Abstract
Alignment of protein structures is a fundamental task in computational molecular biology. Good structural alignments can help detect distant evolutionary relationships that are hard or impossible to discern from protein sequences alone. Here, we study the structural alignment problem as a family of optimization problems and develop an approximate polynomial-time algorithm to solve them. For a commonly used scoring function, the algorithm runs in O(n10/ε6) time, for globular protein of length n, and it detects alignments that score within an additive error of ε from all optima. Thus, we prove that this task is computationally feasible, although the method that we introduce is too slow to be a useful everyday tool. We argue that such approximate solutions are, in fact, of greater interest than exact ones because of the noisy nature of experimentally determined protein coordinates. The measurement of similarity between a pair of protein structures used by our algorithm involves the Euclidean distance between the structures (appropriately rigidly transformed). We show that an alternative approach, which relies on internal distance matrices, must incorporate sophisticated geometric ingredients if it is to guarantee optimality and run in polynomial time. We use these observations to visualize the scoring function for several real instances of the problem. Our investigations yield insights on the computational complexity of protein alignment under various scoring functions. These insights can be used in the design of scoring functions for which the optimum can be approximated efficiently and perhaps in the development of efficient algorithms for the multiple structural alignment problem.
Keywords: protein structural comparison, internal distances matrices
1. Introduction
A fundamental task in structural molecular biology is comparison of protein structures. Evolution conserves protein structure significantly more than protein sequence. Additionally, structural similarity often reflects a common function or origin of proteins (1). In view of this, structural biologists have been making intensive efforts to systematically classify all known protein structures (2, 3), yielding structural databases such as Structural Classification of Proteins (SCOP) (4), Families of Structurally Similar Proteins (FSSP) (5), and CATH (6). Automatic methods for structure comparison are useful for generating such databases. They also may be utilized for classifying newly determined structures, based on similarities with previously classified structures (5). The rapid growth of the Protein Databank (PDB) (7) underscores the need for fast and accurate methods for structure comparison.
The “structural-alignment” problem is the structural analog of the well known sequence-alignment problem. The input to the former consists of two protein structures in three-dimensional space, 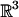 . The desired output is a pair of maximal substructures, one from each protein, that exhibit the highest degree of similarity. A sequential alignment of the two substructures yields a sequence of residue pairs that is called a correspondence. Typically, we simplify the model by comparing the structures using one atom per residue, generally but not necessarily the CA atom; this makes the unit of comparison (a CA atom) correspond to a unit of sequence (a residue). There are two main methods to quantify similarity. The first method computes internal distances between corresponding pairs of atoms in the two proteins and compares these distances in the two proteins under consideration. The second method uses the actual Euclidean distance between corresponding atoms in the two proteins under comparison. To do this, the method must also determine the rigid transformation that optimally positions the two structures vis-á-vis a each other.
. The desired output is a pair of maximal substructures, one from each protein, that exhibit the highest degree of similarity. A sequential alignment of the two substructures yields a sequence of residue pairs that is called a correspondence. Typically, we simplify the model by comparing the structures using one atom per residue, generally but not necessarily the CA atom; this makes the unit of comparison (a CA atom) correspond to a unit of sequence (a residue). There are two main methods to quantify similarity. The first method computes internal distances between corresponding pairs of atoms in the two proteins and compares these distances in the two proteins under consideration. The second method uses the actual Euclidean distance between corresponding atoms in the two proteins under comparison. To do this, the method must also determine the rigid transformation that optimally positions the two structures vis-á-vis a each other.
These two methods of quantifying similarity give rise to two approaches for solving the structural-alignment problem. Investigators subscribing to the first approach have developed heuristic algorithms that compare the internal distance matrices in search of the optimal correspondence. An advantage of these algorithms is that they bypass the need to find an optimal rigid transformation (e.g., refs. 8–14). The most commonly used structural alignment server, dali (9), belongs to this group. Along the second approach, heuristic algorithms have been developed to optimize the correspondence and the rigid transformation simultaneously (e.g., refs. 15–22). Excellent reviews of these and other methods can be found in refs. 3 and 23–25. A prevailing sentiment in both research communities is that structural alignment requires exponential computational resources, and thus, investigations should concentrate on heuristic approaches (13, 24, 26). Indeed, none of the above-mentioned heuristics guarantees finding an optimal alignment with respect to any scoring function.
One of the main tenets of the present work is that measurements of protein coordinates are necessarily noisy, and consequently, there is not much point in seeking exact solutions for the structural-alignment problem. Rather, approximate solutions are called for. Coordinates are merely approximations to a “true” position: proteins are flexible, fluctuating about a mean position, and the physical experiment that provides the coordinates is noisy (27, 28). It follows that in the proper approximate model, the error is additive and not too small. Distinct solutions to the structural-alignment problem that are close to the optimum (depending on measurement errors) are a priori all equally interesting. Furthermore, as noted by Zu-Kang and Sippl (29), multiple correspondences may exist, all equally viable from the biological perspective, and hence all are equally interesting from the computational point of view.
In this article, we present a polynomial-time algorithm that optimizes both the correspondences and rigid transformations (i.e., we operate within the second approach). Our algorithm is not heuristic: it guarantees finding ε-approximations to all solutions of the protein structural-alignment problem. To bound the size of the solution space, we first consider the complexity of searching rotation and translation space. We show that it depends polynomially on the lengths of the proteins, n, and on 1/ε for an approximation parameter ε. On the other hand, the number of possible correspondences grows exponentially with the length. Based on these observations, we suggest an algorithm for structural alignment: search exhaustively the relatively small space of all rigid transformations for an optimal alignment. Because the algorithm is exhaustive, when it fails to find a good alignment, it is certain that none exist. The contribution of this article should be viewed as mostly theoretical rather than practical. We prove that, contrary to common belief (13, 24, 26), finding ε-approximations of the optimal solutions is computationally feasible, albeit too slow to be a useful everyday tool. Furthermore, our approach offers a way to visualize the structural-alignment score as a function of all rigid transformations, which is useful for developing intuitions for better optimization algorithms and heuristics. We display scores for three examples: (i) two structures with a unique good alignment, (ii) two structures with several good alignments, and (iii) two structures that cannot be aligned.
Our solution applies to a broad class of interesting scoring functions, including, most importantly, structal (17), a commonly used score. It can be implemented to optimize the structal score in time complexity O(n10/ε6) for two globular proteins with length that is at most n. We also point out some of the difficulties involved with the (numerous) attempts to solve the structural-alignment problem based on the internal distance matrices of the two proteins.
We introduce the necessary terminology in section 2 and investigate scoring functions in section 3. In section 4, we consider the space of alignments for three specific pairs of proteins and draw our conclusions in section 5.
2. Preliminaries
For the present article, a “protein” is a chain of atoms residing in three-dimensional space. Consider a protein A of n atoms, A = (a1,..., an), with 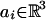 . We assume, without loss of generality, that A is positioned with its center of mass at the origin and is bounded by a box of dimensions XA × YA × ZA. It is known (30) that the volume of a protein is linear in the number of its residues, that is, XA·YA·ZA = O(n). We also let RA denote the radius of the bounding sphere of the protein A. In the special case of “globular” proteins, the size of the protein along all axis XA, YA, ZA is O(n1/3) and RA = O(n1/3). In line with our perspective that this is mostly a theoretical study, we use the O notation freely. Recall the notation g(n) = O[f(n)] means at most cf(n) for a constant c independent of n.
. We assume, without loss of generality, that A is positioned with its center of mass at the origin and is bounded by a box of dimensions XA × YA × ZA. It is known (30) that the volume of a protein is linear in the number of its residues, that is, XA·YA·ZA = O(n). We also let RA denote the radius of the bounding sphere of the protein A. In the special case of “globular” proteins, the size of the protein along all axis XA, YA, ZA is O(n1/3) and RA = O(n1/3). In line with our perspective that this is mostly a theoretical study, we use the O notation freely. Recall the notation g(n) = O[f(n)] means at most cf(n) for a constant c independent of n.
A “subchain” of protein A is a subset of its atoms, arranged by order of appearance in A. Denote the k-long subchain defined by P = (p1, p2,..., pk), where 1 ≤ p1 < p2 < · · · < pk ≤ n, by A(P) = (ap1, ap2,..., apk). A “gap” is two consecutive indices pi, pi+1 such that pi + 1 < pi+1.
Consider two proteins, A of n atoms and B of m atoms, and two subchains, P of protein A and Q of protein B; we assume, without loss of generality, that n ≥ m. We call two subchains P and Q of equal length, |P| = |Q|, a “correspondence.” Thus, a correspondence associates pairs of atoms from two proteins that appear in the same position in their respective subchains. Note that although the two complete proteins can differ in length, the subchains cannot. In the world of protein sequences, the analogous term is alignment; at times, it is used here, too, interchanged with correspondence. The number of gaps in a correspondence, denoted GP,Q, is the sum of the number of gaps in P and Q.
Structural-Alignment Problem. Given two proteins A and B, find two subchains P and Q of equal length such that
A(P) and B(Q) are similar, and
The correspondence length |P| = |Q| is maximal under condition 1.
A protein can be rigidly transformed (i.e., rotated and translated) without affecting its inherent structure. Rotations and translations are each specified by three parameters (31). Because we are interested in the relative position and orientation of the two proteins, we can hold A fixed and only transform B; the rigidly transformed B is denoted by B̂ = (b̂1,..., b̂m). The relative position and orientation of the proteins are useful for solving the protein-alignment problem.
There are various measures of similarity, or deviation, between two subchains. Among the more commonly used are distance root mean squared (dRMS) deviation and coordinate root mean squared (cRMS) deviation, which we define here for completeness.
For subchains P and Q of length k, dRMS is defined as
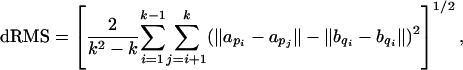 |
and cRMS is defined as
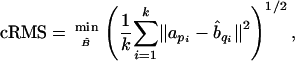 |
where B̂ is the image of protein B under a rigid transformation. The transformation that achieves this minimum can be found in closed form (e.g., by using Kabsch's procedure in ref. 32).
The general structural-alignment problem gives rise to a family of concrete optimization problems, which are specified by the weight given to the (preferably small) deviation of the subchains and the (preferably large) length of the correspondence. Note that cRMS and dRMS measure only deviation and, therefore, must be complemented by a score that favors longer correspondences. An important score that plays a key role here is that used by structal (17). It is closer in spirit to cRMS in that it compares matching pairs of the correspondence and considers the rotated and translated position of the structures. It also penalizes for gaps in the correspondence.
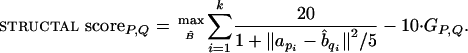 |
When using this score, we seek a rigid transformation and a correspondence that achieves a maximal (rather than minimal) value.
3. Approximate Structural Alignment
We focus on scores that evaluate the similarity of two structures by explicitly applying a rigid transformation to one and then comparing the transformed structure with the other. For such scores, the optimization problem is to find transformations and correspondences of (near) optimal score.
The polynomial-time algorithm we present calculates the optimal score for a substantial number of rotations and translations. It then sifts through these scores to find the best ones, i.e., the pairs (transformation and correspondence) with near globally maximum scores. For the algorithm to run in polynomial time, the following two conditions must hold:
Given a fixed transformation, it should be possible to find in polynomial time an optimal correspondence. We elaborate on this possibility in section 3.1.
The number of rigid transformations under consideration must be bounded by a polynomial. This issue is addressed in section 3.2.
Next, we define guidelines that can be used in designing novel scoring functions for structural alignment; scores that follow these guidelines come hand in hand with a polynomial-time algorithm that finds all near-optimal alignments. Researchers are still far from a thorough understanding of the desirable characteristics of scoring functions for this problem. Some obvious interesting options that are yet to be investigated are variable gap penalties depending on the location of the gap within the structure (e.g., higher penalty inside a helix) and scores that take into account sequence information.
3.1. Separability of Scoring Function. As mentioned above, we are assuming that for a fixed rotation and translation, the optimal correspondence can be determined in polynomial time. This requirement strongly points to scores that can be optimized by using dynamic programming. When applicable, a dynamic programming algorithm finds in polynomial time an optimal solution among an exponential number of potential correspondences. Score functions that are amenable to dynamic programming must satisfy two requirements: (i) optimal substructure, where the restriction of an optimal correspondence to any substructure is itself an optimal correspondence of the substructure, and (ii) the space of relevant subproblems is small (polynomial). For more details, see ref. 33. Scores that satisfy these conditions are called “separable.” The structal score is separable, and the optimal correspondence can be determined by using dynamic programming in O(n2) time and space (17, 34).
3.2. Lipschitz Condition on Scoring Functions. Here, we provide conditions on a scoring function under which its overall behavior can be approximated by evaluating it only polynomially many times. Recall that a scoring function assigns a value to every correspondence and rotation and translation. We present a scheme for approximating all rotations and translations with (local) optimal correspondence that is near the global optimum.
A rigid transformation in 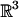 consists of a translation and a rotation. A translation is parameterized by a vector (tx, ty, tz) in
consists of a translation and a rotation. A translation is parameterized by a vector (tx, ty, tz) in 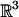 . Here, it suffices to have tx, ty, tz range over [-(XA + XB)/2, (XA + XB)/2], [-(YA + YB)/2, (YA + YB)/2], and [-(ZA + ZB)/2, (ZA + ZB)/2], respectively (recall that XA, YA, ZA and XB, YB, ZB are the sides of the bounding boxes of A and B). There are many ways to parameterize the group of rotations SO(3). For the purpose of our proofs, we represent each rotation by three angles (r1, r2, r3) in the range [0, 2π] (31), which constitutes a 4-fold cover of SO(3). For the purpose of sampling (see section 4), we use the parametrization of rotations by means of quaternions (unit vectors in
. Here, it suffices to have tx, ty, tz range over [-(XA + XB)/2, (XA + XB)/2], [-(YA + YB)/2, (YA + YB)/2], and [-(ZA + ZB)/2, (ZA + ZB)/2], respectively (recall that XA, YA, ZA and XB, YB, ZB are the sides of the bounding boxes of A and B). There are many ways to parameterize the group of rotations SO(3). For the purpose of our proofs, we represent each rotation by three angles (r1, r2, r3) in the range [0, 2π] (31), which constitutes a 4-fold cover of SO(3). For the purpose of sampling (see section 4), we use the parametrization of rotations by means of quaternions (unit vectors in 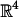 ) (35): a rotation by an angle θ about the normalized vector (nx, ny, nz) is described by (nx cos θ/2, ny cos θ/2, nz cos θ/2, sin θ/2). This representation has the advantage that angular separation of quaternions captures the natural metric of SO(3). In this representation, opposite points in S3 are identified, reflecting the topology of SO(3). Equivalently, it suffices to consider only half of S3.
) (35): a rotation by an angle θ about the normalized vector (nx, ny, nz) is described by (nx cos θ/2, ny cos θ/2, nz cos θ/2, sin θ/2). This representation has the advantage that angular separation of quaternions captures the natural metric of SO(3). In this representation, opposite points in S3 are identified, reflecting the topology of SO(3). Equivalently, it suffices to consider only half of S3.
Assume we have a correspondence between subchains P of protein A and Q of protein B. Fix protein A in space. For every rigid transformation of protein B, one can compute the correspondence-dependent scoring (CDS) function by using the distances between corresponding atom pairs in space. We index the real-valued CDS function by P and Q and denote it ScP,Q.
The CDS function is defined over the space of all rigid transformations. Note that there are exponentially many correspondences and thus exponentially many CDS functions defined in this manner. Specifically, there are 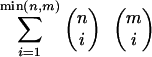 functions. In this article, we are concerned with a scoring function of the form
functions. In this article, we are concerned with a scoring function of the form
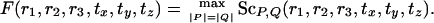 |
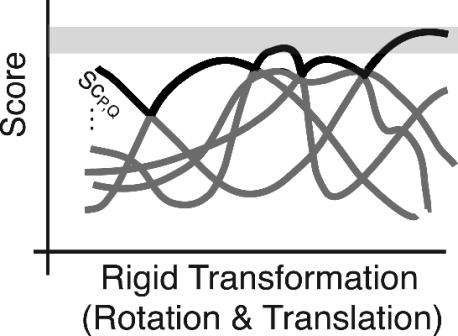
This is the upper envelope of all CDS functions (see Fig. 1).
Fig. 1.
A schematic view of the scoring functions, parameterized by the rigid transformation. The (exponentially many) CDS functions are depicted in dark gray, and their upper envelope is marked in black. We are concerned with all solutions in the light-gray region: top values in the upper envelope.
The following lemma gives conditions on the scoring function. When these conditions hold, it is possible to derive good approximations of all the near-optimal maxima of the function from only polynomially many evaluations of the function.
Definition 3.1: A CDS function ScP,Q satisfies coordinate-wise Lipschitz conditions with values cr and ct if for all rigid transformations 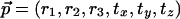 and for all δ > 0,
and for all δ > 0,
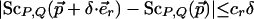 |
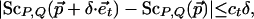 |
where 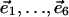 are the standard basis vectors in
are the standard basis vectors in  ,
,  , and
, and 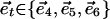 .
.
Lemma 3.1. Let the CDS functions satisfy coordinate-wise Lipschitz conditions with values cr, ct. For every ε > 0, there exists a finite set G = G(ε) of rotations and translations such that
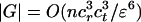 , and
, andFor every choice of a translation and a rotation
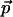 , there is a point
, there is a point 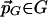 with
with 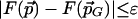 .
.
We refer to the set G as an ε-net for the scoring function.
Proof: The set G(ε) is the product of six sets of equally spaced points in each of its six dimensions. In the three dimensions of rotations, the spacing is δr = ε/3cr; the size of the set in each dimension of rotation is O(cr/ε). Similarly, in the three dimensions of translation, the spacing is δt = ε/3ct, and the size of the set is O[(WA + WB)ct/ε], where W = X, Y, Z, respectively. Taking into account that a protein of n residues satisfies X·Y·Z = O(n), the total size of G follows.
The coordinate-wise Lipschitz condition for each CDS function ScS,T implies that the same condition holds for their upper envelope F. Given a point 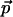 in rotation and translation space, the nearest point
in rotation and translation space, the nearest point 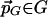 can be reached by moving at most δr/2 along each of the three dimensions of rotation and δt/2 along each of the three dimensions of translation. Because F satisfies Lipschitz condition, the change in value of F induced by each such step is at most δcr/2 in the first case and δct/2 in the latter. Thus, the overall change is at most 3crδr/2 + 3ctδt/2 = ε/2 + ε/2 = ε.
can be reached by moving at most δr/2 along each of the three dimensions of rotation and δt/2 along each of the three dimensions of translation. Because F satisfies Lipschitz condition, the change in value of F induced by each such step is at most δcr/2 in the first case and δct/2 in the latter. Thus, the overall change is at most 3crδr/2 + 3ctδt/2 = ε/2 + ε/2 = ε.
Lemma 3.1 suggests the following algorithm to find all points with near-maximal values of the scoring function F. Let M be the global maximum of F, and call a point 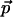 ε-maximal if
ε-maximal if 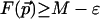 . For every point
. For every point 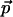 that is ε-maximal, there is a nearby point
that is ε-maximal, there is a nearby point 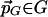 with
with 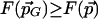 . We would like every ε-maximal point to be accounted for by a nearby point in G. Given ε, evaluate F on all points of G(ε) defined above. Next, select the subset of these points within 2ε from the maximal value found. This algorithm will guarantee finding approximations to all ε-maximal points, satisfying our requirement.
. We would like every ε-maximal point to be accounted for by a nearby point in G. Given ε, evaluate F on all points of G(ε) defined above. Next, select the subset of these points within 2ε from the maximal value found. This algorithm will guarantee finding approximations to all ε-maximal points, satisfying our requirement.
3.2.1. structal-type scores. Consider the family of structal-type scores,
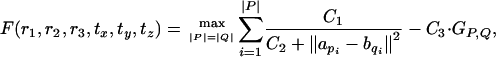 |
where C1, C2, and C3 are positive constants. In the structal score, they are set at C1 = 100, C2 = 5, and C3 = 10. Lemma 3.2 shows that all such functions are well behaved and thus can be approximated by a polynomial-sized net.
structal-type scoring functions can be approximated to ε-accuracy with a net of size O(n8/ε6) for globular proteins and O(n10/ε6) for nonglobular ones. In particular, we show
Lemma 3.2. Any CDS function of the form
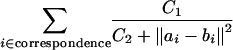 |
satisfies the Lipschitz condition of Lemma 3.1 with ct = O(n) and cr = O(n4/3) in case of globular proteins. For nonglobular proteins, cr = O(n2).
Proof: First consider a single pair of atoms a and b and their contribution to the scoring function. A translation of b by δ along any axis can change ∥a - b∥ by at most δ. A rotation of b by δ around any axis can change ∥a - b∥ by at most Rδ. The function φ(x):= C1/(C2 + x2) has a bounded derivative |φ′(x)| ≤ M(C1, C2) = M. By the mean-value theorem, it follows that a change of δ in any of the six coordinates will result in a change of at most M Rδ + Mδ in the scoring function, the first term being due to rotations and the other to translations. There are at most n contributions to a CDS function (n is the number of atoms in the longer protein), and thus, the total change is bounded by nMRδ from rotations and nMδ from translations. For globular structures, R = O(n1/3), and in general, R = O(n). We have shown that the coordinate-wise Lipschitz condition is satisfied with cr = O(n4/3) for globular proteins and cr = O(n2)for nonglobular ones.
Altogether, finding approximations to all ε-near-optimal points for structal-type scoring functions takes O(n10/ε6) time for globular proteins, which is because of O(n8/ε6) evaluations of the scoring function at the points of G(ε), each evaluation taking O(n2) time. More generally, our scheme is an approximate polynomial algorithm for every separable scoring function that requires only polynomially many evaluation points (see Lemma 3.1). Notice that this scheme gives all near-optimal function values rather than all near-optimal correspondence. It would be interesting to determine whether the task of finding all near-optimal correspondences can be solved efficiently.
3.3. A Closely Related NP-Hard Problem. Associated with every protein chain A of n atoms is an n × n real symmetric matrix D, where D(i, j) is the Euclidean distance in  between the ith and jth atoms of A. This matrix is called “the (internal) distances matrix” and is invariant under rigid and mirror transformations of the protein. The internal distances matrix that corresponds to a subchain S of the protein A is the submatrix (minor) of D consisting of the rows and columns indexed by the elements of S.
between the ith and jth atoms of A. This matrix is called “the (internal) distances matrix” and is invariant under rigid and mirror transformations of the protein. The internal distances matrix that corresponds to a subchain S of the protein A is the submatrix (minor) of D consisting of the rows and columns indexed by the elements of S.
The two representations of a protein, by atomic coordinates and by the internal-distances matrix, are of course closely related. Calculating the distances matrix from the atomic coordinates is easy (and takes quadratic time). It is also known that the coordinates of the protein can be recovered in polynomial time from the distances matrix by using distance geometry (36). This calculation is possible because proteins lie in a three-dimensional Euclidean space. The recovered atomic coordinates are the original ones, modulo a rigid (and possibly a mirror) transformation.
It follows that any algorithm that uses either of these two representations can be converted into one that uses the other. The conversion is straightforward: add a preprocessing and a postprocessing step that translate, in polynomial time, between representations.
The internal-distances matrix representation of proteins may seem attractive, because it limits the search to the correspondences without need to optimize on the rigid transformations. Methods that use the internal-distance matrix representation directly compare pairs of submatrices and optimize a measure that is derived from dRMS deviation. When the correspondence is found, the rigid transformation that optimally superimposes the two substructures can be recovered with Kabsch's procedure (32). It is generally considered a minor problem that the final positioning and orienting of the structures optimizes the cRMS deviation, whereas the correspondence optimizes a different measure (dRMS).
We point out that a correct and efficient solution to the approximate structural-alignment problem must exploit the fact that proteins lie in three-dimensional Euclidean space. In particular, we show that a slightly generalized problem in which the internal distances come from a general metric space (not necessarily Euclidean) is NP-hard. We define a particular scoring function to focus the discussion; a similar argument applies to variants of this scoring function.
Intuitively, the problem is hard because all (exponentially many) pairs of subchains are potential solutions. Notice that if we restrict the number of gaps by a constant, there are only polynomially many potential solutions, which substantially reduces the computational complexity of the problem. The CLIQUE problem is well known to be NP-hard (37): the input is a graph and an integer k, and the output should be either a k-clique or, if there is no such clique, the answer “no.” We reduce the CLIQUE problem to the problem at hand to demonstrate its hardness, that is, we show how an algorithm that finds an ε-approximation to the optimal solution can be used efficiently to solve CLIQUE.
Lemma 3.3. Let DA, DB be distance matrices of two metric spaces (think of them as the internal-distance matrices of chains A and B). Let the score of two subchains P and Q, of equal length |P| = |Q| = k, be
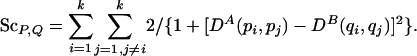 |
For every 0 < ε < 1, it is NP-hard to find subchains that are within ε from the optimal score.
Proof: Given a graph G = (V, E), |V| = n we “construct” two chain structures and use the algorithm for finding correspondences of near-optimal scores. The first structure, denoted SA, has n “atoms” and encodes the graph G: each vertex is associated with an atom (using some ordering), and the distance between two atoms is the length of a shortest path in G between the two corresponding vertices. The internal-distances matrix associated with this structure is an n × n matrix DA, where DA(i, j) is the length of the shortest path from vi to vj. The second structure, denoted SB, has k “atoms;” it encodes a clique of size k. The internal-distances matrix in this case, denoted DB, has zeros on the diagonal and ones elsewhere. If the score is strictly >2(k2 - k) - 1, return the subset of SA (a k-clique); otherwise, return “no.” Because SB has k atoms, the score is a sum of at most k2 - k terms. Also, the distances are integers, which restricts the possible values of the terms in the sum; 2 and 1 are the two largest values. Thus, if a good score is found, it is ≥2(k2 - k). This optimal value is achieved when a k-clique in SA is found; otherwise, there is clearly no k-clique.
An algorithm that solves the approximate structural-alignment problem by using only the metric properties (and not the fact that it is a three-dimensional Euclidean metric) also solves the above generalized problem. Thus, such an algorithm either fails to find optimal approximations or it is inefficient (or that P = NP, which generally is viewed as unlikely).
To summarize, the problem in finding good correspondences is that there are (exponentially) many potential candidates. The number of possibilities can be greatly reduced because the structures lie in three-dimensional Euclidean space and the scores are separable. However, if these restrictions are removed, an exponential blow-up in computational complexity seems unavoidable.
4. Results
We examine the properties of the structal score for structural alignment of various pairs of proteins; we focus on the domain of rotations while optimizing the translation parameters. By the observations discussed above, it suffices to calculate the structal score on a net in the space of transformations. Let R be a net for the space of rotations, T a net for translations, and R × T a net for all rigid transformations. Ideally, we would like to visualize the structal score over R × T to determine all (near) maxima. Visualizing a function of six parameters is, of course, very hard. We thus focus our attention on the three-dimensional space of rotations and define
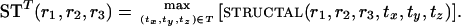 |
The advantage of focusing on STT is that it can be visualized; the disadvantage is that multiple maxima due to translation changes alone are hidden.
To reduce the time for exploring the scoring function over the space of rotations, we heuristically calculate the maximum over a smaller set of translations, denoted T(ot). T(ot) is the set of translations that position an atom from protein A exactly on top of an atom from protein B, |T(ot)| = O(nm). This heuristic speeds up the calculation by a factor of O(n2). The sets T and T(ot) are different; the maximum over T can be higher or lower than the maximum over T(ot). However, we assume that the best translation in T positions at least one atom from A on top of (or close to) an atom from B, implying that STT(ot) and STT reasonably approximate each other. In the supporting information, which is published on the PNAS web site, we show a comparison of the values of STT and STT(ot) for 1,000 random rotations and demonstrate that the two scores indeed approximate each other well.
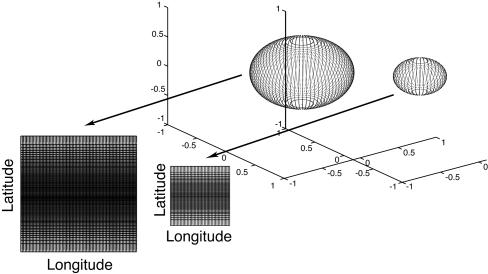
We parameterize the group of rotations by using quaternions (35). Notice that a Cartesian product of longitude and latitude nets is a net for a sphere S2 in R3. Here, we use a longitude net that is uniformly spaced and a latitude net that uniformly spaces its cosine values. This net can be visualized in the plane by placing the longitude values along the x axis and the latitude values along the y axis. Note that unlike the sphere, this display does not show the wrapping around of the longitude values; it also has a distortion around the poles. Fig. 2 shows examples of three-dimensional spheres, overlayed with their net points and a planar layout of the nets. The two-dimensional spheres can be sampled more efficiently (e.g., ref. 38); our sampling scheme allows easy planar visualization of the score function values on the spheres.
Fig. 2.
Visualization of score values for a net of discrete rotations. We model rotations by using quaternions: each rotation is a four-dimensional vector of unit length. We consider a net that covers the space of quaternions, or the unit sphere S3 in 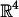 . We use a net that is the union of nets on a discrete set of three-dimensional spheres. For each three-dimensional sphere, we use a Cartesian product of longitude and latitude values and plot it in two dimensions. The width of the two-dimensional plot varies with the radius of its corresponding sphere. The scores are described by using color (specific values and colors are not shown but are shown in Figs. 3 and 4). Notice that there is a distortion associated with this display, especially around the poles. Two spheres, overlaid with their net points and their corresponding two-dimensional plots, are shown.
. We use a net that is the union of nets on a discrete set of three-dimensional spheres. For each three-dimensional sphere, we use a Cartesian product of longitude and latitude values and plot it in two dimensions. The width of the two-dimensional plot varies with the radius of its corresponding sphere. The scores are described by using color (specific values and colors are not shown but are shown in Figs. 3 and 4). Notice that there is a distortion associated with this display, especially around the poles. Two spheres, overlaid with their net points and their corresponding two-dimensional plots, are shown.
We visualize the function STT(ot) for specific pairs of proteins by using short videos. The position in the frame sequence serves as an additional dimension (aside of the two planar coordinates). The data figures show an (ordered) subset of the video frames. The full videos are available in supporting information. Denote a unit quaternion in 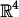 by
by 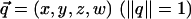 . The set of unit quaternions of a fixed w is a sphere S2 in R3 of radius
. The set of unit quaternions of a fixed w is a sphere S2 in R3 of radius 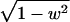 . Each frame in the video has a fixed w value, which determines the width and position of the frame in the sequence. Because we are concerned only with half of S3, w is equally spaced in the interval [-1, 0]. Varying w, we place a net on the corresponding sphere and evaluate the scoring function at all net points. Then, the score values are visualized in the plane as described above by using a color scale ranging from blue (low) to red (high). The number of points on a net varies with the area that it covers, totaling in ≈106 points.
. Each frame in the video has a fixed w value, which determines the width and position of the frame in the sequence. Because we are concerned only with half of S3, w is equally spaced in the interval [-1, 0]. Varying w, we place a net on the corresponding sphere and evaluate the scoring function at all net points. Then, the score values are visualized in the plane as described above by using a color scale ranging from blue (low) to red (high). The number of points on a net varies with the area that it covers, totaling in ≈106 points.
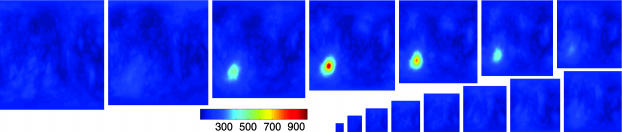
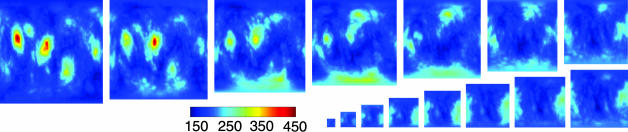
We examine three types of behaviors of the structal-scoring function when aligning pairs of proteins. Fig. 3 shows the score when considering two proteins with a single meaningful maximum: 5rxn and 1brf (SCOP fold classification rubredoxin-like). This figure shows a clear, single, high-scored maximum yielding a good alignment. Fig. 4 shows the scoring function for two proteins with several meaningful maxima: 1mjc and 1shf-a (SCOP fold OB and SH3-like barrel, respectively). This example is listed in the work of Zu-Kang and Sippl (29). A closer examination of the different maxima in Fig. 4 shows that the multiple high-scoring orientations are because of an internal symmetry in the structures 1mjc and 1shf-a; these alignments (sequences and superpositioning of the structures) are given in supporting information. This symmetry, coupled with the two structures being fairly similar to each other, accounts for the multiple orientations that position many atoms from one structure near corresponding atoms from the other. Last, we plot (shown in supporting information) the scores when aligning two structurally different proteins: 1jjd and 1dme (both SCOP fold metallotheionein). In this case, there are only rotations with low scoring alignments, proving that there are no good alignments.
Fig. 3.
Example of a pair of structures with a single meaningful alignment. We plot the STT(ot) score for aligning 5rxn (54 residues) and 1brf (53 residues) over the space of rotations. These proteins have the same SCOP fold classification, rubredoxin-like, and each has three β-strands and three helices. The STT(ot) score function has a single maximum, implying one meaningful way of aligning the pair. The maximal score found is 993, aligning 53 residues to 0.797 Å cRMS.
Fig. 4.
Example of a pair of structures with two meaningful alignments [this example was noted by Zu-Kang and Sippl (29)]. We plot the STT(ot) score for aligning 1mjc (69 residues) and 1shf (59 residues). The two maxima can be seen clearly, as can additional, less significant maxima. One of the two best alignments scores 458, aligning 41 residues to 2.89 Å cRMS; the other scores 454, aligning 38 residues to 2.52 Å cRMS. These proteins are of the same SCOP class, all-β, and a different SCOP fold (OB and SH3-like barrel, respectively).
5. Conclusions
We have presented a polynomial scheme for protein structural alignment. Exploring the space of rigid transformations solves this problem efficiently, because it exploits the fact that proteins reside in a three-dimensional Euclidean space. It seems unclear how to incorporate this crucial information if one phrases the problem through internal-distances matrices. Unless three-dimensionality is taken into account, the problem becomes significantly harder (NP-hard). We found sufficient conditions for a scoring function such that all optima can be found in polynomial time. Devising novel scoring functions that detect biologically significant substructures is still an open area of research.
Experiments with the structal-scoring function on several pairs of proteins suggest that this scoring function is “well behaved” on the domain of rotations. Studying the landscape of various scoring functions can prove valuable for the purpose of developing robust and efficient tools for structural alignment.
Note that an immediate extension of this algorithm solves multiple structural alignment. Namely, sampling the space of rigid transformations and finding the maximum by using dynamic programming can find all approximate global maxima of the upper envelope of the CDS functions. For a fixed, small number of globular proteins, it is a polynomial algorithm [e.g., for three globular proteins, it takes O(n19/ε12) time]. Multiple structural alignment is a wide open problem, and although the direct extension has prohibitive running time, the analysis described in this article offers a means of tackling it.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant CCR-00-86013 and The Sudarsky Center for Computational Biology.
Abbreviations: dRMS, distance root mean squared; cRMS, coordinate root mean squared; CDS, correspondence-dependent scoring.
References
- 1.Chothia, C. & Lesk, A. M. (1986) EMBO J. 5, 823-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibrat, J. F., Madej, T. & Bryant, S. H. (1996) Curr. Opin. Struct. Biol. 6, 377-385. [DOI] [PubMed] [Google Scholar]
- 3.Orengo, C. (1994) Curr. Opin. Struct. Biol. 4, 429-440. [Google Scholar]
- 4.Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995) J. Mol. Biol. 247, 536-540. [DOI] [PubMed] [Google Scholar]
- 5.Holm, L. & Sander, C. (1994) Nucleic Acids Res. 22, 3600-3609. [PMC free article] [PubMed] [Google Scholar]
- 6.Orengo, C. A., Michie, A. D., Jones, S., Jones, D. T., Swindells, M. B. & Thornton, J. M. (1997) Structure (London) 5, 1093-1108. [DOI] [PubMed] [Google Scholar]
- 7.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000) Nucleic Acids Res. 28, 235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor, W. R. & Orengo, C. A. (1989) J. Mol. Biol. 208, 1-22. [DOI] [PubMed] [Google Scholar]
- 9.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123-138. [DOI] [PubMed] [Google Scholar]
- 10.Godzik, A., Skolnick, J. & Kolinski, A. (1993) Protein Eng. 6, 801-810. [DOI] [PubMed] [Google Scholar]
- 11.Yee, D. P. & Dill, K. A. (1993) Protein Sci. 2, 884-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuguchi, K. & Go, N. (1995) Protein Eng. 8, 353-362. [DOI] [PubMed] [Google Scholar]
- 13.Shindyalov, I. N. & Bourne, P. E. (1998) Protein Eng. 11, 739-747. [DOI] [PubMed] [Google Scholar]
- 14.Szustakowski, J. D. & Weng, Z. (2000) Proteins Struct. Funct. Genet. 38, 428-440. [DOI] [PubMed] [Google Scholar]
- 15.Nussinov, R. & Wolfson, H. J. (1991) Proc. Natl. Acad. Sci. USA 88, 10495-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vriend, G. & Sander, C. (1991) Proteins 11, 52-58. [DOI] [PubMed] [Google Scholar]
- 17.Subbiah, S., Laurents, D. V. & Levitt, M. (1993) Curr. Biol. 3, 141-148. [DOI] [PubMed] [Google Scholar]
- 18.Diederichs, K. (1995) Proteins 23, 187-195. [DOI] [PubMed] [Google Scholar]
- 19.Madej, T., Gibrat, J. F. & Bryant, S. H. (1995) Proteins 23, 356-369. [DOI] [PubMed] [Google Scholar]
- 20.May, A. C. W. & Johnson, M. S. (1995) Protein Eng. 8, 873-882. [DOI] [PubMed] [Google Scholar]
- 21.Akutsu, T. (1996) IEICE Trans. Inf. Syst. 12, 1629-1636. [Google Scholar]
- 22.Wu, T. D., Schmidler, S. C., Hastie, T. & Brutlag, D. L. (1998) J. Comput. Biol. 5, 585-595. [DOI] [PubMed] [Google Scholar]
- 23.Lemmen, C. & Lengauer, T. (2000) J. Comput. Aided Mol. Des. 14, 215-232. [DOI] [PubMed] [Google Scholar]
- 24.Eidhammer, I., Jonassen, I. & Taylor, W. R. (2000) J. Comput. Biol. 7, 685-716. [DOI] [PubMed] [Google Scholar]
- 25.Koehl, P. (2001) Curr. Opin. Struct. Biol. 11, 348-353. [DOI] [PubMed] [Google Scholar]
- 26.Godzik, A. (1996) Protein Sci. 5, 1325-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chelvanayagam, G., Roy, G. & Argos, P. (1994) Protein Eng. 7, 173-184. [DOI] [PubMed] [Google Scholar]
- 28.Karplus, M. & Petsko, G. (1990) Nature 347, 631-639. [DOI] [PubMed] [Google Scholar]
- 29.Zu-Kang, F. & Sippl, M. J. (1996) Folding Des. 1, 123-132. [DOI] [PubMed] [Google Scholar]
- 30.Hao, M. H., Rackovsky, S., Liwo, A., Pincus, M. R. & Scheraga, H. A. (1992) Proc. Natl. Acad. Sci. USA 89, 6614-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craig, J. (1986) Introduction to Robotics: Mechanics and Control (Addison–Wesley, Reading, MA).
- 32.Kabsch, W. (1978) Acta Crystallogr. A 34, 827-828. [Google Scholar]
- 33.Cormen, T. H., Leiserson, C. E. & Rivest, R. L. (1990) Introduction to Algorithms (MIT Press, Cambridge, MA).
- 34.Gerstein, M. & Levitt, M. (1996) Proc. Int. Conf. Intell. Syst. Mol. Biol. 4, 59-67. [PubMed] [Google Scholar]
- 35.Shoemake, K. (1985) Comput. Graph. (ACM) 19, 245-254. [Google Scholar]
- 36.Havel, T. F., Kuntz, I. D. & Crippen, G. M. (1983) Bull. Math. Biol. 45, 665-720. [Google Scholar]
- 37.Papadimitriou, C. (1994) Computational Complexity (Addison–Wesley, Reading, MA).
- 38.Saff, E. B. & Kuijlaars, A. B. J. (1997) Math. Intelligencer 19, 5-11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.