Abstract
Aim:
Folic acid (FA)-decorated polyamidoamine dendrimer G4 (G4-FA) was synthesized and studied for targeted delivery of genes to head and neck cancer cells expressing high levels of folate receptors (FRs).
Methods:
Cellular uptake, targeting specificity, cytocompatibility and transfection efficiency were evaluated.
Results:
G4-FA competes with free FA for the same binding site. G4-FA facilitates the cellular uptake of DNA plasmids in a FR-dependent manner and selectively delivers plasmids to FR-high cells, leading to enhanced gene expression.
Conclusion:
G4-FA is a suitable vector to deliver genes selectively to head and neck cancer cells. The fundamental understandings of G4-FA as a vector and its encouraging transfection results for head and neck cancer cells provided support for its further testing in vivo.
Keywords: : folate receptor targeting, gene therapy, head and neck squamous cell carcinoma, PAMAM dendrimer
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide [1]. Although advances have been made in conventional therapy including surgery, chemotherapy and radiation, their impact on improvement of 5-year mortality rate of patients with HNSCC is still minimal [2]. Increased understanding of molecular events involved in HNSCC initiation and progression has led to the development of gene therapy as a potential treatment option [3–8]. To realize gene therapy of HNSCC, it is essential to utilize safe and efficient vectors for delivery. Not much success has been reported for HNSCC gene therapy. Folate receptor alpha (FRα) is typically expressed at very low levels in normal tissues. In contrast, to meet the folate demand of rapidly dividing cells in many cancers including HNSCC, FRα is expressed at significantly elevated levels. Therefore, it can be targeted for developing active tumor-targeting therapies [9–11]. Although FR-targeted drug and gene delivery systems have been developed on the basis of dendrimers [12–17] and many other types of nanoparticle carriers [18–23], few nonviral vectors have been developed for HNSCC [24,25]. Given the scarcity of nonviral vectors for gene therapy of HNSCC and limited clinical outcomes, it is highly desirable to develop and evaluate vectors under the context of HNSCC.
In this work, we report on a study that utilizes polyamidoamine (PAMAM) dendrimer for selective delivery of genes to HNSCC and provides in-depth understanding of how gene delivery and transfection in head and neck squamous cancer cells can be enhanced via FR-targeted PAMAM dendrimers. PAMAM dendrimers bearing amines at the surface are appealing vectors because they not only possess inherent functions for gene delivery [26,27] but offer controlled nanoscale building blocks for construction of defined delivery systems with multifunctionality [28,29]. Although high generation polycationic PAMAM dendrimers tend to be more efficient in gene delivery, their generation-dependent toxicity is more prominent and surface functionalization is needed to maintain cytocompatibility and buffering capacity. Furthermore, large-scale synthesis of functionalized high generation dendrimers under good manufacturing practice conditions is technically challenging and potentially impedes their clinical applications [30–32]. For HNSCCs that are localized to the oral cavity and oropharyngeal region and that are relatively easily accessible, local delivery of therapeutic genes is a possibility, thus eliminating the need of PEGylation of dendrimers, a conventional way to extend circulation lifetime of nanoparticles following systemic delivery. Targeted therapy for localized cancers of HNSCC is preferred as it will spare healthy tissue and promote uptake by tumor cells. With the considerations discussed above, we chose low generation PAMAM dendrimer G4 as the underlying carrier and functionalized with folic acid (FA), a natural ligand of FR. We thoroughly examined the fundamental aspects of the constructed G4-FA conjugates as a vector in connection to HNSCC, including synthesis and characterization, mechanisms and kinetics of cellular uptake, and transfection efficiency.
Methods
Materials
Diaminobutane (DAB) core PAMAM dendrimer generation 4.0 (technical grade) was purchased from NanoSynthons (MI, USA). Dimethyl sulfoxide (DMSO), FA, TFA, formaldehyde solution (37 wt.% in H2O), 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC), N,N-diisopropylethylamine, deuterium oxide (D2O; 99.9 atom% D), deuterated dimethyl sulfoxide (DMSO-d6), and fluorescein isothiocyanate (FITC) were purchased from Sigma-Aldrich (MO, USA). Acetonitrile (ACN), water (HPLC grade), Triton X-100, 4′,6-diamidino-2-phenylindole (DAPI), sodium hydroxide, and phosphate-buffered saline (PBS) were purchased from Fisher Scientific (PA, USA). Ethidium bromide, Dulbecco's modified Eagle medium (DMEM), trypsin-EDTA (0.25%), and penicillin-streptomycin (10,000 U/ml) were purchased from Life Technologies (CA, USA). Label IT® plasmid delivery control Cy3® (referred to as Cy3 plasmid hereafter) was purchased from Mirus Bio (WI, USA). Vectashield mounting medium was purchased from Vector Laboratories (CA, USA). Polyvinylidene difluoride (PVDF) membrane was purchased from Millipore (MA, USA). Western lightning Plus ECL was purchased from Perkin-Elmer (MA, USA). Cosmic calf serum (CS) and pMAX-GFP (pGFP) plasmid were purchased from Lonza (MD, USA). GFP and β-actin antibody were purchased from Santa Cruz Biotechnology (CA, USA). SnakeSkin dialysis tubing with 7,000 molecular weight cut-off (MWCO) and FRα antibody were purchased from Thermo Scientific (IL, USA). Goat anti-rabbit antibody conjugated to horseradish peroxidase and goat anti-mouse antibody conjugated to horseradish peroxidase were purchased from Bio-Rad (CA, USA). WST-1 cell proliferation reagent, protease inhibitor and phosphatase inhibitor cocktail tablets were purchased from Roche Applied Science (NY, USA). Propidium iodide (PI) was purchased from BD Biosciences (CA, USA).
Synthesis
FA (18.6 mg, 42.2 μmol, MW = 441.4 g/mol) was allowed to react with EDC (113.3 mg, 590.9 μmol, MW = 191.71 g/mol) in a mixture of 12 ml of DMF and 4 ml of DMSO for 1 h. The organic reaction mixture was added dropwise to 50 ml of DI water solution containing 100 mg (7. 03 μmol) of PAMAM dendrimer G4 (MW = 14215 g/mol). The reaction mixture was vigorously stirred for 2 days and then dialyzed against DI water using dialysis tubing with MWCO of 7 kDa for 2 days. The resultant G4-FA conjugates were obtained after lyophilization. To prepare FITC-labeled dendrimers, G4-FA and G4 (dissolved in PBS) were reacted with FITC dissolved in DMSO at the feed molar ratio of 1:5 in the dark for a day in the presence of N-diisopropylethylamine in an excess amount. After dialyis against DI water using dialysis tubing with MWCO of 7 kDa for 2 days, the resultant FITC-labeled conjugates were lyophilized. The number of FA and FITC molecules coupled to each dendrimer was quantified by using by using Genesys 6 spectrophotometer (Thermo Scientific).
High performance liquid chromatography
The reverse-phase high performance liquid chromatography (RP-HPLC) system (Waters, MA, USA) consisting of a system Waters 1515 isocratic HPLC pump, a model Waters 717plus autosampler and a model Waters 2487 dual λ absorbance detector was used in this work. An XTerra particle-based RP-HPLC column (length 150 mm, particle size 5 μm, RP18) was purchased from Waters (MA, USA). The mobile phase was H2O:ACN:TFA (750:250:0.38, v/v/v) at a flow rate of 1 ml/min [33].
Proton nuclear magnetic resonance (1H NMR) spectroscopy
1H NMR spectra were recorded on a Varian superconducting fourier-transform NMR spectrometer (Mercury-300).
Polyplex preparation
Dendrimer/DNA plasmid polyplexes at various weight ratios (wt/wt) of dendrimer to plasmid were prepared by vortexing a mixture of dendrimer and DNA plasmid with defined amounts, both of which were predissolved in medium (filtered PBS for size and ζ-potential measurements, DMEM for biological assays) for 10 s followed by equilibration at room temperature for 30 min. The prepared polyplexes were used immediately.
Particle size & ζ-potential measurements
The particle size and ζ-potential of G4 dendrimer and its derivatives (0.5 mg/ml) and polyplexes on the basis of 10 µg/ml of pMAX-GFP plasmid in PBS (filtered through 20 nm filter) were measured at room temperature using a Malvern Zetasizer Nano ZS90 apparatus (Malvern Instruments, Worcestershire, UK).
Gel retardation assay
G4-FA/pMAX-GFP polyplexes formed at various weight ratios (wt/wt) of dendrimer to plasmid (0 μg/2 μg, 0.4 μg/2 μg, 1 μg/2 μg, 2 μg/2 μg, 10 μg/2 μg, 20 μg/2 μg, and 40 μg/2 μg in 500 μl of DMEM) were prepared (Supplementary Table 1). The stability of the prepared polyplexes was examined on the basis of electrophoretic mobility. In general, 20 μl of solution having polyplexes was loaded into a 1% agarose gel containing ethidium bromide (0.5 μg/ml) and subjected to electrophoresis at 100 V for 1 h. The DNA bands were visualized under a UV transilluminator (Alpha Innotech, ProteinSimple, CA, USA) [34].
Cell culture
Multiple cell lines were used in the studies including HN4 cells and HN6 cells, derived from primary squamous cell carcinomas of the head and neck, HN12 cells derived from an HNSCC lymph node metastasis, T98, U87 and U1242 glioblastoma cells and NIH3T3 mouse fibroblasts as an FRα negative control. In general, cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% Cosmic calf serum, 100 units/ml of penicillin and 100 μg/ml of streptomycin at 37°C in 95% air/5% CO2 [35–39].
FRα expression quantification
FRα levels in HN4, HN6, HN12, T98, U87, U1242 and NIH3T3 cells were quantified. Typically, cells were seeded in the six-well plates at a density of 20,000 cells/well. When reaching 80% confluence, they were harvested using cell lysis buffer containing protease and phosphorylase inhibitors and RNA lysis buffer for western blot analysis and real-time PCR, respectively.
Cytocompatibility assessment
HN12 cells were seeded in a 96-well plate at a density of 10,000 cells/well and allowed to attach overnight. For polymer cytocompatibility assessment, the cells were incubated with G4 (0–1000 μg/ml) and G4-FA (0–1000 μg/ml) at 37°C for 48 h. For polyplex cytocompatibility assessment, the cells were incubated with G4/pGFP plasmid and G4-FA/pGFP plasmid polyplexes at 37°C for 48 h, followed by another 48-h culture in a medium supplemented with 10% Cosmic calf serum. At the end of each treatment, the cell viability was determined by WST-1 cell proliferation assay following the manufacturer's protocol [40].
Cellular uptake kinetics of dendrimer carriers alone & plasmid complexed with dendrimers
HN12 cells were seeded in the 60-mm dishes at a density of 50,000 cells/dish. The cells were allowed to attach and grow overnight. Prior to treatment, the medium was removed, and the cells were washed with PBS once. The cells were then incubated with FITC-G4 (5 μg predissolved in 300 μl of DMEM), FITC-G4-FA (5 μg predissolved in 300 μl of DMEM), or Cy3-labeled plasmid complexed with dendrimers (5 μg of G4/1 μg of plasmid or 5 μg of G4-FA/1 μg of plasmid predissovled in 500 μl of DMEM), and the final volume was brought up to 3 ml with DMEM supplemented with 10% serum. The cells were maintained at 37°C for various lengths of time (0, 1, 2, 6 and 24 h). Time point 0 h indicates that HN12 cells were subjected to flow cytometry analysis immediately after dendrimers or dendrimer/plasmid polyplexes were added to the cells. The experiments were conducted on three independent occasions. At end of each treatment, the cells were washed with PBS three-times, resuspended following trypsinization for 5–10 min, centrifuged, washed with PBS once and transferred to microcentrifuge tubes. The processed cells were subjected to flow cytometry analysis by using a Guava EasyCyte mini flow cytometry system (Millipore, MA, USA) [32]. The mean fluorescence intensity values of FITC for quantification of dendrimer carriers and Cy3 for quantification of payload, that is, plasmid, in the cells were reported.
Cellular uptake of FITC-labeled dendrimer carriers and Cy3-labeled plasmid was also monitored by fluorescence imaging. HN12 cells were seeded in 6-well plates at the density of 20,000 cells/well and allowed to attach overnight. The cells received the same treatments as those in flow cytometry studies above. At the end of each treatment, HN12 cells were washed with PBS three-times, fixed with 4% formaldehyde at room temperature for 20 min, washed with PBS three-times, permeated with 0.1% Triton X-100 for 5 min and washed with PBS three-times. The cell nuclei were counterstained with DAPI for 5 min, and the cells were washed with PBS three-times. Fluorescence images were taken on a Zeiss Axiovert 200 inverted fluorescence microscope (Carl Zeiss Microimaging, NY, USA) [34].
Intracellular colocalization studies by confocal microscopy
Confocal microscopy was applied for further illustration of intracellular distribution of FITC-G4-FA complexed with Cy3-plasmid. HN12 were seeded onto microscope glass coverslips (Fisher Scientific, PA, USA) positioned in 6-well plates at a density of 20,000 cells/well and allowed to attach overnight. The spent medium was removed, and the cells were washed with PBS once. The HN12 cells were incubated with FITC-G4-FA/Cy3-plasmid polyplexes (5 µg/1 µg) predissolved in 500 μl of DMEM. The final volume was brought up to 3 ml with DMEM supplemented with 10% serum. After 24 h, the cells were processed for confocal fluorescence imaging following the protocol described above. Fluorescence images at a magnification of 630× were collected on a Zeiss LSM 700 confocal laser scanning microscope.
For comparison, intracellular distribution of targeted dendrimer carrier alone, that is, FITC-G4-FA, was studied using another FR-high cell line, that is, T98. Briefly, T98 cells were seeded onto microscope glass coverslips positioned in 6-well plates at a density of 20,000 cells/well and allowed to attach overnight. The T98 cells were incubated with FITC-G4-FA at 37°C for 24 h and processed for confocal imaging. In particular, the cells were fixed in cold methanol for 15 min, air dried, washed with PBS, blocked in 1.5% normal goat serum for 1 h, incubated with monoclonal Rab5 antibody in the blocking buffer at 4°C overnight, washed with PBS, incubated with Dylight 594-conjugated antirabbit antibody for 1 h and washed with PBS again. T98 cell nuclei were counterstained with DAPI (blue) for 5 min, and the cells were washed with PBS three-times [40].
Effect of free FA on cellular uptake of dendrimers & dendrimer/plasmid polyplexes
HN12 cells were seeded in 60-mm dishes at a density of 50,000 cells/dish. The cells were allowed to attach and grow overnight. To examine how FA affects cellular uptake kinetics of dendrimer carriers alone or complexed with plasmid in a short term, the cells were incubated with final concentrations of FITC-G4 (10 μg/ml), FITC-G4-FA (10 μg/ml), FITC-G4/Cy3 plasmid (5 µg/1 µg/3 ml), FITC-G4-FA/Cy3-plasmid (5 µg/1 µg/3 ml) in the absence or presence of free FA (0.5 mg/ml) at 37°C for 1 h or 2 h. The cells were then processed following the protocols described earlier and subjected to flow cytometry analysis and fluorescence imaging, respectively.
Competitive inhibition assay was conducted to further confirm targeting specificity of FA conjugated to the dendrimer. Briefly, HN12 cells were seeded in 60-mm dishes at a density of 50,000 cells/dish. After overnight culture, they were incubated with FITC-G4-FA (10 μg/ml) in the presence of increasing concentrations of free FA up to 2 mg/ml for 2 h and analyzed by using flow cytometry.
Tumor cell targeting assessment using a co-culture model
An FR-high/FR-low co-culture model was used in this study. Briefly, equal numbers of yellow fluorescent protein (YFP)-expressing HN12 cells (HN12-YFP) (10,000 cells/well) and U87 cells (10,000 cells/well) were seeded in the 6-well plates and cultured overnight. The cells were incubated with G4/Cy3 plasmid or G4-FA/Cy3 plasmid (5 µg/1 µg in 3 ml of medium). Uptake of polyplexes by HN12-YFP and U87 cells was monitored by using fluorescence microscopy. Cy3-positive HN12 and U87 cells at 6 h post-treatment on nine randomly selected fields on fluorescence images and normalized to the total cell number of corresponding cell type in each treatment.
Transfection efficiency evaluation
HN12 cells were seeded at 20,000 cells/well in 6-well plates for fluorescence imaging and western blotting studies and at 50,000 cells/dish in 60-mm dishes for flow cytometry. After they were cultured overnight, the spent medium was removed, and the cells were washed with PBS. The cells were incubated with G4-FA/pGFP(or pYFP) polyplexes or G4/pGFP(or pYFP) polyplexes (5 µg/1 µg in 3 ml DMEM supplemented with 10% serum) at 37°C for 48 h. The spent medium was replaced with fresh medium containing 10% serum and cultured for another 48 h. Afterward, western blotting was applied for quantification of YFP expression levels following the procedure described previously [41]. Flow cytometry was applied to quantify GFP-expressing cell populations. Fluorescent images of the cells post-transfection were taken on a Zeiss Axiovert 200 inverted fluorescence microscope. All the experiments were repeated at least three-times.
Statistical analysis
In cellular uptake kinetics studies, curve fitting models were compared by running Akaike information criterion (AIC) test using GraphPad Prism 5 (CA, USA). Models with lower AIC values are more likely to be correct. All the data were expressed as means ± standard error of the mean. Student's t-test was performed for comparison. A value of p < 0.05 was considered statistically significant.
Results
FRα expression levels
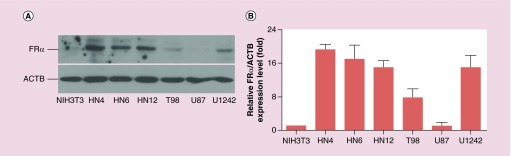
FRα expression levels were evaluated in selected head and neck cancer cell lines and glioblastoma cell lines. The NIH3T3 cells were used as an FRα-negative control. As shown in Figure 1, all three HNSCC (HN4, HN6 and HN12 cells) and glioblastoma (T98 and U1242 cells) express relatively high levels of FRα. In contrast, FRα expression in U87 cells is low. This result was further confirmed by real-time PCR analysis. The mRNA expression level of FRα in U87 cells was significantly lower than those in HN12 and T98 cells (Supplementary Figure 1). HN12 cells were used in the later studies as a representative HNSCC cell line. And U87 cells were cultured with HN12 cells to establish an FR-high/FR-low co-culture model to assess tumor cell targeting of G4-FA.
Figure 1. . Endogenous expression levels of folate receptor α as determined by western blot analysis conducted on four independent occasions (n = 4).
A representative image is shown. FRα: Folate receptor α.
Synthesis & characterization of dendrimer derivatives & polyplexes
G4-FA and FITC-labeled conjugates were successfully synthesized (Supplementary Figure 2) and fully characterized for assessment of purity and compositions. The 1H NMR spectra confirmed the presence of the aromatic proton peaks of FA (8.5, 7.6, 6.7 ppm) and G4 (multiple methylene proton peaks between 3.6 and 2.1 ppm) in the resulting G4-FA conjugates (Supplementary Figure 3). The HPLC chromatogram confirmed the purity of G4-FA and FITC-labeled conjugates after dialysis (Supplementary Figure 4). By calculating the area under the curve, the purity of G4-FA, FITC-G4-FA and FITC-G4 conjugates was about 97, 97 and 95%, respectively. UV-Visible spectroscopy analysis revealed that an average of 2.3 and 2.2 FITC molecules were successfully conjugated onto each G4-FA conjugate and G4 dendrimer, respectively. The molecular weight of dendrimer and its derivatives are summarized in Supplementary Table 2.
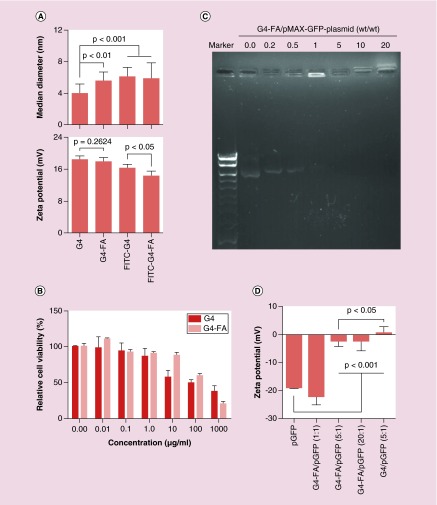
DLS results showed the ζ-potential of G4-FA, FITC-G4-FA and FITC-G4 conjugates remained positive, an essential feature for polyplexation. Of note, the ζ-potential of G4-FA conjugates was not significantly different from that of G4 dendrimer; however, the ζ-potential of FITC-G4-FA conjugates was significantly lower than that of FITC-G4 conjugates (Figure 2A). The size of G4-FA, FITC-G4-FA and FITC-G4 conjugates significantly increased compared with that of G4 dendrimer (Figure 2A), reflecting the surface modification on the dendrimer. The ζ-potential of G4 after conjugation with FA and/or FITC reduced slightly but remained relatively high. Our study confirmed dose-dependent toxicity of both G4 dendrimer and G4-FA conjugates (Figure 2B). However, G4-FA has improved cytocompatibility. The half maximal inhibitory concentrations (IC50) of G4 dendrimer and G4-FA conjugates were 67 μg/ml (4.7 μM) and 159 μg/ml (10.1 μM), respectively. A weight ratio of 1:1 or higher is sufficient in stably complexing DNA plasmid (Figure 2C). G4-FA/pGFP polyplexes become less negative with increasing weight ratio (Figure 2D).
Figure 2. . Characterization of dendrimers and dendrimer/plasmid polyplexes.
(A) The size and ζ-potential of G4 dendrimer and its derivatives. (B) The cytocompatibility of G4 dendrimer and G4-FA in HN12 cells as determined by WST-1 assay. (C) The complexation stability of G4-FA/GFP plasmid at various weight ratios. (D) The ζ-potential of GFP plasmid and its polyplexes formed with dendrimers. Sample size (n) was in the range of 4–9 for the above experiments.
Cellular uptake of G4-FA via FR-mediated endocytosis
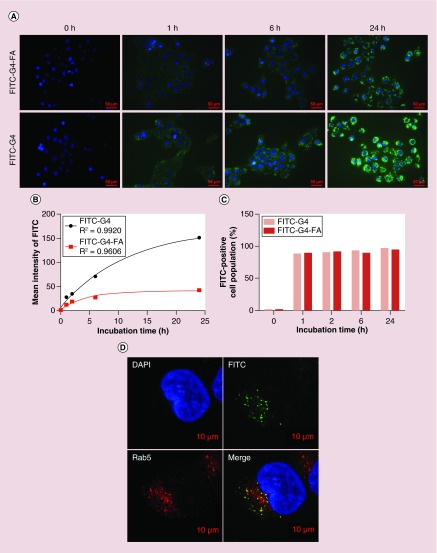
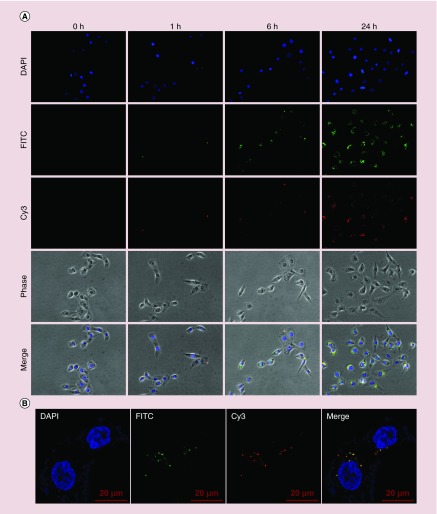
The cellular uptake efficiency of nanoparticles directly affects therapeutic effects [42]. We studied the cellular uptake of FITC-labeled G4 dendrimer and G4-FA conjugates in FR-high HN12 cells. Fluorescence images showed, both FITC-G4-FA and FITC-G4 conjugates were time-dependently taken up by HN12 cells (Figure 3A). Notably, stronger FITC fluorescence was observed in the cells treated with FITC-G4 conjugates likely due to their higher positive ζ-potential (Figure 2A). We quantified the cellular uptake kinetics of FITC-G4 and FITC-G4-FA conjugates in HN12 cells by using flow cytometry analysis (Supplementary Figure 5 and plotted in Figure 3B). The cellular uptake of FITC reached a plateau within 24 h in the cells treated with FITC-G4-FA conjugates. HN12 cells kept taking up FITC-G4 conjugates within 24 h. Over 90% cells were FITC-positive following both FITC-G4 and FITC-G4-FA conjugates treatment (Figure 3C). It is known that polycationic dendrimers, for example, G4, enters early endosomes and is then routed toward the late endosomes and lysosomes [43,44]. Similarly, FITC-G4-FA conjugates were localized in endosome (stained with Rab5) as illustrated in another FRα-high T98 cells (Figure 3D).
Figure 3. . Cellular uptake and intracellular distribution of dendrimer carriers in HN12 cells.
(A) Fluorescence images of cellular uptake of conjugates at selected time points. (B) Cellular uptake kinetics as determined by flow cytometry (Supplementary Figure 5). (C) Quantification of FITC-positive cell populations as determined by flow cytometry. (D) Colocalization of FITC-G4-FA with early endosomes (Rab5) observed in FR-high cell line (T98 cells) at 24 h.
FR: Folate receptor.
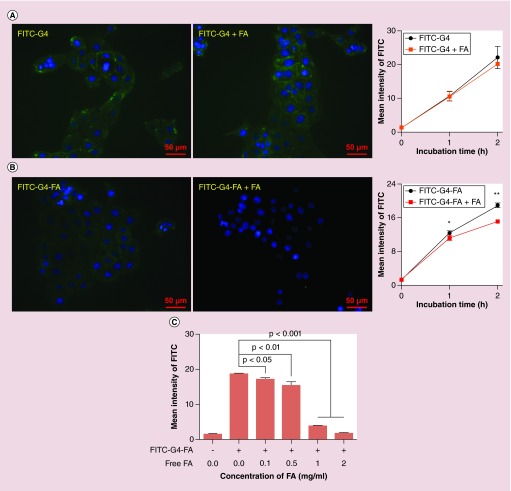
Cellular uptake of FITC-G4 was not influenced by free FA at the concentration of 0.5 mg/ml as illustrated by fluorescence imaging (Figure 4A & Supplementary Figure 6A). In contrast, uptake of FITC-G4-FA was significantly reduced by the addition of free FA (Figure 4B & Supplementary Figure 6B). More pronounced uptake inhibition of FITC-G4-FA was observed when FA concentration was increased to 1 and 2 mg/ml (Supplementary Figure 7 and plotted in Figure 4C).
Figure 4. . Effects of free folic acid on cellular uptake of dendrimer carriers in HN12 cells.
(A) Fluorescence images following 1-h incubation are presented. (B) Cellular uptake of FITC-G4-FA was determined by using flow cytometry and plotted upon Supplementary Figure 6. Fluorescence images following 1-h incubation are presented. (C) Competitive inhibition assay on FITC-G4-FA by flow cytometry and plotted in Supplementary Figure 7 (n = 6).
*p < 0.05; **p < 0.01.
FA: Folic acid.
G4-FA mediates uptake of plasmids in an FR-dependent manner & shows high tumor selectivity
We have previously shown G4/Cy3 plasmid polyplexes were gradually internalized into 293T cells in 24 h [34]. In the current study, time lapse imaging and colocalization results qualitatively show a time-dependent internalization of G4-FA/plasmid polyplexes (Figure 5A). Both dendrimer and plasmid were internalized by more than 90% of HN12 cells in at 24-h post-transfection, indicating 24-h is a proper incubation period for plasmid uptake. Confocal microscopy was applied to closely examine intracellular distribution of G4-FA and plasmid. It was clearly observed that polyplexes (yellow) had started the dissociation process to release individual vector (green) and plasmid (red) as shown in Figure 5B.
Figure 5. . Cellular uptake and intracellular distribution of FITC-G4-FA/Cy3-plasmid polyplexes in HN12 cells.
(A) Cellular uptake of FITC-G4-FA/Cy3-plasmid polyplexes in HN12 cells over a period of 24 h was monitored with fluorescence microscopy (200×). (B) Intracellular distribution of FITC-G4-FA/Cy3-plasmid polyplexes in HN12 cells at 24-h incubation illustrated in the confocal images (630×).
We further quantified the cellular uptake of G4-FA/Cy3-plasmid and G4/Cy3-plasmid polyplexes by using flow cytometry. The cellular uptake of Cy3-plasmid mediated with G4-FA took place at a lower rate (Supplementary Figure 8 and plotted in Figure 6A). The uptake inhibition of G4/Cy3-plasmid polyplexes and G4-FA/plasmid polyplexes by free FA was observed (Supplementary Figure 9 and plotted in Figure 6B & C).
Figure 6. . Effects of free folic acid on cellular uptake of dendrimer/plasmid polyplexes in HN12 cells.
(A) Cellular uptake kinetics of Cy3 plasmid mediated by G4 and G4-FA in HN12 cells as determined by flow cytometry and plotted upon Supplementary Figure 8. (B) Effect of FA on G4-mediated cellular uptake of Cy3 plasmid in the first 2 h by flow cytometry and plotted upon Supplementary Figure 9A. (C) Effect of FA on G4-FA-mediated cellular uptake of Cy3 plasmid in the first 2 h by flow cytometry and plotted in Supplementary Figure 9B.
FA: Folic acid.
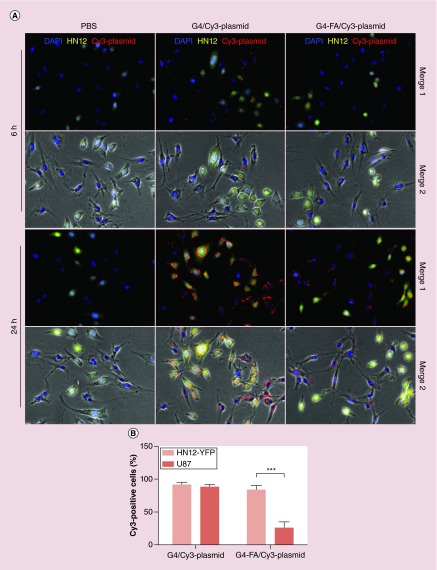
The fluorescence images obtained in the HN12-YFP/U87 (FR-high/FR-low) co-culture model study qualitatively illustrate that G4-FA promoted preferential plasmid uptake in FR-high cells, that is, HN12-YFP (Figure 7A). Cy3 plasmid was nonselectively taken up by both HN12-YFP and U87 cell populations following incubation with G4/Cy3 plasmid polyplexes. At 6 h, nearly 89% of U87 cells were positive in Cy3 channel (Figure 7B). In contrast, the percentage of Cy3-positive U87 cells reduced to 25% in the co-culture model following incubation with G4-FA/Cy3 plasmid. A longer incubation inevitably led to more plasmid uptake in both cell populations regardless of vector type as shown in the images taken at 24 h. However, it is noteworthy that the relative level of Cy3 positive dots in U87 cells incubated with G4-FA polyplexes was significantly lower than in U87 incubated with G4 polyplexes. This study further validates that G4-FA is more selective than G4 in delivering plasmid to FR-high cells.
Figure 7. . G4-FA promotes preferential cellular uptake of plasmid in folate receptor-high cells.
(A) Cellular uptake of polyplexes in an HN12-YFP/U87 (FR-high/FR-low) co-culture model monitored by using fluorescence microscopy. Blue, DAPI; yellow, HN12-YFP cells; and red, Cy3-plasmid. (B) Relative percentages of HN12-YFP and U87 cells presenting Cy3 plasmid in the co-culture model following 6-h incubation with G4-FA/Cy3 plasmid and G4/Cy3 plasmid polyplexes.
DAPI: 4′,6-diamidino-2-phenylindole; FR: Folate receptor.
G4-FA shows high gene transfection efficiency
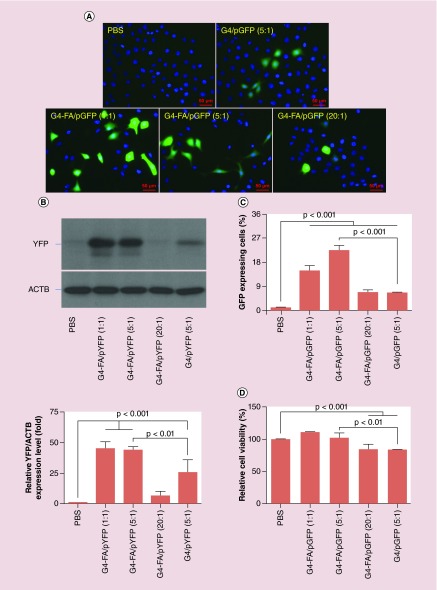
The in vitro gene transfection efficiency of G4-FA was evaluated using HN12 cells with GFP and YFP plasmid as reporters. To ascertain whether the use of the FA targeting moiety would result in improved gene transfection, G4-mediated gene transfection was evaluated for comparison. The HN12 cells, following various transfection treatments, are shown in Figure 8A. G4-FA is superior to nontargeting G4 in terms of overall amount of transgene expression and proportions of transfected cells according to western blotting (Figure 8B) and flow cytometry analysis (Figure 8C & Supplementary Figure 10). In particular, G4-FA resulted in 72% more protein expression and 250% more GFP-positive HN12 cells when complexed with plasmid at the complexation ratio of 5:1. Complexation ratio was found to affect transfection efficiency of G4-FA. G4-FA seems to be able to transfect more HN12 cells and induce stronger GFP expression at low ratios, that is, 1:1 (14.9%) and 5:1 (22.6%) compared with a high ratio of 20:1 (6.8%). Taken together, G4-FA displayed the highest efficiency in gene transfection of HN12 cells when it was complexed with plasmid at a weight ratio of 5:1. According to post-transfection cell viability assessment (Figure 8D), no toxicity effect was found for G4-FA when it was complexed with plasmid at weight ratios of 1:1 and 5:1. G4-FA showed toxicity at the weight ratio of 20:1 in gene transfection.
Figure 8. . Enhanced gene transfection in HN12 cells by G4-FA.
(A) Fluorescence images of HN12 cells receiving different treatments. (B) Quantification of YFP expression levels by using western blot analysis. (C) Quantification of GFP-expressing cell populations as determined by flow cytometry and plotted in Supplementary Figure 10. (D) Post-transfection cell viability assessment as determined by WST-1 assay.
Discussion
Generally, higher ζ-potential of nanoparticles can result in higher nonspecific cellular uptake [45,46]. Thus, the cellular uptake of FITC-G4 conjugates is expected to be slightly higher than that of FITC-G4-FA conjugates. This was confirmed by us in this work. Gel retardation assay and ζ-potential measurement results suggest that although G4-FA/pGFP polyplexes can be stably formed at a weight ratio of 1:1, the number of G4-FA conjugates (molar ratio of 134:1) was not enough to shield the plasmid. At weight ratios of 5 and 20 (molar ratio of 671 and 2683), the plasmid can be sufficiently shielded by G4-FA conjugates in complexation, nearly neutralizing the surface charge of polyplexes. In contrast, the ζ-potential of G4/pGFP polyplexes at a weight ratio of 5 was determined to be 1 mV.
Cellular uptake studies revealed that the mean FITC intensity of the cells treated with FITC-G4 was higher than those treated with FITC-G4-FA, but the FITC-positive cell population was similar between two groups. These results indicate the cellular uptake of G4 is through nonspecific absorptive endocytosis and G4-FA enters cells via receptor-mediated endocytosis. The saturated receptors on the cell surface may limit cellular uptake of G4-FA, thus resulting in the plateau in the uptake kinetics curve. G4-FA is able to compete with free FA for the same binding site. It appears to be more important to understand the intracellular trafficking pattern of DNA plasmids mediated with vectors. Therefore, we assessed the distribution of the polyplexes in HN12 cells at various time points post-transfection using fluorescence microscopy. FITC-labeled G4-FA conjugates and a Cy3-labeled plasmid were employed for in vitro trafficking of vector and plasmid, respectively. Further analysis of uptake of plasmid mediated with dendrimers confirmed that, suggesting G4-FA/Cy3-plasmid polyplexes enter cells likely following receptor-mediated endocytosis. Stronger uptake inhibition of G4-FA/Cy3-plasmid by free FA was noticed when compared with uptake of FITC-G4-FA alone. This was attributed to significant reduction of nonspecific cellular uptake as a result of ζ-potential shift from positive to negative. This may also explain why free FA inhibits uptake of G4/Cy3-plasmid to a certain extent.
We established a co-culture model, which contained FR-high HN12 cells and FR-low U87 cells. To distinguish HN12 cells from U87 cells, we used HN12 cells expressing YFP (HN12-YFP) in the co-culture model. FR-dependent cellular uptake of G4-FA/plasmid polyplexes was confirmed. G4-FA delivers significantly more plasmids to FR-high cells.
Increasing vector to plasmid weight ratio does not necessarily augment gene transfection efficiency for G4-FA. As described previously, free FA works as a competitive ligand to inhibit uptake of G4-FA/Cy3-plasmid polyplexes in HN12 cells. It is likely that increasing vector to plasmid ratio yields excess amount of G4-FA conjugates in the polyplexes, which, in turn, function as competitive ligands to bind FR, and subsequently inhibit uptake of G4-FA/plasmid polyplexes in HN12 cells. In addition, higher complexation ratios tend to make unpacking of polyplexes slower and cause transfection to be less efficient. However, we also observed that G4 facilitates higher cellular uptake of plasmid but shows much lower transfection efficiency in HN12 cells. We attribute this to reduced cell activity and apoptosis induced by nonspecific absorptive endocytosis of G4. It is known that dendrimers function as a proton sponge to facilitate the escape from endosomes and lysosomes. We found most Cy3-labeled plasmids and G4-FA were surrounding nuclei after 24-h transfection, but they were still bound, indicating the dissociation occurred slowly. Further experiments are needed to understand the intracellular dissociation kinetics of dendrimer and plasmid in the future, and a controlled release strategy may further enhance transfection efficiency using G4-FA conjugates to FRα-high cells.
Conclusion
In conclusion, FA-decorated PAMAM dendrimer generation 4 (i.e., G4-FA) shows improved cytocompatibility and is able to form stable complexes with DNA plasmids at weight ratios of 1:1 and above. G4-FA competes with free FA for the same binding site, that is, FR, on the surface of head and neck cancer cells, for example, HN12 cells. G4-FA was also found to facilitate the cellular uptake of DNA plasmid in a FR-dependent manner. Transfection efficiency of G4-FA was much higher than that of G4. G4-FA seems most efficient in transfecting HN12 cells and maintains good cytocompatibility when it is complexed with plasmid at a weight ratio of 5:1. The fundamental understanding of G4-FA as a vector and the encouraging transfection results gained in this work provided support for its further testing in vivo.
Executive summary.
Not much success has been reported for head and neck squamous cell carcinoma (HNSCC) gene therapy. Given the scarcity of nonviral vectors for gene therapy of HNSCC and limited clinical outcomes, it is highly desirable to develop and evaluate vectors under the context of HNSCC.
G4-FA and FITC-labeled G4-FA (FITC-G4-FA) conjugates were synthesized on the basis of polyamidoamine dendrimer G4 and characterized in terms of particle size, ζ-potential, cytotoxicity, and complexation with plasmid.
G4-FA conjugates and G4-FA/plasmid polyplexes were taken up by FR-high HN12 cells via an FRα-mediated endocytosis, while at a rate slower than nonspecific absorptive endocytosis of G4 and G4/plasmid polyplexes.
In a co-culture model of HN12-YFP/U87 (FR-high/FR-low), G4-FA/plasmid polyplexes were preferentially taken up by FRα-high cells than FRα-low cells.
FA-decoration of polyamidoamine dendrimer enhanced transfection efficiency by maintaining good cytocompatibility and increasing transgene expression level and transfected cell population.
This work concludes that G4-FA is a suitable vector to deliver genes selectively to head and neck cancer cells.
Supplementary Material
Footnotes
Financial & competing interests disclosure
This work was supported, in part, by the National Science Foundation (CAREER award CBET0954957), and National Institutes of Health (R01EY024072 and R01DE024381). Confocal microscopy was performed at the VCU Department of Anatomy & Neurobiology Microscopy Facility, supported, in part, by funding from NIH-NINDS Center Core Grant P30NS047463 and, in part, by funding from NIH-NCI Cancer Center Support Grant P30CA016059. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Harrington K, Alvarez-Vallina L, Crittenden M, et al. Cells as vehicles for cancer gene therapy: the missing link between targeted vectors and systemic delivery? Hum. Gene Ther. 2002;13(11):1263–1280. doi: 10.1089/104303402760128504. [DOI] [PubMed] [Google Scholar]
- 2.Thomas SM, Grandis JR. The current state of head and neck cancer gene therapy. Hum. Gene Ther. 2009;20(12):1565–1575. doi: 10.1089/hum.2009.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ameyar-Zazoua M, Guasconi V, Ait-Si-Ali S. siRNA as a route to new cancer therapies. Expert Opin. Biol. Ther. 2005;5(2):221–224. doi: 10.1517/14712598.5.2.221. [DOI] [PubMed] [Google Scholar]
- 4.Cabanillas R, Rodrigo JP, Astudillo A, Dominguez F, Suarez C, Chiara MD. P53 expression in squamous cell carcinomas of the supraglottic larynx and its lymph node metastases: new results for an old question. Cancer. 2007;109(9):1791–1798. doi: 10.1002/cncr.22646. [DOI] [PubMed] [Google Scholar]
- 5.Ndoye A, Dolivet G, Hogset A, et al. Eradication of p53-mutated head and neck squamous cell carcinoma xenografts using nonviral p53 gene therapy and photochemical internalization. Mol. Ther. 2006;13(6):1156–1162. doi: 10.1016/j.ymthe.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Cardinali M, Jakus J, Shah S, Ensley JF, Robbins KC, Yeudall WA. p21(WAF1/Cip1) retards the growth of human squamous cell carcinomas in! vivo . Oral Oncol. 1998;34(3):211–218. doi: 10.1016/s1368-8375(97)00083-3. [DOI] [PubMed] [Google Scholar]
- 7.Morris JC, Wildner O. Therapy of head and neck squamous cell carcinoma with an oncolytic adenovirus expressing HSV-tk. Mol. Ther. 2000;1(1):56–62. doi: 10.1006/mthe.1999.0014. [DOI] [PubMed] [Google Scholar]
- 8.Kim SK, Fan Y, Papadimitrakopoulou V, et al. DPC4, a candidate tumor suppressor gene, is altered infrequently in head and neck squamous cell carcinoma. Cancer Res. 1996;56(11):2519–2521. [PubMed] [Google Scholar]
- 9.Parker N, Turk MJ, Westrick E, Lewis JD, Low PS, Leamon CP. Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay. Anal. Biochem. 2005;338(2):284–293. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 10.Elnakat H, Ratnam M. Distribution, functionality and gene regulation of folate receptor isoforms: implications in targeted therapy. Adv. Drug Deliv. Rev. 2004;56(8):1067–1084. doi: 10.1016/j.addr.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Ross JF, Chaudhuri PK, Ratnam M. Differential regulation of folate receptor isoforms in normal and malignant tissues in vivo and in established cell lines. Physiologic and clinical implications. Cancer. 1994;73(9):2432–2443. doi: 10.1002/1097-0142(19940501)73:9<2432::aid-cncr2820730929>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 12.Myc A, Kukowska-Latallo J, Cao P, et al. Targeting the efficacy of a dendrimer-based nanotherapeutic in heterogeneous xenograft tumors in vivo . Anticancer Drugs. 2010;21(2):186–192. doi: 10.1097/CAD.0b013e328334560f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arima H, Arizono M, Higashi T, et al. Potential use of folate-polyethylene glycol (PEG)-appended dendrimer (G3) conjugate with alpha-cyclodextrin as DNA carriers to tumor cells. Cancer Gene Ther. 2012;19(5):358–366. doi: 10.1038/cgt.2012.9. [DOI] [PubMed] [Google Scholar]
- 14.Wang M, Hu H, Sun Y, et al. A pH-sensitive gene delivery system based on folic acid-PEG-chitosan-PAMAM-plasmid DNA complexes for cancer cell targeting. Biomaterials. 2013;34(38):10120–10132. doi: 10.1016/j.biomaterials.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Kang C, Yuan X, Li F, et al. Evaluation of folate-PAMAM for the delivery of antisense oligonucleotides to rat C6 glioma cells in vitro and in vivo . J. Biomed. Mater. Res. A. 2010;93A(2):585–594. doi: 10.1002/jbm.a.32525. [DOI] [PubMed] [Google Scholar]
- 16.Sunoqrot S, Bugno J, Lantvit D, Burdette JE, Hong S. Prolonged blood circulation and enhanced tumor accumulation of folate-targeted dendrimer-polymer hybrid nanoparticles. J. Control. Release. 2014;191:115–122. doi: 10.1016/j.jconrel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silpe JE, Sumit M, Thomas TP, et al. Avidity modulation of folate-targeted multivalent dendrimers for evaluating biophysical models of cancer targeting nanoparticles. ACS Chem. Biol. 2013;8(9):2063–2071. doi: 10.1021/cb400258d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhury A, Das S, Bunte RM, Chiu GN. Potent therapeutic activity of folate receptor-targeted liposomal carboplatin in the localized treatment of intraperitoneally grown human ovarian tumor xenograft. Int. J. Nanomedicine. 2012;7:739–751. doi: 10.2147/IJN.S26172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao P, Wang H, Yu M, et al. Paclitaxel-loaded, folic-acid-targeted and TAT-peptide-conjugated polymeric liposomes: in vitro and in vivo evaluation. Pharm. Res. 2010;27(9):1914–1926. doi: 10.1007/s11095-010-0196-5. [DOI] [PubMed] [Google Scholar]
- 20.Qiu LY, Yan L, Zhang L, Jin YM, Zhao QH. Folate-modified poly(2-ethyl-2-oxazoline) as hydrophilic corona in polymeric micelles for enhanced intracellular doxorubicin delivery. Int. J. Pharm. 2013;456(2):315–324. doi: 10.1016/j.ijpharm.2013.08.071. [DOI] [PubMed] [Google Scholar]
- 21.Werner ME, Copp JA, Karve S, et al. Folate-targeted polymeric nanoparticle formulation of docetaxel is an effective molecularly targeted radiosensitizer with efficacy dependent on the timing of radiotherapy. ACS Nano. 2011;5(11):8990–8998. doi: 10.1021/nn203165z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arote RB, Hwang SK, Lim HT, et al. The therapeutic efficiency of FP-PEA/TAM67 gene complexes via folate receptor-mediated endocytosis in a xenograft mice model. Biomaterials. 2010;31(8):2435–2445. doi: 10.1016/j.biomaterials.2009.11.106. [DOI] [PubMed] [Google Scholar]
- 23.Kukowska-Latallo JF, Candido KA, Cao Z, et al. Nanoparticle targeting of anticancer drug improves therapeutic response in animal model of human epithelial cancer. Cancer Res. 2005;65(12):5317–5324. doi: 10.1158/0008-5472.CAN-04-3921. [DOI] [PubMed] [Google Scholar]
- 24.Gleich LL. Gene therapy for head and neck cancer. Laryngoscope. 2000;110(5 Pt 1):708–726. doi: 10.1097/00005537-200005000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Villaret D, Glisson B, Kenady D, et al. A multicenter Phase II study of tgDCC-E1A for the intratumoral treatment of patients with recurrent head and neck squamous cell carcinoma. Head Neck. 2002;24(7):661–669. doi: 10.1002/hed.10107. [DOI] [PubMed] [Google Scholar]
- 26.Kesharwani P, Iyer AK. Recent advances in dendrimer-based nanovectors for tumor-targeted drug and gene delivery. Drug Discov. Today. 2014;20(5):536–547. doi: 10.1016/j.drudis.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee CC, Mackay JA, Frechet JM, Szoka FC. Designing dendrimers for biological applications. Nat. Biotechnol. 2005;23(12):1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 28.Xu L, Zhang H, Wu Y. Dendrimer advances for the central nervous system delivery of therapeutics. ACS Chem. Neurosci. 2014;5(1):2–13. doi: 10.1021/cn400182z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H. Nanoparticle-mediated brain-specific drug delivery, imaging, and diagnosis. Pharm. Res. 2010;27(9):1759–1771. doi: 10.1007/s11095-010-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Liu C, Laurini E, et al. Efficient delivery of sticky siRNA and potent gene silencing in a prostate cancer model using a generation 5 triethanolamine-core PAMAM dendrimer. Mol. Pharm. 2012;9(3):470–481. doi: 10.1021/mp2006104. [DOI] [PubMed] [Google Scholar]
- 31.Posocco P, Liu X, Laurini E, et al. Impact of siRNA overhangs for dendrimer-mediated siRNA delivery and gene silencing. Mol. Pharm. 2013;10(8):3262–3273. doi: 10.1021/mp400329g. [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Liu X, Rocchi P, Qu F, Iovanna JL, Peng L. Arginine-terminated generation 4 PAMAM dendrimer as an effective nanovector for functional siRNA delivery in vitro and in vivo . Bioconjug. Chem. 2014;25(3):521–532. doi: 10.1021/bc4005156. [DOI] [PubMed] [Google Scholar]
- 33.Myc A, Majoros IJ, Thomas TP, Baker JR., Jr Dendrimer-based targeted delivery of an apoptotic sensor in cancer cells. Biomacromolecules. 2007;8(1):13–18. doi: 10.1021/bm060815l. [DOI] [PubMed] [Google Scholar]
- 34.Yuan Q, Yeudall WA, Yang H. PEGylated polyamidoamine dendrimers with bis-aryl hydrazone linkages for enhanced gene delivery. Biomacromolecules. 2010;11(8):1940–1947. doi: 10.1021/bm100589g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quick QA, Gewirtz DA. An accelerated senescence response to radiation in wild-type p53 glioblastoma multiforme cells. J. Neurosurg. 2006;105(1):111–118. doi: 10.3171/jns.2006.105.1.111. [DOI] [PubMed] [Google Scholar]
- 36.Patel V, Ensley JF, Gutkind JS, Yeudall WA. Induction of apoptosis in head-and-neck squamous carcinoma cells by gamma-irradiation and bleomycin is p53-independent. Int. J. Cancer. 2000;88(5):737–743. doi: 10.1002/1097-0215(20001201)88:5<737::aid-ijc9>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Teh MT, Ji Y, et al. EPS8 upregulates FOXM1 expression, enhancing cell growth and motility. Carcinogenesis. 2010;31(6):1132–1141. doi: 10.1093/carcin/bgq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golding SE, Rosenberg E, Adams BR, et al. Dynamic inhibition of ATM kinase provides a strategy for glioblastoma multiforme radiosensitization and growth control. Cell Cycle (Georgetown, Tex.) 2012;11(6):1167–1173. doi: 10.4161/cc.11.6.19576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan Q, Lee E, Yeudall WA, Yang H. Dendrimer-triglycine-EGF nanoparticles for tumor imaging and targeted nucleic acid and drug delivery. Oral Oncol. 2010;46(9):698–704. doi: 10.1016/j.oraloncology.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L, Zolotarskaya OY, Yeudall WA, Yang H. Click hybridization of immune cells and polyamidoamine dendrimers. Adv. Healthc. Mater. 2014;3(9):1430–1438. doi: 10.1002/adhm.201300515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu L, Bai Q, Rodriguez-Agudo D, et al. Regulation of hepatocyte lipid metabolism and inflammatory response by 25-hydroxycholesterol and 25-hydroxycholesterol-3-sulfate. Lipids. 2010;45(9):821–832. doi: 10.1007/s11745-010-3451-y. [DOI] [PubMed] [Google Scholar]
- 42.Oh N, Park JH. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int. J. Nanomedicine. 2014;9(Suppl. 1):51–63. doi: 10.2147/IJN.S26592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitchens KM, Kolhatkar RB, Swaan PW, Eddington ND, Ghandehari H. Transport of poly(amidoamine) dendrimers across Caco-2 cell monolayers: Influence of size, charge and fluorescent labeling. Pharm. Res. 2006;23(12):2818–2826. doi: 10.1007/s11095-006-9122-2. [DOI] [PubMed] [Google Scholar]
- 44.Kitchens KM, Foraker AB, Kolhatkar RB, Swaan PW, Ghandehari H. Endocytosis and interaction of poly (amidoamine) dendrimers with Caco-2 cells. Pharm. Res. 2007;24(11):2138–2145. doi: 10.1007/s11095-007-9415-0. [DOI] [PubMed] [Google Scholar]
- 45.Chen H, Wang L, Yeh J, et al. Reducing non-specific binding and uptake of nanoparticles and improving cell targeting with an antifouling PEO-b-PgammaMPS copolymer coating. Biomaterials. 2010;31(20):5397–5407. doi: 10.1016/j.biomaterials.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He C, Hu Y, Yin L, Tang C, Yin C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials. 2010;31(13):3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.