Abstract
Altered cellular metabolism is an emerging hallmark of cancer. Accumulating recent evidence links long non-coding RNAs (lncRNAs), a still poorly understood class of non-coding RNAs, to cancer metabolism. Here we review the emerging findings on the functions of lncRNAs in cancer metabolism, with particular emphasis on how lncRNAs regulate glucose and glutamine metabolism in cancer cells, discuss how lncRNAs regulate various aspects of cancer metabolism through their cross-talk with other macromolecules, explore the mechanistic conceptual framework of lncRNAs in reprogramming metabolism in cancers, and highlight the challenges in this field. A more in-depth understanding of lncRNAs in cancer metabolism may enable the development of novel and effective therapeutic strategies targeting cancer metabolism.
Keywords: AMPK, cancer metabolism, c-Myc, long non-coding RNAs, NBR2, mTOR, tumor suppression
Introduction
To support increased proliferation and growth, cancer cells generally reprogram their metabolism to generate ATP rapidly, to facilitate the biosynthesis of macromolecules, and to maintain appropriate redox homeostasis. Compared to normal cells, cancer cells are associated with different metabolic features, such as excessive glucose uptake, more reliance on aerobic glycolysis, increased glutamine uptake and glutaminolysis, and altered lipid metabolism [1]. Mutations in several metabolic enzymes, most notably isocitrate dehydrogenase 1/2, have been identified in various forms of human cancers, hence evidencing a direct link between metabolism and cancer [2]. However, overall mutation rates for metabolic enzymes are relatively low in human cancers. Instead, studies in the past decade have revealed that altered metabolism in cancer is mainly regulated by signaling pathways involved in cell proliferation and growth, which in turn control the metabolism network through various transcriptional and post-translational regulatory mechanisms [3]. Principal among these are the phosphoinositide 3-kinase (PI3K) pathway, which couples extracellular growth factor stimulation to cell proliferation and growth; the mammalian target of rapamycin complex1 (mTORC1, also known as mechanistic TORC1) pathway, which senses and integrates nutrient cues to control cell growth; the liver kinase B1 (LKB1)/AMP-activated protein kinase (AMPK)–mediated energy sensing pathway; and several transcription factors that regulate cell growth and metabolism, including p53, hypoxia-inducible factor 1 (HIF1), and c-Myc [4, 5].
Recent advances in transcriptome analysis have prompted the realization that the human genome encodes an unappreciated large number of non-coding RNAs (ncRNAs), RNA transcripts that do not appear to be translated into proteins [6]. Many ncRNAs are expressed in temporally and spatially controlled manners and have potential transcriptional, post-transcriptional, and epigenetic regulatory functions [7]. Based on the size of the transcripts, ncRNAs are classified into small ncRNAs (shorter than 200 nucleotides) and long ncRNAs (lncRNAs; longer than 200 nucleotides). Small ncRNAs include microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), and small nucleolar RNAs (snoRNAs) [8–10], and it is widely appreciated that small ncRNAs, particularly miRNAs, regulate a broad spectrum of biological processes, including cancer metabolism [11–14]. The functions of lncRNAs in an array of biological processes have been established [15]; however, their roles in the regulation of metabolism and energy homeostasis remain largely elusive. In this perspective, we first briefly introduce the biology of lncRNAs and summarize their classification and known biochemical functions. Then we review the recent findings on the functions of lncRNAs in cancer metabolism, with particular emphasis on how lncRNAs regulate glucose and glutamine metabolism in cancer cells. Finally, we discuss the future prospects for studying lncRNAs in cancer metabolism.
Brief introduction to lncRNAs
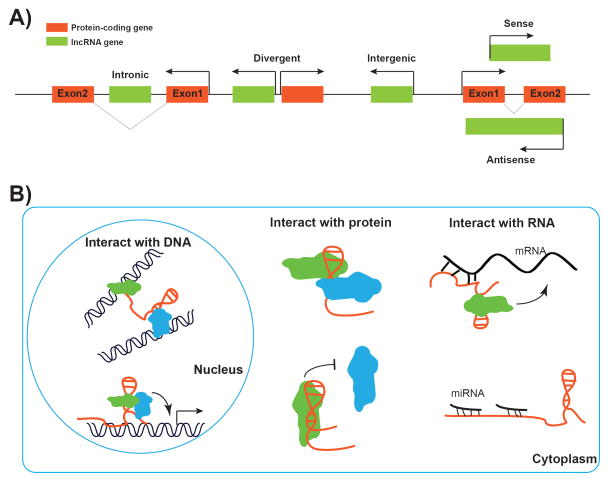
One of the most surprising findings from genomic studies in the past decade is the realization that there is extensive transcription from non-protein-coding regions in the human genome [16]. It is now widely appreciated that there are numerous ncRNAs, whether short or long, encoded in our genome. lncRNAs refer to a loosely classified group of RNA transcripts that are longer than 200 nucleotides and have no apparent protein-coding potential. Similar to protein-coding transcripts, lncRNAs are generally transcribed by RNA polymerase II, 5′-capped, spliced, and polyadenylated [17]. In recent years, high-throughput sequencing technologies, coupled with newly developed computational approaches for transcriptome assembly and annotation, have identified tens of thousands of lncRNAs in the human genome, significantly outnumbering protein-coding genes [18–20]. Since lncRNAs generally lack distinct sequence features and share poor primary sequence conservation, it is difficult to provide categorical definitions of lncRNAs based on their sequence information. Instead, the current classification of lncRNAs is largely based on their genomic location relative to nearby protein-coding genes. Specifically, lncRNAs that are located within the introns of protein-coding genes are referred to as intronic lncRNAs, while lncRNAs that do not overlap with any protein-coding gene are referred to as intergenic lncRNAs. Some lncRNA/mRNA pairs are transcribed from shared promoters by bidirectional transcription: such lncRNAs are classified as divergent lncRNAs. An lncRNA that overlaps with the sense or antisense strand of another protein-coding gene is classified as a sense or antisense lncRNA, respectively (Fig. 1A).
Figure 1.
Classification and mechanisms of lncRNA. A: According to their proximity to protein-coding genes, lncRNAs can be classified into intronic, divergent, intergenic, sense, and antisense lncRNAs. B: Mechanisms of lncRNA in regulation of gene expression. In the nucleus, lncRNAs can interact with chromatin/DNA to regulate gene expression through mediating long-range chromatin interactions or recruitment of regulators to specific genomic loci (left panel). In cytoplasm, lncRNAs can promote or repress the assembly of a protein complex through protein scaffold function or harming protein-protein interaction (middle panel). Furthermore, lncRNAs can interact with mRNA directly and regulate its stability, splicing, or translation or function as sponge to sequester other RNAs, especially miRNAs (right panel).
The expression of most lncRNAs is highly tissue-specific, and is regulated under many physiological and pathological conditions, and dysregulation of lncRNA expression is associated with many human diseases, including cancer [21]. Many studies have shown that lncRNAs regulate a diverse array of biological processes, including cell proliferation, differentiation, survival, and migration. lncRNAs exert their biological functions via their interactions with other cellular macromolecules [22]. For example, via binding to the chromatin/DNA, lncRNAs can function as guides of protein-DNA interactions or enhancers to their neighboring genes. In addition, through interacting with proteins, lncRNAs can function as scaffolds for protein-protein interactions or decoys to proteins (lncRNAs can titrate away or “sponge” other proteins). Finally, through binding to mRNA or other ncRNAs, lncRNAs can regulate mRNA splicing, RNA stability, protein translation, or sequestration of miRNAs (Fig. 1B). For example, lncRNAs can cross-talk with other small RNAs, such as miRNAs, through competing endogenous RNA (ceRNA) mechanisms [23]. According to this model, lncRNAs can function as molecular sponges and titrate miRNAs away from binding to the corresponding miRNA target protein-coding transcripts, which in turn regulate the protein levels of miRNA targets at post-transcriptional levels. Thus, lncRNA, miRNAs, and protein-coding RNAs may form complex regulatory networks to regulate various aspects of biology. Given the enormous flexibility of RNA molecules, many more biochemical and biological functions of lncRNAs are expected to be revealed in future studies.
How do lncRNAs control cancer metabolism?
Glucose and glutamine are the two most abundant nutrients catabolized in significant quantities in cancer cells. Glucose, the most important source of carbon, not only generates energy, but also provides precursors for synthesis of various macromolecules. Glutamine, on the other hand, provides both carbon and nitrogen sources for bioenergetic and biosynthetic processes [5, 24]. Many oncogenic signaling pathways regulate glucose and/or glutamine metabolism, and many cancer cells exhibit oncogene-dependent addictions to glucose or glutamine in culture. In this section, we discuss the emerging functions of lncRNAs in cancer metabolism, focusing on the metabolism of glucose and glutamine. We also discuss the emerging function of lncRNAs in the regulation of mitochondria, the central organelle in energy metabolism.
LncRNAs regulate glucose metabolism
Glucose is the primary energy source for cells. The usage of glucose by cancer cells is significantly different from that of normal cells. Specifically, normal non-proliferating cells, under aerobic conditions, generate ATP primarily through oxidative phosphorylation and switch to glycolysis for ATP generation only under nonaerobic conditions; in contrast, most cancer cells mainly rely on glycolysis to generate ATP even under aerobic conditions, which is known as aerobic glycolysis, or the Warburg effect [3]. The rationale for cancer cells using a less efficient way of generating ATP is not entirely clear, but it has been suggested that, through aerobic glycolysis, a significant fraction of glucose will be shunted into other biosynthetic pathways for the generation of fatty acids, amino acids, and nucleotides, which are the essential building blocks to support cancer cell growth. Many oncogenic signaling pathways promote the Warburg effect through transcriptional or post-translational regulation of various metabolic enzymes involved in glycolysis [3].
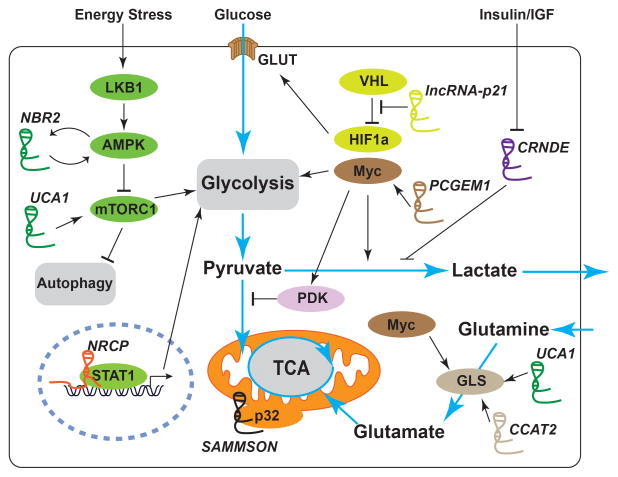
As mentioned earlier, c-Myc and HIF1 are two critical transcription factors that regulate the expression of many genes involved in glycolysis and promote glycolysis. Accordingly, several lncRNAs have been identified to regulate glucose metabolism in cancer cells through c-Myc or HIF1 (Fig. 2). PCGEM1 was identified as an lncRNA that regulates glucose metabolism through its binding to c-Myc [25]. Mechanistically, PCGEM1 functions as a co-activator of c-Myc and regulates the expression of many genes that encode enzymes involved in glucose uptake and glycolysis. In prostate cancer cells, PCGEM1 promotes glucose uptake for aerobic glycolysis, coupling with the pentose phosphate shunt to facilitate biogenesis of nucleotides and lipids [25]. Another example is lncRNA-p21, which was initially identified as an lncRNA induced by p53 [26]. A subsequent study revealed that lncRNA-p21 could also be induced by hypoxia, and is essential for hypoxia-induced glycolysis mediated by HIF-1α [27]. Mechanistically, lncRNA-p21 can bind to both HIF-1α and VHL proteins, disrupts the VHL-HIF-1α interaction, and attenuates VHL-mediated HIF-1α ubiquitination and degradation, leading to the accumulation of HIF-1α protein. Such a positive feedback loop between HIF-1α and lncRNA-p21 presumably further enhances glycolysis under hypoxia [27]. As master transcription factors to regulate glucose metabolism, c-Myc and HIF not only are subjected to tight regulation by lncRNAs, but also regulate the expression of other lncRNAs [28, 29], thus forming complex regulatory networks to regulate glucose metabolism. It will be important to further characterize c-Myc/HIF -regulated lncRNAs and their potential roles in the regulation of glucose metabolism.
Figure 2.
Schematic illustration of selected lncRNAs involved in the regulation of cancer metabolism. By regulating the master regulators/signaling pathways, lncRNAs can reprogram major cancer metabolism pathways. Detailed mechanisms of action of these lncRNAs are described in the main text.
Other lncRNAs have been identified to regulate glycolysis through different mechanisms (Fig. 2). ceruloplasmin (NRCP) was identified to be an lncRNA highly expressed in ovarian cancer, and NRCP knockdown resulted in significantly increased apoptosis, decreased cell proliferation, and defective glycolysis [30]. Mechanistically, NRCP functions as an intermediate binding partner between STAT1 and RNA polymerase II and promotes STAT1-RNA polymerase II interaction, leading to increased expression of downstream target genes such as glucose-6-phosphate isomerase, Aldolase A, and Aldolase C [30]. CRNDE is an lncRNA highly expressed in colorectal adenomas [31] and regulated by insulin/IGF signaling [32]. It has been shown that CRNDE promotes the Warburg effect, though the underlying molecular mechanism remains elusive [32]. Finally, UCA1, an lncRNA upregulated in blacker cancer, promotes aerobic glycolysis via regulating the expression of hexokinase 2, a rate-limiting enzyme in glycolysis. Mechanistically, UCA1 may regulate hexokinase 2 expression through the mTOR–STAT3/miR143 pathway [33]. Collectively, these studies revealed that diverse lncRNAs regulate glycolysis in cancer cells through impinging on different transcriptional machineries involved in transcriptional regulation of gene targets in functioning glycolysis pathway.
ATP is the “molecular unit of currency” of intracellular energy transfer: ATP stores the chemical energy metabolized from glucose (as well as from other nutrients), and the energy released from ATP hydrolysis is used to drive essentially all cellular processes. As such, the ATP level in our cells needs to be carefully monitored, and this energy checkpoint is mainly mediated by the energy sensor AMPK, a Ser/Thr kinase complex [34]. In response to energy stress, the ATP concentration drops and, correspondingly, AMP concentration rises. AMP binds to AMPK and leads to AMPK allosteric activation. AMPK is also phosphorylated by an upstream kinase, LKB1, leading to further activation of AMPK. Once activated, AMPK phosphorylates a wide range of downstream targets to restore energy balance in response to energy stress. One prominent effect upon AMPK activation is the suppression of protein and lipid syntheses, as these anabolic processes consume a large amount of energy. Since protein and lipid syntheses are required for tumor growth, the LKB1-AMPK-mediated energy-sensing pathway suppresses tumor development in many cancers [35]. Indeed, LKB1 is a well-established tumor suppressor mutated in various forms of human cancers [36]. Our recent study uncovered an lncRNA-involved mechanism to regulate AMPK activation in response to energy stress [37, 38] (Fig. 2). NBR2 (neighbor of BRCA1 gene 2) was discovered as a glucose starvation-induced lncRNA through LKB1-AMPK signaling. Furthermore, functional studies revealed that NBR2 directly interacts with the kinase domain of AMPK and promotes AMPK kinase activity. Overexpression of NBR2 in cancer cells led to AMPK activation and proliferation suppression, while NBR2 knockdown downregulated glucose starvation-induced AMPK activation, leading to defective autophagy/apoptosis and enhanced tumor development [37]. Interestingly, NBR2 originally was identified as a non-coding gene that resides adjacent to the tumor suppressor gene BRCA1 [39], and both genes are transcribed from a shared promoter by bidirectional transcription [51], but its function in cancer biology had remained elusive until our recent study [37]. NBR2 represents one of the first identified lncRNAs that can directly regulate kinase function, although the exact biochemical mechanism by which NBR2 promotes AMPK kinase activity awaits further investigation. Notably, recent studies revealed that many metabolic enzymes are capable of interacting with RNAs [40,41]. Thus, NBR2-AMPK model may provide a conceptual framework to further identify and study other novel lncRNAs that can directly regulate kinase or metabolic enzyme activities.
LncRNAs regulate glutamine metabolism
Glutamine, the second principal growth-supporting metabolite, is the most abundant amino acid in both cell culture medium and blood in vivo, and serves as an essential metabolite to support cancer cell proliferation and growth. In the first step of glutamine metabolism, glutaminase catalyzes the hydrolysis of glutamine to glutamate and ammonia. Glutamate then serves as the precursor for several metabolism processes: first, glutamate can be further metabolized to α-ketoglutarate, a tricarboxylic acid cycle intermediate, by either glutamate dehydrogenase or amino acid transaminases. Second, glutamate contributes to reduced nitrogen for biosynthesis of nucleotides and other nonessential amino acids. Finally, glutamate is also a precursor of glutathione, a major cellular antioxidant [24].
Glutaminase controls the rate-limiting step in glutamine metabolism and thus is subjected to tight regulation. There are two glutaminase isozymes, GLS and GLS2. GLS exists in at least two isoforms, namely KGA (glutaminase kidney isoform) and GAC (glutaminase isoform c) [24]. Accumulating evidences revealed that lncRNAs modulate glutamine metabolism through regulation of different glutaminases (Fig. 2). As discussed previously, UCA1 was identified as an lncRNA that regulates glucose metabolism [33]. A recent study revealed that UCA1 also regulates glutamine metabolism [42]. The study showed that the expression of UCA1 and GLS2 are positively correlated in bladder cancer tissues and cell lines, and that UCA1 overexpression induces GLS2 expression at both the mRNA and protein levels and promotes glutaminolysis in human bladder cancer cells. Mechanistically, it was proposed that UCA1 serves as a molecular sponge to interfere with the function of miR-16, which is known to target GLS2 [42]. Colon cancer-associated transcript 2 (CCAT2), an lncRNA associated with increased risk for various types of cancers, was also recently found to modulate glutamine metabolism in colon cancer cells [43]. Mechanistic studies showed that CCAT2 interacts with the CFIm complex, a protein complex that regulates mRNA cleavage and polyadenylation, and regulates the alternative splicing and the poly(A) site selection of GLS mRNA, resulting in the preferential expression of the more aggressive splice isoform GAC. Because glutamine serves as the precursor for several metabolism processes and glutamine metabolism involves various metabolic enzymes, future studies are expected to identify novel lncRNAs that regulate other aspects involved in glutamine metabolism.
LncRNAs link to mitochondrial function
Mitochondria play a central role in metabolic homeostasis [44]. In addition to producing ATP, mitochondria generate important biosynthetic intermediates and reactive oxygen species that propagate various cellular signaling processes. Dysregulation of mitochondrial function has been linked to various metabolic disorders and cancer. Emerging data suggest that lncRNAs also play important roles in maintaining mitochondrial function (Fig. 2). SAMMSON, a recently annotated lncRNA, is found to be specifically upregulated in melanoma and required for melanoma growth and survival [45]. SAMMSON primarily localizes in the cytoplasm, a certain fraction co-localizing with mitochondria. An RNA pulldown assay showed that p32, an important regulator of mitochondrial homeostasis and metabolism, is a major binding protein to SAMMSON. Furthermore, upon SAMMSON silencing, p32 rapidly shuttled into the nucleus, suggesting that SAMMSON interacts with p32 to maintain the appropriate mitochondrial localization and function of p32. Through interaction with p32, SAMMSON regulates the maturation of mitochondrial 16S rRNA, the expression of mitochondrial-encoded proteins, and the maintenance of mitochondrial membrane potential and oxidative phosphorylation [45]. It will be of interest to identify other mitochondria-localized lncRNAs and study their roles in cancer metabolism in future studies.
Future challenges in studying lncRNAs in cancer metabolism
Recent studies have revealed the important functions of lncRNAs in cancer metabolism. However, compared to our ample understanding of the regulation of cancer metabolism by protein-coding genes, our knowledge of lncRNAs in cancer metabolism is still limited: several challenges and important questions remain to be addressed in future studies. First, compared to the annotation of protein-coding genes, it is still challenging to annotate lncRNAs accurately. In addition, the expression levels of most lncRNA genes are relatively low, and experimental approaches to predict or study the biological function of the majority of lncRNAs remain scant. Second, the conservation of lncRNAs across different organisms is generally low. With the current computational tools, most of the lncRNAs identified from human cells/tissues do not appear to have corresponding orthologue genes in invertebrate organisms, and in many cases, even in mouse [46]. Such features significantly limit our capability to use relevant model systems, such as genetically engineered mouse models, to study the lncRNAs identified in humans. Correspondingly, it may be difficult to translate findings of lncRNAs made from other model organisms into human setting if such lncRNAs are not conserved among different organisms.
Finally, the major conceptual frameworks for our current understanding of lncRNAs have been mainly built upon the studies of nucleus-localized lncRNAs. Accordingly, most of the lncRNAs discussed in this review are proposed either to regulate gene transcription or to serve as decoys to other macromolecules, consistent with the commonly proposed functions for lncRNA, as summarized in Fig. 1B. In this regard, NBR2 is a remarkable example, because it represents one of the first cytoplasm-localized lncRNAs that directly regulate kinase function [37]. Given that biological pathways regulating cancer metabolism mainly occur in the cytoplasm or intracellular organelles, future studies on lncRNAs in cancer metabolism will undoubtedly uncover other novel lncRNA-involved regulatory mechanisms that will go beyond the current conceptual frameworks. The hypothesis that some lincRNAs can be transported and function in intracellular organelles (such as mitochondria and lysosomes) to control metabolism is of particular interest, and remains to be tested. In addition, essentially all currently studied lncRNAs are proposed to exert their biological effects through interactions with proteins or other nucleic acids (DNA/RNA). We envision that some linRNAs involved in the regulation the cancer metabolism may directly interact with other metabolites, such as sugars, lipids, and amino acids. Various protein-based nutrient sensors function to sense and respond to intracellular or extracellular nutrient availability [47]. The aforementioned AMPK protein complex directly interacts with AMP and serves as the energy sensor [34]. Recent studies also identified several amino acid sensors that sense different forms of amino acids to regulate mTORC1 activation and cell growth. For example, the Sestrin family of proteins directly interact with leucine and function as leucine sensor to regulate mTORC1 [48], whereas CASTOR family of proteins directly interact with arginine and may function as arginine sensor to control mTORC1 activation [49]. Notably, in bacteria, riboswitches serve as RNA-based sensors for various intracellular metabolites, in which a regulatory segment of mRNA molecules can directly bind to certain metabolites and modulate the synthesis of the proteins encoded by the corresponding mRNAs, thus coordinating the nutrient availability to the production of the proteins that function in the corresponding metabolism pathways [50]. Although riboswitches mainly occur in bacteria, it is conceivable that there may exist lncRNA-based sensors that can directly interact with metabolites and sense the nutrient availability in mammals. Future studies will be directed to address these exciting questions and hypotheses.
Conclusion and prospects
LncRNAs are emerging as an important class of regulators in cancer metabolism. The discoveries and functional studies of lncRNAs involved in cancer metabolism have expanded our knowledge of the mechanisms of reprogramming of cancer metabolism, as exemplified by the studies described above. We envision the future studies will address the technical and conceptual challenges discussed above, and will reveal many more exciting discoveries on how lncRNAs control cancer metabolism. Given the highly dynamic expression patterns of lncRNAs in cancers and emerging important functions of lncRNAs in cancer metabolism, future studies on lncRNAs in cancer metabolism may identify novel biomarkers or therapeutic targets for cancer treatment.
Acknowledgments
We thank Sunita Patterson for manuscript editing. We also are grateful for funding support from MD Anderson Cancer Center, Cancer Prevention Research Institute of Texas (RP130020), National Institutes of Health (CA181196 and CA190370), Ellison Medical Foundation (AG-NS-0973-13), and Gabrielle’s Angel Foundation for Cancer Research. B. G. is a Kimmel Scholar and an Ellison Medical Foundation New Scholar.
Abbreviations
- AMPK
AMP-activated protein kinase
- CCAT2
Colon cancer-associated transcript 2
- ceRNAs
competing endogenous RNAs
- GAC
glutaminase isoform c
- GLS
glutaminase
- HIF1
hypoxia-inducible factor 1
- KGA
glutaminase kidney isoform
- LKB1
the liver kinase B1
- lncRNAs
long non-coding RNAs
- miRNAs
microRNAs
- mTORC1
mammalian target of rapamycin complex 1 or mechanistic target of rapamycin complex 1
- NBR2
neighbor of BRCA1 gene 2
- ncRNAs
non-coding RNAs
- piRNAs
piwi-interacting RNAs
- PI3K
phosphoinositide 3-kinase
- snoRNAs
small nucleolar RNAs
Footnotes
Conflict of interest
The authors have no conflict of interest to declare.
References
- 1.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102:932–41. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 5.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–98. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–37. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Dupuis-Sandoval F, Poirier M, Scott MS. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA. 2015;6:381–97. doi: 10.1002/wrna.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: Its biogenesis and functions. Annu Rev Biochem. 2015;84:405–33. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- 10.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 11.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–50. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumortier O, Hinault C, Van Obberghen E. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 2013;18:312–24. doi: 10.1016/j.cmet.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Chan B, Manley J, Lee J, Singh SR. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015;356:301–8. doi: 10.1016/j.canlet.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Jin LH, Wei C. Role of microRNAs in the Warburg effect and mitochondrial metabolism in cancer. Asian Pac J Cancer Prev. 2014;15:7015–9. doi: 10.7314/apjcp.2014.15.17.7015. [DOI] [PubMed] [Google Scholar]
- 15.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 16.Djebali S, Davis CA, Merkel A, Dobin A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 18.Cabili MN, Trapnell C, Goff L, Koziol M, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyer MK, Niknafs YS, Malik R, Singhal U, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yotsukura S, duVerle D, Hancock T, Natsume-Kitatani Y, et al. Computational recognition for long non-coding RNA (lncRNA): Software and databases. Brief Bioinform. 2016 doi: 10.1093/bib/bbv114. in press. [DOI] [PubMed] [Google Scholar]
- 21.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salmena L, Poliseno L, Tay Y, Kats L, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–84. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung CL, Wang LY, Yu YL, Chen HW, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci USA. 2014;111:18697–702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huarte M, Guttman M, Feldser D, Garber M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Kim T, Jeon YJ, Cui R, Lee JH, et al. Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/dju505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhry H, Schodel J, Oikonomopoulos S, Camps C, et al. Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2. EMBO Rep. 2014;15:70–6. doi: 10.1002/embr.201337642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rupaimoole R, Lee J, Haemmerle M, Ling H, et al. Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 2015;13:2395–402. doi: 10.1016/j.celrep.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham LD, Pedersen SK, Brown GS, Ho T, et al. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2:829–40. doi: 10.1177/1947601911431081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta. 2014;1843:372–86. doi: 10.1016/j.bbamcr.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Li X, Wu S, Xue M, et al. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 2014;105:951–5. doi: 10.1111/cas.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–62. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–75. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. 2008;27:6908–19. doi: 10.1038/onc.2008.342. [DOI] [PubMed] [Google Scholar]
- 37.Liu X, Xiao ZD, Han L, Zhang J, et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016;18:431–42. doi: 10.1038/ncb3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Xiao ZD, Gan B. An lncRNA switch for AMPK activation. Cell Cycle. 2016 doi: 10.1080/15384101.2016.1184515. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu CF, Brown MA, Nicolai H, Chambers JA, et al. Isolation and characterisation of the NBR2 gene which lies head to head with the human BRCA1 gene. Hum Mol Genet. 1997;6:1057–62. doi: 10.1093/hmg/6.7.1057. [DOI] [PubMed] [Google Scholar]
- 40.Beckmann BM, Horos R, Fischer B, Castello A, et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat Commun. 2015;6:10127. doi: 10.1038/ncomms10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castello A, Fischer B, Eichelbaum K, Horos R, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 42.Li HJ, Li X, Pang H, Pan JJ, et al. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn J Clin Oncol. 2015;45:1055–63. doi: 10.1093/jjco/hyv132. [DOI] [PubMed] [Google Scholar]
- 43.Redis RS, Vela LE, Lu W, Ferreira de Oliveira J, et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol Cell. 2016;61:520–34. doi: 10.1016/j.molcel.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng Z, Ristow M. Mitochondria and metabolic homeostasis. Antioxid Redox Signal. 2013;19:240–2. doi: 10.1089/ars.2013.5255. [DOI] [PubMed] [Google Scholar]
- 45.Leucci E, Vendramin R, Spinazzi M, Laurette P, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–22. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 46.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–10. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfson RL, Chantranupong L, Saxton RA, Shen K, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–8. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chantranupong L, Scaria SM, Saxton RA, Gygi MP, et al. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell. 2016;165:153–64. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao ZD, Liu X, Zhuang L, Gan B. NBR2: a former junk gene emerges as a key player in tumor suppression. Mol Cell Oncol. 2016;3(4) doi: 10.1080/23723556.2016.1187322. [DOI] [PMC free article] [PubMed] [Google Scholar]