Abstract
The x-ray crystal structure of an oligomeric miniprotein has been determined to a 1.2-Å resolution by means of multiwavelength anomalous diffraction phasing with selenomethionine analogs that retain the biophysical characteristics of the native peptide. Peptide 1, comprising α and β secondary structure elements with only 21 aa per monomer, associates as a discrete tetramer. The peptide adopts a previously uncharacterized quaternary structure in which α and β components interact to form a tightly packed and well defined hydrophobic core. The structure provides insight into the origins of the unusual thermal stability of the oligomer. The miniprotein shares many characteristics of larger proteins, including cooperative folding, lack of 1-anilino-8-naphthalene sulfonate binding, and limited deuterium exchange, and possesses a buried surface area typical of native proteins.
Oligomerization is a fundamental strategy for generating protein structural and functional complexity in nature. Many large proteins are believed to have arisen from oligomeric precursors that, by means of gene duplication and fusion, have become one encoded protein (1). Some examples are the ribosome anti-association factor eIF6 (2), the periplasmic binding proteins (3, 4), and the eightfold β/α barrel (5). Functionally, many proteins (6) are inactive in the monomeric state but are active as a dimer or higher-order oligomer; examples include GCN4 (7) and MCP-1 (8).
We are interested in studying the structural and functional advantages conferred by protein oligomerization. We thus set out to create miniproteins that would adopt a defined oligomeric state. Miniprotein models have been used to investigate biological processes and to model larger proteins through the incorporation of catalytic or other functionality (9–11). Oligomeric miniproteins serve as minimal model systems for quaternary structure formation. Moreover, they constitute platforms for probing whether more complex oligomeric structures might ultimately support function.
The strategy for oligomerization was inspired by “domain swapping,” or the exchange of association partners, an important evolutionary mechanism for protein oligomerization (12–15). This strategy has previously been used for the design of a coiled-coil domain-swapped dimer (16). The prototypic ββα (BBA) motif BBA5 (Fig. 1) was chosen as the building block because it represents a discretely folded and structured miniprotein motif (17, 18). This motif includes interactions between secondary structural elements, thus presenting an opportunity to favor similar interactions between monomers within an oligomer. Because it has been reported that shortening the linker between domains favors domain-swapped oligomers (19, 20), we introduced a bias toward oligomerization in the BBA motif by varying the length of the 3-aa hinge region between secondary structural elements (21). Our approach for the discovery of homooligomers of peptides with ββα supersecondary structure was based on a complementary design and screening process (21, 22).
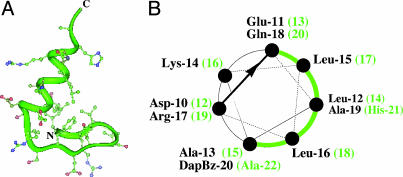
Fig. 1.
Comparison of BBA5 and peptide 1. (A) The monomeric precursor BBA5. (B) Helical wheel diagram of peptide 1. The hydrophobic face of the helix is indicated in green. Green text corresponds to BBA5 numbering.
BBAT1 (21, 22), a homooligomeric peptide, and the Gly9dAla analog, 1 (23) (Table 1), share many characteristic features of larger proteins, including cooperative folding, lack of 1-anilino-8-naphthalene sulfonate binding, and limited deuterium exchange. Circular dichroism and 2D NMR experiments established that there are close contacts between residues in the helix and hairpin segments, that there are interhelical interactions, and that each monomer is in a symmetrical environment. Key features include the hinge region and the constrained β-hairpin motif, established by a β-turn-forming dPro-Ser sequence (24, 25). While solution-state biophysical experiments clearly revealed the presence of a defined oligomeric state, the exact nature of the structure and the forces contributing to the observed native-protein-like stability remained unknown. A detailed knowledge of the atomic structure of the oligomer would provide insight into the forces that contribute to oligomerization and a model for understanding quaternary structure.
Table 1. Sequences of peptides BBA5, BBAT1, 1, 2, and 3, with hairpin and helix portions as indicated.
| Hairpin
|
Helix
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | ||
| BBA5 | Ac | Y | R | V | p | S | Y | D | F | SRS | D | E | L | A | K | L | L | R | Q | H | A | G | NH2 |
| BBAT1 | Ac | Y | R | I | p | S | Y | D | F | G | D | E | L | A | K | L | L | R | Q | A | Z | G | NH2 |
| 1 | Ac | Y | R | I | p | S | Y | D | F | a | D | E | L | A | K | L | L | R | Q | A | Z | G | NH2 |
| Ala13SeMet (2) | Ac | Y | R | I | p | S | Y | D | F | a | D | E | L | SM | K | L | L | R | Q | A | Z | G | NH2 |
| Gln18SeMet (3) | Ac | Y | R | I | p | S | Y | D | F | a | D | E | L | A | K | L | L | R | SM | A | Z | G | NH2 |
Ac, acetyl; p, d-proline; a, d-alanine; SM, SeMet; Z, DapBz.
We report herein the structures of the homooligomeric miniprotein, 1, and of two analogs, 2 and 3. This represents the first structure of a mixed α/β oligomeric miniprotein. The structure reveals that the stoichiometry of the oligomer is tetrameric, rather than trimeric, as originally proposed (21). The architecture of this oligomeric peptide is highly reminiscent of a native protein and supports a water-exclusive core. Additionally, the unique role of unnatural amino acids in conferring native secondary structure in a short peptide sequence (21 residues per monomer) is defined.
Materials and Methods
Further experimental details can be found in Supporting Text, which is published as supporting information on the PNAS web site.
Peptide Synthesis. Peptides were prepared by standard fluoren-9-ylmethoxycarbonyl-based solid-phase peptide synthesis and purified by preparative reverse-phase HPLC. Purified selenomethionine (SeMet)-containing peptides were oxidized with 3% H2O2 in 10 mM phosphate (pH 7.2) for 20 min and then immediately repurified by preparative HPLC. Peptides were characterized by electrospray mass spectroscopy and analytical HPLC.
Analytical Ultracentrifugation. Dialyzed peptide solutions were spun at 25°C, using two-sector epon centerpieces, in a Beckman XL-I analytical ultracentrifuge at 40,000, 45,000, and 50,000 rpm for ≈24 h at each speed. Data were analyzed by using the programs nonlin (26) and sedphat (27).
Partial Specific Volume Determination. The densities of a concentrated solution of peptide 1 and of four dilutions thereof (80%, 60%, 40%, and 20% of the stock solution) were measured at 20°C by using a DMA 60 density meter (Anton Paar, Graz, Austria) at the National Analytical Ultracentrifugation Facility (University of Connecticut, Storrs), to determine the partial specific volume.
Crystallization. Crystals were grown by using vapor diffusion with hanging-drop geometry by mixing 1.5 μl of protein (≥8 mg/ml) with 1.5 μl of reservoir solution. Peptides 1, 2, and 3 crystallize overnight as small rods from 100 mM Hepes-Na (pH 7.5)/10% vol/vol i-propanol/20% wt/vol polyethylene glycol 4000.
Data Collection and Phasing. Peptides 2 and 3. Multiwavelength anomalous diffraction (MAD) data sets were collected at beam-line X12C of the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY). Data were processed with the denzo and scalepack package (28) (Table 2 and Table 4, which is published as supporting information on the PNAS web site). Heavy-atom sites were located by using solve (29) with an initial figure of merit of 0.62 and 0.56 for the data sets from peptides 2 and 3, respectively. Initial electron-density maps showed connected helical density.
Table 2. Key data collection and refinement statistics for peptides 1, 2, and 3.
| Selenomethionine MAD
|
|||||||
|---|---|---|---|---|---|---|---|
| Native 1 | 2 | 3 | |||||
| Unit cell, Å, ° | a = 38.94, b = 56.17 c = 32.32, α = 90 β = 90, γ = 90 | a = 50.03, b = 46.87, c = 31.43 α = 90, β = 98.61, γ = 90 | a = 57.91, b = 21.33, c = 31.90 α = 90, β = 121.32, γ = 90 | ||||
| Space group | P21212 | C2 | C2 | ||||
| Wavelength, Å | 0.900 | 0.978240 | 0.977939 | 0.949359 | 0.978031 | 0.97902 | 0.949320 |
| Resolution, Å | ∞–1.2 | ∞–1.5 | ∞–1.5 | ∞–1.5 | ∞–1.5 | ∞–1.5 | ∞–1.5 |
| Total/unique reflections | 78,662 | 32,445 | 32,558 | 34,820 | 13,425 | 13,435 | 14,451 |
| 21,067 | 17,229 | 17,273 | 18,363 | 7,811 | 7,814 | 8,373 | |
| Completeness, % | 92.5 (54.7) | 76.0 (24.9) | 76.0 (24.9) | 80.4 (32.1) | 74.4 (16.1) | 74.7 (16.6) | 80.0 (26.9) |
| I/σ(I) | 26.8 (3.7) | 33.8 (7.7) | 32.3 (6.2) | 30.6 (5.5) | 33.6 (7.1) | 32.1 (6.6) | 30.7 (5.5) |
| Rmerge,* % | 3.4 (22.9) | 3.5 (6.6) | 3.4 (8.6) | 3.0 (11.0) | 3.5 (8.5) | 3.6 (9.6) | 3.5 (10.1) |
| f′/f″ (e-) | — | –10.0/3.4 | –7.5/5.7 | –2.5/3.0 | –10.0/3.4 | –7.5/5.7 | –2.5/3.0 |
| Refinement Statistics | |||||||
| Resolution, Å | 8–1.2 | 50–1.5 | 50–1.5 | ||||
| No. of reflections working/test set | 17,629 | 15,512 | 6,983 | ||||
| 1,977 | 1,604 | 786 | |||||
| Rwork†/Rfree‡, % | 18.8§/28.5§ | 16.1/19.9 | 16.9/18.3 | ||||
| 18.2¶/27.8¶ | |||||||
Values for the outermost shell (1.22–1.20 Å for 1, 1.55–1.50 Å for 2, and 1.55–1.50 for 3) are shown in parentheses.
Rmerge = ΣhklΣi|Ihkl,i-〈Ihkl〉I|/ΣhklΣi|Ihkl,i|, where 〈Ihkl〉 is the mean intensity of the multiple Ihkl,i observations for symmetry related reflections
Rwork = Σhkl|Fobs – Fcalc|/Σhkl|Fobs|
Rfree = Σhkl ΣT|Fobs – Fcalc|/Σhkl|Fobs|, where the test set T includes 10% of the data
Values calculated by using all data
Values calculated by using data for which F > 4σ(F)
Peptide 1. A data set was collected at beamline 14-BMC of the BIOCARS facility (Argonne National Laboratory, Argonne, IL). Data processing was performed as for 2 and 3. Starting phases were obtained by molecular replacement with molrep (30) with the refined tetramer of 3, including side chains, as a search model, resulting in an initial correlation coefficient of 0.53 and R factor of 0.43.
Refinement. Iterative rounds of minimization and simulated annealing by slow-cool torsional molecular dynamics, individual B-factor refinement, and manual rebuilding were used to refine all structures with the program cns (31), using an MLHL target (for 2 and 3) or an MLF target (for 1). Topology and parameter files were created for nonstandard groups by using bond lengths and angles from the literature. For statistical cross-validation purposes, 10% of the data were excluded from refinement (32). The refinement for peptide 1 was continued by using shelxl (33), extending the resolution from 1.5 Å to 1.2 Å. Anisotropic B-factor refinement resulted in a drop in both R and Rfree. Analysis of the Ramachandran plot defined by procheck (34) showed a good final model for all structures, with 92% of residues in the most favorable regions and 8% of residues in additionally allowed regions for the structures of 1, 2, and 3. All d-proline residues fell in the generously allowed regions and were excluded from the percentages calculated above. The refined structure of 1 contained 722 protein atoms and 118 solvent atoms per asymmetric unit. The refined structure of 2 contained 744 protein atoms, 4 isopropanol atoms, and 134 solvent atoms per asymmetric unit. The refined structure of 3 contained 364 protein atoms, 4 isopropanol atoms, and 69 solvent atoms per asymmetric unit. Final model statistics are summarized in Table 2.
We attribute the relatively high value of Rfree for the structure of peptide 1 to an inability to overcome the model bias due to a lack of independent phase information inherent in a molecular replacement solution. The high Rfree value does not appear to be due to poor data quality, an incorrect space group, incorrect register, misplaced waters, or any other single large error (see Supporting Text for further discussion). Composite omit maps showed no additional information relative to SigmaA-weighted 2Fo - Fc maps. Attempts to improve the maps with the Prime-and-Switch (35) protocol in solve and shelxe were unsuccessful, presumably because of the relatively low solvent content of crystals of 1.
Results and Discussion
Short peptides that have protein-like properties reduce protein complexity to the essential features and are thus invaluable minimal models. Pioneering efforts in protein design (36, 37) have resulted in the development of structures comprising α (38, 39), β (40, 41), and mixed α/β (17, 18) secondary structure. It has been more difficult to use smaller peptide motifs to mimic defined tertiary and quaternary structure. Nonetheless, native-like physical properties, including cooperative folding and discrete tertiary structure, have been achieved in some higher-order structures including the helical bundles and coiled-coil motifs (36, 39, 42). The final adopted folds of these monomeric and oligomeric constructs are reminiscent of the organization found in native proteins. Because there is no clear natural prototype for the development of small oligomeric motifs of mixed α and β secondary structure, the design strategy we adopted involved patterning oligomerization after the phenomenon of domain swapping, wherein forces present in the monomers could be exploited for tertiary and quaternary structure stabilization.
The oligomeric structure of the miniprotein peptide 1 (23) was determined by x-ray crystallography. The incorporation of a d-alanine at the hinge position (residue 9), in place of the original glycine in BBAT1, imparts a greater thermodynamic stability because of a more limited sampling of conformational space. Although both peptides demonstrate cooperative folding, the transition point for thermal denaturation (Tm) of peptide 1 was considerably higher than that of BBAT1 (64°C vs. 40°C). Peptide 1 is also more soluble than BBAT1, a trait that is advantageous for crystallization trials. The peptide crystallizes readily and diffracts to a very high resolution.
Obtaining phases for peptide 1 proved unexpectedly challenging. Extensive experimentation with molecular replacement models and algorithms, using the NMR-derived structure of the monomeric precursor BBA5 (17, 18), failed to provide phase information. Although very high-resolution data (≈1.2 Å) were available, direct-methods approaches failed, presumably because of the relatively large size of the target. However, the structure was too small for the heavy atom soaks used to phase macro-molecules, and the intrusion of a heavy atom into the small unit cell resulted in crystal damage, high mosaicity, and significant nonisomorphism. Similarly, synthetic analogs brominated or iodinated at aromatic residues did not appear to retain the native structure, nor could they be crystallized. We ultimately succeeded in determining the structure of 1 through a selenomethionine-based MAD approach with analogs of 1, followed by molecular replacement with this data to generate the structure of the native peptide 1.
Because the sequence of 1 does not contain any methionine residues to guide in the placement of SeMet substitutions, it was necessary to search for analogs adopting the native structure. We sought two criteria from a suitable derivative: (i) the same solution-phase physical properties as peptide 1, and (ii) homogeneity of the selenomethionine oxidation state. Six peptides were prepared having SeMet substitutions at hydrophobic positions along the BBA helix (Fig. 2). Because these peptides could not be maintained in the pure reduced form, it was most straightforward to oxidize the SeMet residues to the corresponding selenoxide. Upon oxidation, two of these peptides, 2 and 3 (with SeMet at positions 13 and 18, respectively), satisfy both criteria (Fig. 1 and Table 3). In peptides 2 and 3 the oxidized SeMet residues are located at the hydrophobic/hydrophilic interface of the helix, an area that can accommodate a large hydrophobic residue with polar character.
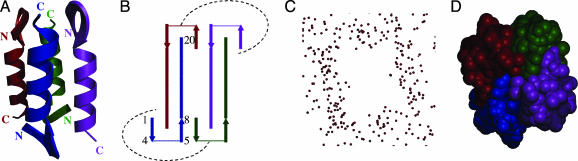
Fig. 2.
Tetrameric structure of peptide 1.(A) Ribbon diagram of tetramer; side view. (B) Topology diagram. The dashed line indicates intermonomer interaction. (C) Crystallographic waters, with void in the shape of the tetramer. (D) CPK model of peptide 1 indicating space occupied by tetramer.
Table 3. Results of equilibrium analytical ultracentrifugation experiments indicating stoichiometries of peptides 1, 2, and 3 derived by using experimental and calculated partial specific volumes.
| Calculated ν̄†
|
Measured ν̄‡§
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Peptide | Monomer MW | Sigma* | ν̄ | MW | Stoichiometry | ν̄ | MW | Stoichiometry |
| 1¶ | 2,558 | 1.778 | 0.740 | 9,928 | 3.9 | 0.754 | 10,517 | 4.1 |
| 2¶ | 2,681 | 1.982 | 0.740 | 11,088 | 4.1 | 0.754 | 11,721 | 4.4 |
| 3∥ | 2,624 | 1.863 | 0.744 | 10,569 | 4.0 | 0.754 | 11,016 | 4.2 |
MW, molecular weight.
Sigma is related to the molecular weight by the equation M=σRT/(1–ν̄ρ)ω2r and determined by using the program nonlin
Partial specific volume calculated by using sednterp (25°C)
ν̄ of all four peptides are assumed to be approximately equal
Extrapolated from data at 20°C to 25°C by using the formula ν̄(T) = ν̄(20°C) – [4.25·10-4 (T – 298.15)]
Single species model, B ≠ 0
Single species model, B = 0
Peptides 1, 2, and 3 crystallize in unique unit cells and accordingly make distinct crystal contacts. Consequently, we chose to pursue structure determination of both 2 and 3 by means of MAD phasing. The oligomeric state of the molecule would then be determined independently from two different sets of crystal conditions, ensuring an unbiased interpretation of the true structure. MAD data sets were collected, and experimental phases were obtained to a 1.5-Å resolution. The structures of 2 and 3 were fully refined. The refined, experimentally phased structure of 2 was used to obtain molecular replacement phases for peptide 1. The crystal structures for the crystallographically independent tetramers of the three peptides were found to be highly similar, with rms deviations of <0.52 Å, confirming that the substitution of an oxidized SeMet at an interface position is structurally benign.
The crystal structures of peptides 1, 2, and 3 fully account for the biophysical data collected for this family of peptides. Each monomer is in a symmetrical environment, as suggested by the degeneracy of the NMR signals in spectra of BBAT1. The residues that were predicted to be in close proximity based on NOESY spectroscopy are in close contact in the crystal structures. The oligomeric BBA has significant interdomain contacts between helical and hairpin structural elements, and the nature and location of these hydrophobic contacts is the same as those found in BBA5, as envisaged by the domain-swapping oligomerization strategy (Figs. 5 and 6, which are published as supporting information on the PNAS web site). In addition, there are new interhelical interactions present in the oligomer that are not found in the monomeric precursor BBA5. These interactions account for the characteristic CD signature (43) observed for peptides BBAT1, 1, 2, and 3. Whereas oligomeric BBA peptides such as 1 and BBAT1 were originally assigned as trimeric in aqueous solution on the basis of sedimentation equilibrium analytical ultracentrifugation (AUC) experiments, the oligomerization state for peptides 1, 2, and 3 in the crystalline state (having 28%, 29%, and 23% solvent, respectively, present in the crystal form) is clearly tetrameric. Sedimentation equilibrium AUC is a standard method for the determination of molecular weight. However, there are cases of unexpectedly low molecular weights being obtained for designed miniproteins, particularly due to charge-based nonideality under salt-free conditions (44–46).
This discrepancy could be due to the use of an incorrect estimation of the partial specific volume, data collection conditions, or data analysis protocols. The partial specific volume is generally calculated as the weighted average of tabulated values per amino acid residue (47, 48), a method that generally results in very low errors in the molecular weights obtained (48). The partial specific volume of peptide 1 was measured directly by densitometry to obtain confirmation that the tabulated values regularly used for macromolecules were appropriate for this miniprotein family containing nonstandard amino acids. This measurement resulted in a value of 0.752 cm3/g at 20°C in 10 mM sodium phosphate (pH 7.2), a value in excellent agreement with the calculated value of 0.738 cm3/g. We found that optimized experimental conditions, including longer solution columns, obtained by using a two-sector velocity cell rather than a standard six-sector equilibrium cell, and a higher salt content, resulted in improved data and more ideal behavior of the peptide. Analysis of these data, using corrected values for the partial specific volume and including nonideality in the model, resulted in stoichiometries consistent with the crystallographic finding of a tetramer, as shown in Table 3.
The tetramer consists of four monomers arranged in an antiparallel topology (Fig. 3 Right A and B). Each monomer within the tetramer is flanked by two monomers oriented in the opposite direction and makes helix–helix, helix–hairpin, and hairpin–helix interactions with these adjacent monomers. This interaction between the secondary structural elements is allowed by the fact that each monomer is approximately linear. A consequence of the linearity of the structural elements in the monomer is that the symmetry of the oligomeric structure is constrained to multimers of even number. The monomers associate with a slight right-handed superhelical twist, the reverse hand of what is commonly seen with coiled coils (39).
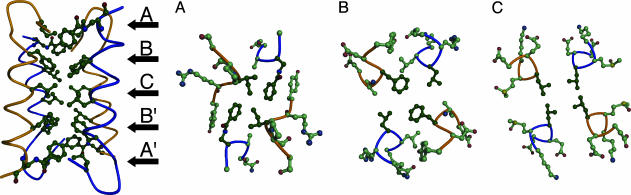
Fig. 3.
The hydrophobic core of tetrameric peptide 1. (Left) Side view of the tetramer, highlighting the positions of the five palindromic core layers. (Right) Cross-sectional slices through layers A (Ile-3/DapBz-20) (A), B (Phe-8/Leu-16) (B), and C (Leu-12/Leu-12) (C). Residues comprising the core are in bold.
The well defined central hydrophobic core of the tetramer is formed by the residues from one monomer in proximity to core residues from all three of the other monomers. Those residues on the periphery of the hydrophobic interface interact with residues of the antiparallel monomer nearest them. The hydrophilic residues are solvent-exposed. The tetramer possesses approximate, but not exact, fourfold symmetry: Each monomer is slightly closer to one neighbor than to the other.
The crystallographic packing also suggests that the peptide associates as a tetramer before crystallization. Each tetramer is enveloped by a sheath of waters of crystallization (Fig. 2C). Yet there is a clearly demarcated zone of water exclusion in the central core region of the tetramer, indicating that no waters can pass into this hydrophobic core. In addition, Connolly surface diagrams (49) calculated by using the tetrameric structure show that solvent is excluded from the hydrophobic core.
The tetrameric structure is highly protein-like in the degree of complexity and tightly organized packing of side chains. Moreover, the relationships between surface area, volume, and molecular weight fit well to a series of empirical relationships developed by Chothia for monomeric and oligomeric proteins of varying sizes (Table 5, which is published as supporting information on the PNAS web site) (50).
The tight packing of the hydrophobic core accounts for the cooperative unfolding and high melting temperatures of these peptides. This packing is remarkable in light of the very short linear sequence (21 aa in total, with 8 residues in the β-hairpin and 12 residues in the helix). Moreover, the packing is highly specific, in contrast to the nonspecific molten globule core common to many designed peptide oligomers, where hydrophobic residues form a core but lack specific interactions amongst residues in the core (39). In many designed coiled-coil peptides specificity has been achieved by introducing a buried polar residue to mimic the role of buried asparagine in GCN4 (51) in setting the register of the association and in preventing nonspecific interactions (52, 53). Interestingly, the hydrophobic core does not contain any buried polar residues that may confer packing specificity. It appears that the hairpin region in the oligomeric peptides serves to set the register of the monomers with respect to one another. The relative prevalence of bulky and conformationally restricted aromatic residues in the core (vide infra) may be another determinant of specificity. A third contribution may arise from the fact that a loss of registration in such a small peptide would result in too great an energetic cost because a greater relative percentage of the surface area would be exposed to solvent.
The hydrophobic interface of peptide 1 is defined by five palindromic layers (Fig. 3) including residues from both the α-helix and the β-sheet: Ile-3/DapBz-20 (layer A) (DapBz, benzoylated l-α,β-diaminopropionic acid), Phe-8/Leu-16 (layer B), Leu-12/Leu-12 (layer C), Leu-16/Phe-8 (layer B′), and DapBz-20/Ile-3 (layer A′). Two of the five residues involved in these contacts (Phe and DapBz) are aromatic amino acids. The three inner layers, composed of residues from the α-helix, roughly resemble an antiparallel four-membered coiled coil. Whereas in an antiparallel tetrameric coiled coil the angles at which side chains reach into the core, as measured by the vector connecting Cα and Cβ atoms, are regular, in the BBA tetramer each of three unique layers differs with regard to the Cα–Cβ vectors. There is no clear hydrophobic patterning analogous to a coiled-coil heptad repeat, although every fourth residue of the helix is a hydrophobic residue forming the core. Ile-3, located on the β-sheet just before the type II′ turn, does not conform to this repeat pattern.
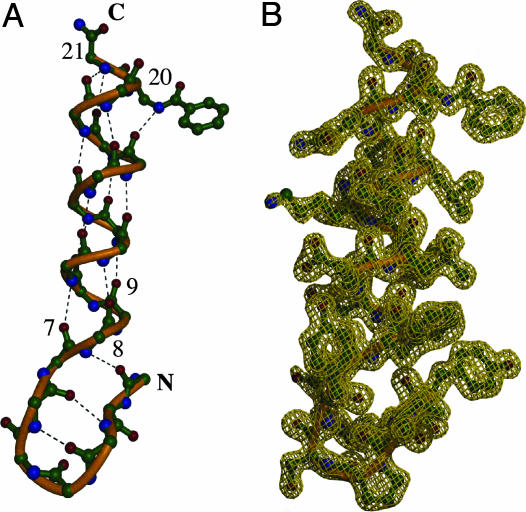
The β-hairpin in the monomer is very similar to that in BBA5 (18), as was predicted based on their high sequence identity (the sole difference is a Val3Ile substitution). A dual role is played by the last two residues of the hairpin (Asp-7 and Phe-8), which simultaneously participate in the β-hairpin and nucleate the α-helix via (i, i + 4) hydrogen bonds from their carbonyl groups to the amides of residues one turn above (Fig. 4A). Indeed, the dihedral angles of Phe-8 are α-helical.
Fig. 4.
Monomer of BBA. (A) Hydrogen bonding within a (BBA)4 monomer. Residues 1–8 form a β-hairpin, and residues 7 and 8 also nucleate the helix through i,i + 4 hydrogen bonding. Residue 9, a dAla, is part of the first turn of the helix. The DapBz side chain at position 20 and the glycine at position 21 are H-bond acceptors that cap the α-helix. (B) Monomer model of peptide 2 in an electron-density map.
The nonstandard dihedral angles of d-alanine allow it to contribute to the first turn of the helix and to compactly reverse the direction of the hairpin, producing a linear monomer. The dihedral angles (φ = -51.8° ± 2.5°, Ψ = -52.1° ± 2.0°) fall in an allowed region corresponding to the right-handed α-helical region for an l-amino acid on a Ramachandran plot. These dihedrals fall in the “inverted α-left” region (-60°, -45°), defined by Mitchell and Smith (54), allowed for d-amino acids in an l-amino acid environment. Intriguingly, this region, although allowed, is sparsely populated in known structures of amino acids in ldl environments. (Similarly, the αL region is sparsely populated in l-amino acid environments.) Although the d-alanine residue does not access dihedral angles that an l-amino acid cannot, the conformational space available to it in the inverted α-left region is quite limited, which may impart significant structural specificity and account for the observed stabilization of 1 relative to BBAT1.
The unnatural amino acid DapBz is a remnant of the fluorescence-quenching screen used for the identification of oligomerization (22). As the last completely ordered residue of the helix, DapBz plays a key structural role as a helix cap. Notably, the amide nitrogen of the benzoyl group forms an (i, i + 4) hydrogen bond to the carbonyl of Leu-16 that is the penultimate structural feature of the helix, as shown in Fig. 4. The C-terminal glycine is flexible, exhibiting alternate conformations in several of the monomers. However, the glycine amide nitrogen does form an (i, i + 4) hydrogen bond to the carbonyl groups of residues Arg-17 and Gln-18.
Conclusions
The high-resolution crystal structure of peptide 1 reveals a compact tetramer exhibiting the hallmarks of native protein structure. The oligomer size, intermediate between a protein and a small molecule, presented unique challenges with regard to obtaining phase information. The structure was ultimately solved by means of MAD phasing using SeMet analogs, which retained the biophysical characteristics of the native peptide. The intermediate size also necessitated experimental determination of the partial specific volume to accurately interpret sedimentation equilibrium results. The initial domain-swapping strategy for oligomerization resulted in a unique quaternary arrangement in which α and β components participate to form a thermally stable structure with a tightly packed and well defined hydrophobic core. The unique d-alanine residue allows for an economy in the design by producing a sharp change in orientation at the hinge region, affording a roughly linear monomer. In addition, the DapBz simultaneously fulfills the role of a helix cap and a hydrophobic core residue. Together, the overall design affords a highly compact oligomeric miniprotein, with only 21 aa per monomer, that manifests many of the physical characteristics of native proteins.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of Jeffrey W. Lary and Prof. James L. Cole (National Analytical Ultracentrifugation Facility, University of Connecticut). We also acknowledge the use of the X12C beamline at the National Synchrotron Light Source. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38. We thank Prof. George M. Sheldrick for helpful suggestions and Prof. Catherine L. Drennan and Dr. Geoffrey F. Stamper for valuable assistance in the early stages of this project. This work was supported by National Science Foundation Grant CHE 9996335 (to B.I.) and National Institutes of Health Training Grant HL07291 (to E.P.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DapBz, benzoylated l-α,β-diaminopropionic acid; MAD, multiwavelength anomalous diffraction; SeMet, selenomethionine.
Data deposition: The coordinates of the refined structures have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1SN9 (1), 1SNA (2), and 1SNE (3)].
References
- 1.Taylor, W. R., May, A. C. W., Brown, N. P. & Aszódi, A. (2001) Rep. Prog. Phys. 64, 517-590. [Google Scholar]
- 2.Groft, C. M., Beckmann, R., Sali, A. & Burley, S. K. (2000) Nat. Struct. Biol. 7, 1156-1164. [DOI] [PubMed] [Google Scholar]
- 3.Dassa, E., Saurin, W., Hofnung, M., Kuan, G. & Saier, M. H., Jr. (1995) Res. Microbiol. 146, 271-278. [DOI] [PubMed] [Google Scholar]
- 4.Saurin, W. & Dassa, E. (1994) Protein Sci. 3, 325-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lang, D., Thoma, R., Henn-Sax, M., Sterner, R. & Wilmanns, M. (2000) Science 289, 1546-1550. [DOI] [PubMed] [Google Scholar]
- 6.Klemm, J. D., Schreiber, S. L. & Crabtree, G. R. (1998) Annu. Rev. Immunol. 16, 569-592. [DOI] [PubMed] [Google Scholar]
- 7.Hope, I. A. & Struhl, K. (1987) EMBO J. 6, 2781-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, Y. & Rollins, B. J. (1995) Mol. Cell. Biol. 15, 4851-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson, D. E., Farid, R. S., Moser, C. C., Urbauer, J. L., Mulholland, S. E., Pidikiti, R., Lear, J. D., Wand, A. J., DeGrado, W. F. & Dutton, P. L. (1994) Nature 368, 425-431. [DOI] [PubMed] [Google Scholar]
- 10.Shogren-Knaak, M. A. & Imperiali, B. (1999) Bioorg. Med. Chem. 7, 1993-2002. [DOI] [PubMed] [Google Scholar]
- 11.Broo, K. S., Nilsson, H., Nilsson, J., Flodberg, A. & Baltzer, L. (1998) J. Am. Chem. Soc. 120, 4063-4068. [Google Scholar]
- 12.Bennett, M. J., Schlunegger, M. P. & Eisenberg, D. (1995) Protein Sci. 4, 2455-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, Y. & Eisenberg, D. (2002) Protein Sci. 11, 1285-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heringa, J. & Taylor, W. R. (1997) Curr. Opin. Struct. Biol. 7, 416-421. [DOI] [PubMed] [Google Scholar]
- 15.Bennett, M. J., Choe, S. & Eisenberg, D. (1994) Proc. Natl. Acad. Sci. USA 91, 3127-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogihara, N. L., Ghirlanda, G., Bryson, J. W., Gingery, M., DeGrado, W. F. & Eisenberg, D. (2001) Proc. Natl. Acad. Sci. USA 98, 1404-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Struthers, M. D., Cheng, R. P. & Imperiali, B. (1996) Science 271, 342-345. [DOI] [PubMed] [Google Scholar]
- 18.Struthers, M. D., Ottesen, J. J. & Imperiali, B. (1998) Fold. Des. 3, 95-103. [DOI] [PubMed] [Google Scholar]
- 19.Pei, X. Y., Holliger, P., Murzin, A. G. & Williams, R. L. (1997) Proc. Natl. Acad. Sci. USA 94, 9637-9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albright, R. A., Mossing, M. C. & Matthews, B. W. (1996) Biochemistry 35, 735-742. [DOI] [PubMed] [Google Scholar]
- 21.Mezo, A. R., Ottesen, J. J. & Imperiali, B. (2001) J. Am. Chem. Soc. 123, 1002-1003. [DOI] [PubMed] [Google Scholar]
- 22.Mezo, A. R., Cheng, R. P. & Imperiali, B. (2001) J. Am. Chem. Soc. 123, 3885-3891. [DOI] [PubMed] [Google Scholar]
- 23.McDonnell, K. A. & Imperiali, B. (2002) J. Am. Chem. Soc. 124, 428-433. [DOI] [PubMed] [Google Scholar]
- 24.Hutchinson, E. G. & Thornton, J. M. (1994) Protein Sci. 3, 2207-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haque, T. S., Little, J. C. & Gellman, S. H. (1994) J. Am. Chem. Soc. 116, 4105-4106. [Google Scholar]
- 26.Johnson, M. L., Correia, J. C., Yphantis, D. A. & Halvorson, H. R. (1981) Biophys. J. 36, 575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuck, P. (2003) Anal. Biochem. 320, 104-124. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vagin, A. & Teplyakov, A. (1997) J. Appl. Crystallogr. 30, 1022-1025. [Google Scholar]
- 31.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 32.Brünger, A. T. (1997) Methods Enzymol. 277, 366-396. [DOI] [PubMed] [Google Scholar]
- 33.Sheldrick, G. M. (1984) Acta Crystallogr. A 40, Suppl. S, C440-C440. [Google Scholar]
- 34.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 35.Terwilliger, T. C. (2001) Acta Crystallogr. D 57, 1763-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeGrado, W. F., Summa, C. M., Pavone, V., Nastri, F. & Lombardi, A. (1999) Annu. Rev. Biochem. 68, 779-819. [DOI] [PubMed] [Google Scholar]
- 37.Venkatraman, J., Shankaramma, S. C. & Balaram, P. (2001) Chem. Rev. 101, 3131-3152. [DOI] [PubMed] [Google Scholar]
- 38.Hill, R. B., Raleigh, D. P., Lombardi, A. & DeGrado, W. F. (2000) Acc. Chem. Res. 33, 745-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohn, W. D. & Hodges, R. S. (1998) Trends Biotechnol. 16, 379-389. [Google Scholar]
- 40.Kortemme, T., Ramirez-Alvarado, M. & Serrano, L. (1998) Science 281, 253-256. [DOI] [PubMed] [Google Scholar]
- 41.Ottesen, J. J. & Imperiali, B. (2001) Nat. Struct. Biol. 8, 535-539. [DOI] [PubMed] [Google Scholar]
- 42.Bryson, J. W., Betz, S. F., Lu, H. S., Suich, D. J., Zhou, H. X., O'Neil, K. T. & DeGrado, W. F. (1995) Science 270, 935-941. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, N. E., Zhu, B.-Y., Kay, C. M. & Hodges, R. S. (1992) Biopolymers 32, 419-426. [DOI] [PubMed] [Google Scholar]
- 44.de Alba, E., Santoro, J., Rico, M. & Jiménez, M. A. (1999) Protein Sci. 8, 854-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carulla, N., Woodward, C. & Barany, G. (2002) Protein Sci. 11, 1539-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schenk, H. L. & Gellman, S. H. (1998) J. Am. Chem. Soc. 120, 4869-4870. [Google Scholar]
- 47.Cohn, E. J. & Edsall, J. T. (1965) in Proteins, Amino Acids and Peptides as Ions and Dipolar Ions, eds. Cohn, E. J. & Edsall, J. T. (Hafner, New York), pp. 370-381.
- 48.Durchschlag, H. (1986) in Thermodynamic Data for Biochemistry and Biotechnology, ed. Hinz, H.-J. (Springer, Berlin), pp. 45-128.
- 49.Connolly, M. L. (1983) Science 221, 709-713. [DOI] [PubMed] [Google Scholar]
- 50.Janin, J., Miller, S. & Chothia, C. (1988) J. Mol. Biol. 204, 155-164. [DOI] [PubMed] [Google Scholar]
- 51.Harbury, P. B., Zhang, T., Kim, P. S. & Alber, T. (1993) Science 262, 1401-1407. [DOI] [PubMed] [Google Scholar]
- 52.Lumb, K. J. & Kim, P. S. (1995) Biochemistry 34, 8642-8648. [DOI] [PubMed] [Google Scholar]
- 53.Schneider, J. P., Lear, J. D. & DeGrado, W. F. (1997) J. Am. Chem. Soc. 119, 5742-5743. [Google Scholar]
- 54.Mitchell, J. B. O. & Smith, J. (2003) Proteins Struct. Funct. Genet. 50, 563-571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.