner, M. (1997) J. Biomed. Opt. 2, 364-374. [DOI] [PubMed] [Google Scholar]
- 39.Ferns, M., Deiner, M. & Hall, Z. (1996) J. Cell Biol. 132, 937-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallace, B. G. (1994) J. Cell Biol. 125, 661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace, B. G. (1992) Science 257, 50-56.1621094 [Google Scholar]
- 42.Borges, L. S. & Ferns, M. (2001) J. Cell Biol. 153, 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen, Y., Kamat, V., Dougherty, E. R., Bittner, M. L., Meltzer, P. S. & Trent, J. M. (2002) Bioinformatics 18, 1207-1215. [DOI] [PubMed] [Google Scholar]
- 44.Xuan, Z., Wang, J. & Zhang, M. Q. (2003) Genome Biol. 4, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen, H., Herndon, M. E. & Lawler, J. (2000) Matrix Biol. 19, 597-614. [DOI] [PubMed] [Google Scholar]
- 46.Wilczak, N., de Vos, R. A. & De Keyser, J. (2003) Lancet 361, 1007-1011. [DOI] [PubMed] [Google Scholar]
- 47.Aoyama, S., Shirakawa, O., Ono, H., Hashimoto, T., Kajimoto, Y. & Maeda, K. (2003) Psychiatry Clin. Neurosci. 57, 545-547. [DOI] [PubMed] [Google Scholar]
- 48.Toyooka, K., Iritani, S., Makifuchi, T., Shirakawa, O., Kitamura, N., Maeda, K., Nakamura, R., Niizato, K., Watanabe, M., Kakita, A., et al. (2002) J. Neurochem. 83, 797-806. [DOI] [PubMed] [Google Scholar]
- 49.Siomi, M. C., Siomi, H., Sauer, W. H., Srinivasan, S., Nussbaum, R. L. & Dreyfuss, G. (1995) EMBO J. 14, 2401-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loeb, J. A. & Fischbach, G. D. (1995) J. Cell Biol. 130, 127-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedrosa-Domellof, F., Gohlsch, B., Thornell, L. E. & Pette, D. (1993) FEBS Lett. 335, 239-242. [DOI] [PubMed] [Google Scholar]
- 52.Pedrosa-Domellof, F. & Thornell, L. E. (1994) J. Histochem. Cytochem. 42, 77-88. [DOI] [PubMed] [Google Scholar]
- 53.Kucera, J. & Walro, J. M. (1995) Anat. Embryol. (Berlin) 192, 149-158. [DOI] [PubMed] [Google Scholar]
- 54.Kucera, J., Walro, J. M. & Gorza, L. (1992) J. Histochem. Cytochem. 40, 293-307. [DOI] [PubMed] [Google Scholar]
- 55.Pedrosa, F., Butler-Browne, G. S., Dhoot, G. K., Fischman, D. A. & Thornell, L. E. (1989) Histochemistry 92, 185-194. [DOI] [PubMed] [Google Scholar]
- 56.Ecob-Prince, M., Hill, M. & Brown, W. (1989) J. Neurol. Sci. 90, 167-177. [DOI] [PubMed] [Google Scholar]
- 57.Booth, F. W. & Kelso, J. R. (1973) Can. J. Physiol. Pharmacol. 51, 679-681. [DOI] [PubMed] [Google Scholar]
- 58.Thomason, D. B. & Booth, F. W. (1989) Adv. Myochem. 2, 79-82. [PubMed] [Google Scholar]
- 59.Wicks, K. L. & Hood, D. A. (1991) Am. J. Physiol. 260, C841-C850. [DOI] [PubMed] [Google Scholar]
- 60.Edman, K. A., Radzyukevich, T. & Kronborg, B. (2002) J. Physiol. 541, 905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lecker, S. H. (2003) Curr. Opin. Clin. Nutr. Metab. Care 6, 271-275. [DOI] [PubMed] [Google Scholar]
- 62.Maier, A. & Mayne, R. (1995) Dev. Dyn. 202, 284-293. [DOI] [PubMed] [Google Scholar]
- 63.Schroder, J. M., Bodden, H., Hamacher, A. & Verres, C. (1989) Muscle Nerve 12, 221-232. [DOI] [PubMed] [Google Scholar]
- 64.Maier, A. & Mayne, R. (1987) Am. J. Anat. 180, 226-236. [DOI] [PubMed] [Google Scholar]
- 65.Klein, R., Silos-Santiago, I., Smeyne, R. J., Lira, S. A., Brambilla, R., Bryant, S., Zhang, L., Snider, W. D. & Barbacid, M. (1994) Nature 368, 249-251. [DOI] [PubMed] [Google Scholar]
- 66.Kucera, J., Fan, G., Jaenisch, R., Linnarsson, S. & Ernfors, P. (1995) J. Comp. Neurol. 363, 307-320. [DOI] [PubMed] [Google Scholar]
- 67.Bagri, A. & Tessier-Lavigne, M. (2002) Adv. Exp. Med. Biol. 515, 13-31. [PubMed] [Google Scholar]
- 68.Bagnard, D., Lohrum, M., Uziel, D., Puschel, A. W. & Bolz, J. (1998) Development (Cambridge, U.K.) 125, 5043-5053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.