Abstract
Schizophrenia is a devastating neurodevelopmental disorder with a complex genetic etiology. Widespread cortical gray matter loss has been observed in patients and prodromal samples. However, it remains unresolved whether schizophrenia-associated cortical structure variations arise due to disease etiology or secondary to the illness. Here we address this question using a partitioning-based heritability analysis of genome-wide SNP and neuroimaging data from 1,750 healthy individuals. We find that schizophrenia-associated genetic variants explain a significantly enriched proportion of trait heritability in eight brain phenotypes (FDR=10%). In particular, intracranial volume (ICV) and left superior frontal gyrus thickness exhibit significant and robust associations with schizophrenia genetic risk under varying SNP selection conditions. Cross disorder comparison suggests that the neurogenetic architecture of schizophrenia-associated brain regions is, at least in part, shared with other psychiatric disorders. Our study highlights key neuroanatomical correlates of schizophrenia genetic risk in the general population. These may provide fundamental insights into the complex pathophysiology of the illness, and a potential link to neurocognitive deficits shaping the disorder.
Keywords: Schizophrenia, SNP heritability analysis, linear mixed effects model, genomic partitioning strategy, superior frontal gyrus, intracranial volume
Introduction
Schizophrenia is a common and devastating brain disorder with a strong genetic basis (heritability h2 ~ 80%)(1). Neuroimaging studies have reported widespread brain anatomical changes as potential pathogenic features of the disorder(2). Such brain alterations have been linked to various clinical symptoms, encompassing cognitive impairment(3), auditory hallucinations(4–7), and social dysfunction(8). Nevertheless, a major question remains unanswered: which of the structural changes reflect a shared genetic basis with disease etiology – and thus are potential endophenotypes(9), or arise secondary to disease progression or treatment-related mechanisms(10). Studies of patients, although unquestionably informative, have encountered multiple challenges, including confounding effects of drugs and comorbidity conditions, and genetic, clinical heterogeneity of the patient population.
The emerging availability of large-scale cohorts of unaffected individuals with neuroimaging and genotype data provides a unique opportunity to gain insight into the shared genetic basis of brain anatomy and disease. As with many other complex disorders, schizophrenia is highly polygenic(11) and genetic vulnerability to the disorder lies on a continuum across health and disease(12). Therefore, examining the relationship between normal variation in brain anatomy and genetic liability to schizophrenia has the potential to reveal neuroanatomical correlates of schizophrenia disease risk. Traditionally, effects of individual schizophrenia risk variants or aggregated polygenic disease burden have been studied on a prioritized list of brain imaging traits(13–15). These works have yielded key discoveries. For example, accumulating evidence suggests that dysregulated prefrontal cortex function(14, 16–18) and white matter connectivity(19, 20) index inherited schizophrenia disease risk.
Here, we use a partitioning-based heritability analysis to investigate the shared genetic basis of brain anatomy and schizophrenia risk using genome-wide single nucleotide polymorphism (SNP) data. Linear mixed effects models(21) provide a powerful means to quantify heritability of major psychiatric disorders(22–24) and brain structural traits(25, 26). By utilizing the genetic similarity between unrelated individuals, this method can estimate the heritability of a phenotype without family or twin data. Furthermore, by partitioning the genome-wide variants into biologically meaningful categories, SNP heritability analysis has illustrated the genetic architecture of various human traits(27–30). We use the genomic partitioning strategy to identify brain phenotypes for which normal variation is significantly modulated by schizophrenia-risk variants(27, 28). Our central hypothesis is that independently ascertained schizophrenia-associated variants will explain a significantly enriched proportion of the phenotypic variation for brain traits that share a genetic basis with the disease. Here, we tested this hypothesis using neuroimaging and genome-wide SNP data from 1,750 healthy young adults(31–34). By identifying key neuroanatomical correlates of schizophrenia genetic risk in the general population, we ultimately aim to provide key insights into the complex pathophysiology of the illness, and a potential link to neurocognitive deficits shared across diverse psychiatric disorders.
Materials and Methods
Study subjects
Our neuroimaging genetics sample consists of 2,795 healthy subjects collected in two independent studies: the Harvard/MGH Brain Genomics Superstruct Project (GSP; N=1,817)(32, 33) and the Queensland Twin Imaging Study (QTIM; N=978)(31, 34). The GSP project recruited over 3,000 native English-speaking healthy adults between October 2008 and March 2013 from Harvard University, Massachusetts General Hospital (MGH), and the surrounding Boston communities. Participants were assessed for their physical health, past and present history of psychiatric illness, medication usage, family history of psychiatric illness, and alcohol/tobacco/caffeine usage. Participants were excluded if they reported a history of head trauma, current/past Axis I pathology or neurological disorder, current/past psychotropic medication usage, acute physical illness, and/or loss of consciousness. Data analyses in this study were restricted to 1,340 young adults (18–35 years old) of self-identified non-Hispanic European ancestry (age, 21.54±3.19 years old; female, 53.18%; right-handedness, 91.74%). All participants provided written informed consent in accordance with guidelines set by the Partners Health Care Institutional Review Board or the Harvard University Committee on the Use of Human Subjects in Research.
The Queensland Twin Imaging Study (QTIM) is a neuroimaging project that studies genetic influences on brain structure and function using healthy young adult twins. Each member of twin pairs and their siblings were assessed using extensive diagnostic interviews to exclude anyone with a history of brain disorders, diseases, injuries, or substance abuse. Written informed consent was obtained from all participants. We restricted analysis to self-identified Caucasian subjects with both structural MRI scans and genome-wide SNP data. We also included only unrelated individuals in the QTIM to minimize potential biases that may arise if family and unrelated population samples are used together in subsequent heritability analysis. Between each of the twin pairs, one subject was selected at random. The final study samples consisted of 515 unrelated subjects (age, 22.99±3.21 years; female 63.35%). All subjects were right-handed. The QTIM study was approved by the Human Research Ethics Committees of the QIMR Berghofer Medical Research Institute, University of Queensland, and Uniting Health Care, Wesley Hospital.
Imaging data acquisition and processing
For GSP, high-resolution structural brain MR images were collected on matched 3T Tim Trio scanners (Siemens, Erlangen, Germany) at Harvard University and MGH using the 12 or 32 channel phased array head coil. Structural data included a high-resolution multi-echo T1-weighted magnetization-prepared gradient-echo image (multi-echo MP-RAGE; TR=2200 ms, TI=1100 ms, TE=1.54 ms for image 1 to 7.01 ms for image 4, FA=7º, 1.2×1.2×1.2 mm and FOV=230). An optimized study reference template was generated from 700 subjects available through the existing dataset. FreeSurfer (v.4.5.0)(35) was used to process the structural brain MRI scans and to compute global and regional morphological measurements. In the QTIM cohort, structural MRI scans were obtained on a single 4-tesla scanner (Bruker Medspec). T1-weighted 3D structural images were acquired with an inversion recovery rapid gradient echo sequence (MPRAGE; TR=1500 ms; TE=3.35 ms; flip angle=8°; slice thickness=0.9 mm, with a 256 acquisition matrix; with a final voxel resolution=0.9 × 0.9 × 0.9 mm). Quality control procedures for imaging data were described in detail in our previous work(32, 34). In brief, images were screened for artefacts, acquisition problems, processing errors and excessive motion. Each image was examined on a per-slice basis along each principal axis. Typical data quality issues included electronic noise resulting in bright lines through multiple slices, motion artefacts appearing as hazy bands across the image, poor head positioning resulting in wraparound artefacts, distortions from dental work, and limited image contrast. Anatomical scans were then processed through FreeSurfer 5.3.0 in a fully automated manner, without manual corrections. The resulting data were visually inspected for errors. In both datasets, average cortical thickness and surface area measurements were extracted from 68 cortical brain regions (34 regions per hemisphere) defined by the Desikan-Killiany atlas(36). We also included three global brain phenotypes: intracranial volume (ICV), total cortical surface area, and average cortical thickness. We did not include regional volumetric measures as they are a mixture of, and largely genetically correlated with cortical thickness and surface area measurements(37).
Genotyping and data quality control
SNP genotyping was conducted using Illumina OmniExpress BeadChip and Human610-Quad BeadChip for GSP and QTIM respectively. Both datasets were processed with the same genetics protocol for standardized and stringent quality control (QC)(38). SNP markers were excluded if they have genotyping call rates < 99.8%, minor allele frequency (MAF) < 1%, and Hardy-Weinberg equilibrium p < 1×10−2. Subjects were excluded by checking sex discrepancy, cryptic relatedness (π >0.125), or low genotyping call rates (< 99%). PLINK multidimensional scaling analysis was used to exclude population outliers with respect to HapMap 3 European reference samples. To adjust for potential sub-population stratification, we performed principal component analysis (PCA) using EIGENSOFT (web resources). After these QC steps, 460,656 SNPs remained for 1,308 subjects in GSP. Genotyping call rate was 0.99967. In QTIM, 500,332 SNPs remained for 442 independent subjects. Genotyping call rate was 0.999726.
Genome-wide heritability analyses of brain anatomical measures
The narrow-sense heritability, h2g, is defined for each trait as the proportion of total phenotypic variance in the population that is explained by additive genetic factors. As previously described(21), the linear mixed-effects modeling method estimates h2g by: (1) calculating the kinship matrix using the genotypic relatedness matrix (GRM) from high-density SNP data; and (2) estimating variance explained by the GRM jointly with a residual component using the restricted maximum likelihood (REML) strategy. Therefore, h2g can be formulated as: , where var(g) and var(e) are the genetic and residual variance components estimated by the REML approach. The variation of the variance component estimates is estimated from the inverse Fisher information and propagated using a first-order Taylor expansion. In the present study, we used GCTA(21) (web resources) to calculate the GRM and to estimate heritability h2g for examined brain traits conditioned on age, sex, handedness, scanner, console, coil types, and top 4 principal components (PCAs) of ancestry in each study. All analyses of surface area and global brain volumetric measures were adjusted for ICV to account for unexplained variability due to head size(39). Heritability estimates were set between zero and one. As previously noted, only unrelated subjects were used in the heritability analysis in both GSP and QTIM. To further prevent an effect of cryptic relatedness, subjects were excluded if corresponding elements in the GCTA GRM > 0.025. Heritability estimate results from the GSP and QTIM samples were meta-analyzed using a fixed-effects inverse-variance-weighted approach. The power analysis using the GCTA-GREML Power Calculator (web resource) indicates that using our sample size of 1,750 subjects, we have the power of 80.49% to estimate the SNP-heritability of 0.8 at the type I error rate of 3.60×10−4 (α=0.05/139; Bonferroni corrected for examining 139 measures).
Genomic partitioning of heritability using schizophrenia GWAS loci
We obtained the latest genome-wide SNP analysis results of schizophrenia(40) from the Psychiatric Genomics Consortium (PGC; web resource). The association summary statistics consisted of the meta analysis results of 9,270,274 autosomal SNPs using 36,989 cases and 113,075 controls. SNPs were filtered by imputation quality (INFO<0.8) and minor allele frequency (MAF<0.01). In each dataset, genotyped SNPs were ranked based on disease-association p-values from the PGC study, and the top 10% of the genotyped SNPs were retained as disease-associated SNPs. Two GRMs were then calculated based on the set of selected SCZ-associated SNPs and the set of remaining genotype SNPs, respectively. Joint analysis of the two genetic components was performed to estimate the genome-wide SNP heritability as a sum of h2SCZ-GWAS (attributed to by SCZ-associated SNPs) and h2C(SCZ-GWAS) (attributed to a complementary, unselected genotype SNP set). As described by Yang et al.(21), joint analysis of the two GRMs as random effects prevents inflated estimates of local SNP heritability. As in the genome-wide SNP heritability analysis, age, sex, handedness, site, scanner, coil, and top 4 PCAs were included as covariates in the REML. We provide the detailed summary of the partitioning-based SNP heritability analysis in the Supplementary Method.
Enrichment analysis
We tested whether observed partial heritability h2SCZ-GWAS is higher than what would be expected under uniform contribution of genotyped SNPs using the Monte Carlo simulation approach detailed in Toro et al(41). First, we calculated the SNP-set heritability expected under no enrichment as f × h2g, the multiplication of the fraction f of the genome corresponding to each SNP set and the genome-wide heritability estimate h2g. The f was calculated as the proportion of the number of SNPs n in a given set over the total number of N genotype SNPs. The Z score was constructed to test whether the observed partial heritability h2SCZ-GWAS is significantly larger than f × h2g as below: . We calculated the empirical significance of enrichment by comparing the observed Z score to those from 100,000 random permutations, where we assigned n randomly selected genotype SNPs as the SCZ-GWAS SNP set and the remaining unselected SNPs as the other set, and applied partitioning-based heritability analysis. To adjust for multiple testing across brain regions, we used the false discovery rate (FDR) approach.
Genomic partitioning of heritability using other psychiatric disorder GWAS loci
We obtained genome-wide association statistics for five psychiatric disorders(42) from the Psychiatric Genomics Consortium (PGC; web resource). The disorders included: bipolar disorder (BIP; N=11,810), major depressive disorder (MDD; N=16,610), attention deficit hyperactivity disorder (ADHD; N=5,422), autism spectrum disorder (ASD; N=10,263), and anorexia nervosa (N=17,767). As a control dataset, we also examined GWAS results from Crohn’s disease obtained from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC; N=20,883; web resource). The heritability estimation and enrichment analysis procedure was identical to that of schizophrenia.
RESULTS
Genome-wide linear mixed effects modeling nominates 35 brain anatomical traits with significant SNP heritability
Our study sample consisted of 2,795 unrelated healthy subjects of European ancestry recruited in two neuroimaging genetics studies: the Harvard/Massachusetts General Hospital (MGH) Genomic SuperStruct Project (GSP; N=1,817)(32, 33) and the Queensland Twin Imaging Study (QTIM; N=978)(31, 34). After unified and stringent quality control (QC) of genotype and imaging data, we retained a total of 1,750 unrelated young adult subjects (age 18–35). We first estimated the genome-wide SNP heritability h2g of 139 brain anatomical measures. Target regions of interest (ROI) included 3 global and 136 left/right lateral measurements for cortical thickness and surface areas (34 regions per hemisphere) defined by the Desikan-Killiany atlas(36). Estimated heritability varied across brain structures (Supplementary Table 1). Among 3 global traits, significant SNP heritability was found for intracranial volume (ICV; h2g = 0.880; likelihood ratio test (LRT) p = 1.11×10−4) and overall mean cortical thickness (h2g = 0.796; p = 5.43×10−4). Total surface area showed modest but nominally significant heritability after adjusting for ICV (h2g = 0.443; p = 3.51×10−2), suggesting a distinct genetic component aside from ICV. Considerable correlation was found for the SNP heritability of lateral measures between left and right hemispheres (Pearson correlation r = 0.43; p = 6.85×10−4). Overall, the top 35 out of 139 traits exhibited significant SNP heritability based on a false discovery rate (FDR) of 10% (Table 1), and thus were forwarded to subsequent partitioning analyses. Using LD-pruned SNPs did not significantly change the results (Supplementary Table 2).
Table 1.
Genome-wide SNP heritability estimates of 35 brain ROIs with FDR < 10%
| Lobe | Phenotype | H2 | SE | P | FDR |
|---|---|---|---|---|---|
| Global | Intracranial volume | 0.880 | 0.238 | 1.11E−04 | 0.004 |
| Global | Overall mean cortical thickness | 0.796 | 0.244 | 5.43E−04 | 0.011 |
| Frontal | Left precentral gyrus thickness | 0.718 | 0.249 | 1.97E−03 | 0.021 |
| Frontal | Left rostral anterior cingulate cortex thickness | 0.737 | 0.243 | 1.20E−03 | 0.017 |
| Frontal | Left superior frontal gyrus thickness | 0.597 | 0.246 | 7.69E−03 | 0.056 |
| Frontal | Right lateral orbital frontal cortex thickness | 0.483 | 0.240 | 2.22E−02 | 0.091 |
| Frontal | Right pars opercularis surface area | 0.545 | 0.252 | 1.54E−02 | 0.076 |
| Frontal | Right paracentral lobule thickness | 0.494 | 0.252 | 2.51E−02 | 0.099 |
| Frontal | Right precentral gyrus thickness | 0.731 | 0.244 | 1.36E−03 | 0.017 |
| Occipital | Left cuneus cortex thickness | 0.550 | 0.244 | 1.21E−02 | 0.071 |
| Occipital | Left lateral occipital cortex thickness | 0.498 | 0.248 | 2.23E−02 | 0.091 |
| Occipital | Right cuneus cortex thickness | 0.723 | 0.251 | 1.97E−03 | 0.021 |
| Parietal | Left inferior parietal cortex thickness | 0.566 | 0.248 | 1.12E−02 | 0.071 |
| Parietal | Left postcentral gyrus thickness | 0.501 | 0.249 | 2.21E−02 | 0.091 |
| Parietal | Left posterior-cingulate cortex thickness | 0.601 | 0.246 | 7.26E−03 | 0.056 |
| Parietal | Left precuneus cortex surface area | 0.555 | 0.262 | 1.70E−02 | 0.079 |
| Parietal | Left precuneus cortex thickness | 0.896 | 0.245 | 1.29E−04 | 0.004 |
| Parietal | Left superior parietal gyrus surface area | 0.558 | 0.251 | 1.32E−02 | 0.071 |
| Parietal | Left superior parietal gyrus thickness | 0.903 | 0.241 | 8.86E−05 | 0.004 |
| Parietal | Right postcentral gyrus thickness | 0.760 | 0.246 | 1.00E−03 | 0.016 |
| Parietal | Right precuneus cortex surface area | 0.547 | 0.246 | 1.31E−02 | 0.071 |
| Parietal | Right precuneus cortex thickness | 0.965 | 0.243 | 3.59E−05 | 0.003 |
| Parietal | Right superior parietal gyrus thickness | 0.941 | 0.239 | 4.01E−05 | 0.003 |
| Parietal | Right supramarginal gyrus thickness | 0.769 | 0.240 | 6.67E−04 | 0.012 |
| Temporal | Left banks superior temporal sulcus thickness | 0.680 | 0.242 | 2.42E−03 | 0.024 |
| Temporal | Left entorhinal cortex thickness | 0.587 | 0.249 | 9.27E−03 | 0.064 |
| Temporal | Left fusiform gyrus surface area | 0.566 | 0.259 | 1.44E−02 | 0.074 |
| Temporal | Left insula cortex surface area | 0.561 | 0.251 | 1.26E−02 | 0.071 |
| Temporal | Left superior temporal gyrus surface area | 0.658 | 0.244 | 3.47E−03 | 0.032 |
| Temporal | Left transverse temporal cortex thickness | 0.555 | 0.245 | 1.18E−02 | 0.071 |
| Temporal | Right entorhinal cortex surface area | 0.651 | 0.251 | 4.78E−03 | 0.042 |
| Temporal | Right insula cortex surface area | 0.878 | 0.252 | 2.46E−04 | 0.006 |
| Temporal | Right middle temporal gyrus surface area | 0.610 | 0.244 | 6.20E−03 | 0.051 |
| Temporal | Right temporal pole surface area | 0.524 | 0.249 | 1.76E−02 | 0.079 |
| Temporal | Right transverse temporal cortex thickness | 0.536 | 0.254 | 1.73E−02 | 0.079 |
Genomic partitioning of SNP heritability shows a significantly enriched contribution of schizophrenia GWAS loci for several brain phenotypes
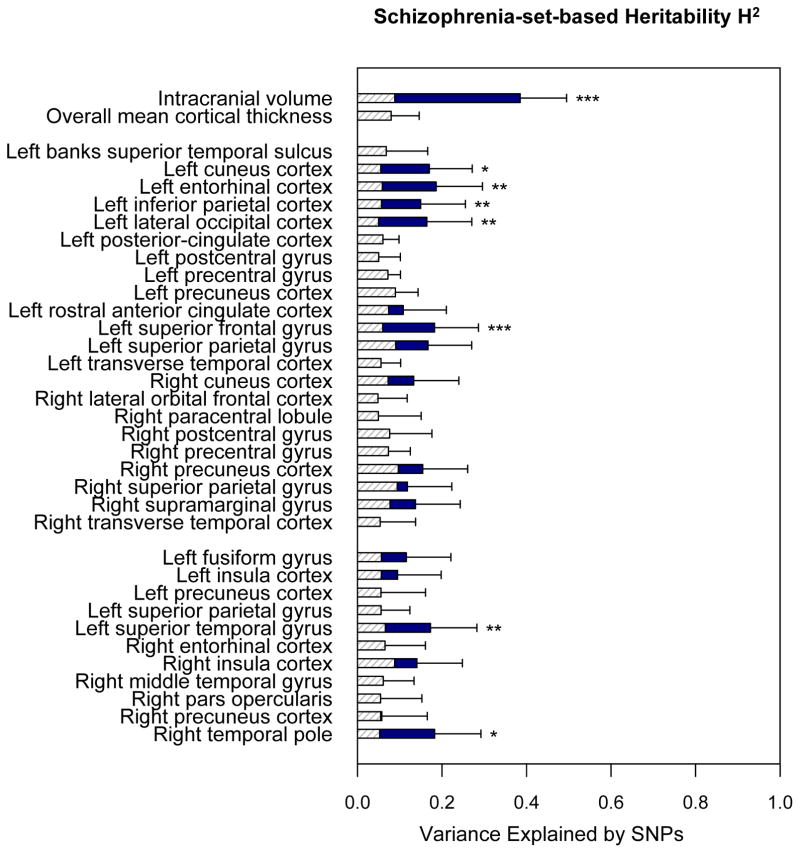
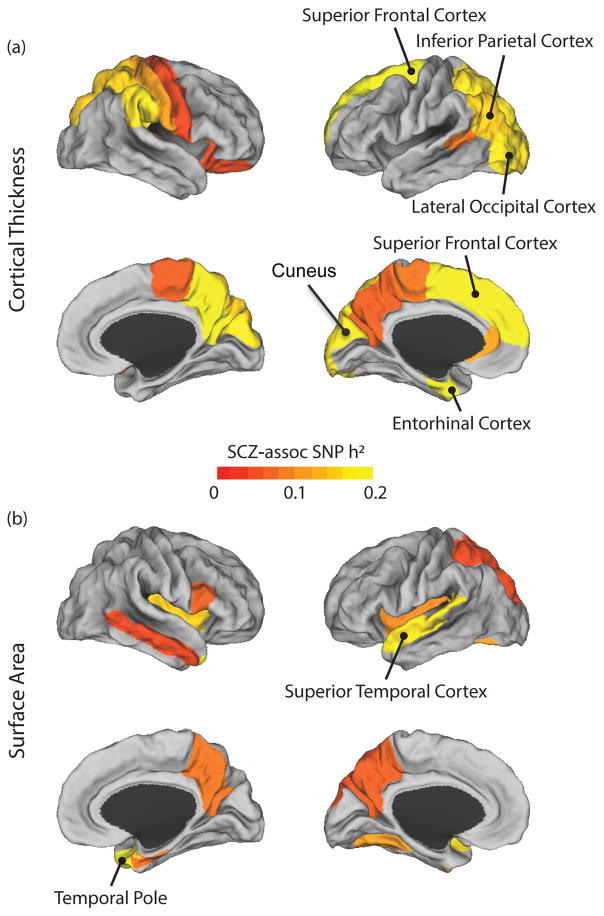
Next, we examined how much of the genome-wide SNP heritability of these 35 brain anatomical traits is attributable to independently ascertained schizophrenia-associated genetic variants. Using genome-wide association results from the largest Psychiatric Genomics Consortium (PGC) schizophrenia study, we divided genotyped SNPs in our dataset into two mutually exclusive sets: the top 10% of schizophrenia-associated SNPs and the remaining 90% of SNPs. For each brain measure, we estimated genome-wide SNP heritability using a joint analysis of the two SNP sets. Finally, increased contribution of a schizophrenia-associated SNP set was quantified using Monte Carlo simulations as previously described (see Methods)(22, 27, 28). As summarized in Figure 1 and Table 2, eight anatomical traits showed significantly enriched contribution of the schizophrenia-associated SNP set at FDR q = 0.1: (1) ICV (enrichment p = 2.61×10−5); (2) left superior frontal thickness (p = 7.25×10−4); (3) left entorhinal cortex thickness (p = 1.02×10−3); (4) right temporal pole surface area (p = 2.77×10−3); (5) left superior temporal surface area (p = 3.98×10−3); (6) left inferior parietal thickness (p = 7.36×10−3); (7) left lateral occipital cortex thickness (p = 9.28×10−3); and (8) left cuneus cortex thickness (p = 1.64×10−2). Figure 2 shows the spatial distribution of the schizophrenia-associated SNP-based heritability estimates for the 35 brain phenotypes: cortical thickness (Fig. 2A) and surface area (Fig. 2B) across the entire cerebral cortex. We also confirmed that partitioned heritability estimates for ICV and superior frontal cortex thickness show consistently enriched contribution of schizophrenia-associated SNPs across varying SNP selection thresholds (10–50%; Supplementary Table 3).
Figure 1.
Estimates of SNP heritability explained by independently ascertained schizophrenia-associated SNPs compared to the null expectation. The disease-associated SNP selection procedure is as follows: all genotyped SNPs were ranked based on the results of the recent genome-wide case/control association study of schizophrenia, obtained from the Psychiatric Genomics Consortium. Based on the rank, disease-associated SNPs were selected to consist of 10% of the genotyped SNPs in our dataset, and the remaining 90% of SNPs were assigned as non-associated SNPs. GCTA was used to perform a joint analysis of two set-based genetic components. The y-axis represents 35 brain anatomical traits separated into three groups: (1) global anatomical measures; (2) cortical thickness; and (3) surface area measurements. The x-axis represents estimated schizophrenia-set-based SNP heritability for a respective trait. Estimated heritability is displayed by a solid navy-color bar, while the null expectation (i.e., estimates calculated based on the proportion of SNPs with respect to genome-wide SNP heritability) is shown by a dashed gray-line bar. Error bars represent the standard errors of the estimates. Significance of enrichment was calculated using a Monte-Carlo simulation. Brain traits with enriched schizophrenia-set-based heritability at FDR q ≤ 0.1 are highlighted with asterisk (*** uncorrected p<0.001; ** p <0.01; * p<0.05).
Table 2.
Enrichment analysis results of schizophrenia-associated SNP-set-based heritability for 35 brain traits
| Trait | Whole genome-based heritability | Disorder-associated SNP set heritability | Enrichment analysis results | ||||
|---|---|---|---|---|---|---|---|
| H2(G) | H2(d) | SE(d) | P(d) | e(H2(d)) | P(H2(d)>e(H2(d)) | FDR | |
| Intracranial volume | 0.88 | 0.385 | 0.11 | 2.28E−04 | 0.088 | 2.61E−05 | 0.001 |
| Overall mean cortical thickness | 0.796 | 0.043 | 0.103 | 3.46E−01 | 0.08 | 1.00E+00 | 1.000 |
| Left banks superior temporal sulcus thickness | 0.68 | 0.066 | 0.1 | 2.69E−01 | 0.068 | 5.28E−01 | 0.812 |
| Left cuneus cortex thickness | 0.55 | 0.17 | 0.102 | 5.02E−02 | 0.055 | 1.64E−02 | 0.072 |
| Left entorhinal cortex thickness | 0.587 | 0.186 | 0.11 | 4.81E−02 | 0.059 | 1.02E−03 | 0.012 |
| Left fusiform gyrus surface area | 0.566 | 0.115 | 0.105 | 1.15E−01 | 0.057 | 9.68E−02 | 0.302 |
| Left inferior parietal cortex thickness | 0.566 | 0.149 | 0.106 | 7.53E−02 | 0.057 | 7.36E−03 | 0.043 |
| Left insula cortex surface area | 0.561 | 0.095 | 0.103 | 1.78E−01 | 0.056 | 2.30E−01 | 0.504 |
| Left lateral occipital cortex thickness | 0.498 | 0.164 | 0.106 | 6.16E−02 | 0.05 | 9.28E−03 | 0.046 |
| Left postcentral gyrus thickness | 0.501 | 0.001 | 0.101 | 5.00E−01 | 0.05 | 1.00E+00 | 1.000 |
| Left posterior-cingulate cortex thickness | 0.601 | 0.001 | 0.098 | 2.80E−01 | 0.06 | 1.00E+00 | 1.000 |
| Left precentral gyrus thickness | 0.718 | 0.001 | 0.102 | 2.86E−01 | 0.072 | 1.00E+00 | 1.000 |
| Left precuneus cortex surface area | 0.555 | 0.055 | 0.105 | 2.97E−01 | 0.055 | 5.05E−01 | 0.812 |
| Left precuneus cortex thickness | 0.896 | 0.04 | 0.103 | 3.41E−01 | 0.09 | 1.00E+00 | 1.000 |
| Left rostral anterior cingulate cortex thickness | 0.737 | 0.108 | 0.102 | 1.36E−01 | 0.074 | 2.26E−01 | 0.504 |
| Left superior frontal gyrus thickness | 0.597 | 0.182 | 0.104 | 6.39E−02 | 0.06 | 7.25E−04 | 0.012 |
| Left superior parietal gyrus surface area | 0.558 | 0.02 | 0.104 | 4.31E−01 | 0.056 | 1.00E+00 | 1.000 |
| Left superior parietal gyrus thickness | 0.903 | 0.167 | 0.103 | 4.96E−02 | 0.09 | 4.35E−02 | 0.169 |
| Left superior temporal gyrus surface area | 0.658 | 0.173 | 0.11 | 5.80E−02 | 0.066 | 3.98E−03 | 0.028 |
| Left transverse temporal cortex thickness | 0.555 | 0.001 | 0.102 | 2.65E−01 | 0.055 | 1.00E+00 | 1.000 |
| Right cuneus cortex thickness | 0.723 | 0.133 | 0.107 | 9.27E−02 | 0.072 | 9.85E−02 | 0.302 |
| Right entorhinal cortex surface area | 0.651 | 0.056 | 0.104 | 2.99E−01 | 0.065 | 4.94E−01 | 0.812 |
| Right insula cortex surface area | 0.878 | 0.141 | 0.107 | 8.92E−02 | 0.088 | 1.21E−01 | 0.302 |
| Right lateral orbital frontal cortex thickness | 0.483 | 0.015 | 0.102 | 4.34E−01 | 0.048 | 1.00E+00 | 1.000 |
| Right middle temporal gyrus surface area | 0.61 | 0.032 | 0.102 | 3.64E−01 | 0.061 | 1.00E+00 | 1.000 |
| Right paracentral lobule thickness | 0.494 | 0.045 | 0.105 | 3.35E−01 | 0.049 | 5.41E−01 | 0.812 |
| Right pars opercularis surface area | 0.545 | 0.047 | 0.105 | 3.12E−01 | 0.055 | 5.57E−01 | 0.812 |
| Right postcentral gyrus thickness | 0.76 | 0.073 | 0.103 | 2.38E−01 | 0.076 | 4.11E−01 | 0.799 |
| Right precentral gyrus thickness | 0.731 | 0.022 | 0.103 | 4.16E−01 | 0.073 | 1.00E+00 | 1.000 |
| Right precuneus cortex surface area | 0.547 | 0.058 | 0.107 | 2.98E−01 | 0.055 | 5.29E−01 | 0.812 |
| Right precuneus cortex thickness | 0.965 | 0.154 | 0.107 | 7.96E−02 | 0.097 | 1.04E−01 | 0.302 |
| Right superior parietal gyrus thickness | 0.941 | 0.118 | 0.105 | 1.25E−01 | 0.094 | 3.11E−01 | 0.641 |
| Right supramarginal gyrus thickness | 0.769 | 0.137 | 0.106 | 1.23E−01 | 0.077 | 1.15E−01 | 0.302 |
| Right temporal pole surface area | 0.524 | 0.182 | 0.11 | 4.83E−02 | 0.052 | 2.77E−03 | 0.024 |
| Right transverse temporal cortex thickness | 0.536 | 0.034 | 0.104 | 3.78E−01 | 0.054 | 1.00E+00 | 1.000 |
Figure 2.
Schizophrenia-associated SNP heritability estimates of average cortical thickness (a) and surface area (b) measurements of 35 anatomical traits that are significantly heritable. Eight cortical structural traits that show significantly enriched contribution of the schizophrenia-associated SNPs at an FDR of 0.1 are labeled.
The neurogenetic architecture of schizophrenia-associated brain phenotypes is partly shared with other psychiatric disorders
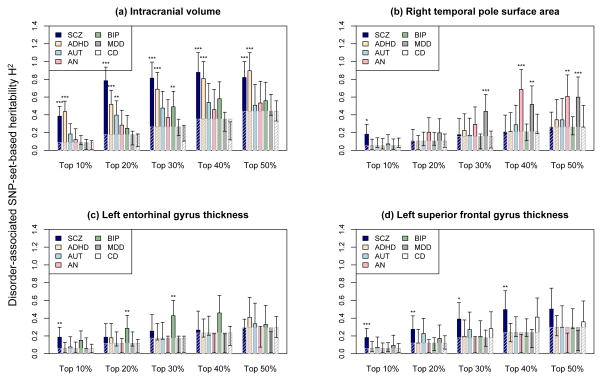
There is compelling evidence that major psychiatric disorders share genetic vulnerability(22, 42). We examined whether the eight brain phenotypes for which we found significant enrichment of schizophrenia-associated genetic variants, are also correlated with disease risk for other psychiatric disorders. This comparative analysis used publicly available GWAS results of five psychiatric disorders: (1) anorexia (AN); (2) attention deficit hyperactivity disorder (ADHD); (3) autism spectrum disorders (ASD); (4) bipolar disorder (BIP); and (5) major depressive disorder (MDD). We also included Crohn’s disease (CD) as a non-psychiatric control dataset. Partitioning and enrichment analyses were conducted for each disorder as previously described. After correcting for multiple testing (FDR q ≤ 0.1), four brain phenotypes showed significant association with the examined psychiatric disorders. Figure 3 displays the comparative analysis results for the four brain phenotypes (all results in Supplementary Table 4). ICV showed significantly enriched association with multiple disorders. As we observed with schizophrenia, ICV showed enrichment for ADHD-associated SNPs under all SNP selection thresholds (top 10–50%; enrichment p ≤ 5.60×10−5). Genetic variants associated with another neurodevelopmental disorder ASD also showed a relationship to ICV (top 20%; p = 2.61×10−3). Right temporal pole surface area was enriched for SNPs associated with MDD (top 30–50%; p ≤ 3.15×10−3) and AN (top 50%; p = 1.60×10−3), while left entorhinal cortex thickness showed enrichment for BIP risk SNPs (top 20, 30%; p ≤ 3.52×10−3). Finally, in contrast to the other three brain regions, left superior frontal gyrus thickness was associated only with schizophrenia genetic risk (top 10–40%; p ≤ 6.99×10−3). As expected(22), Crohn’s disease-associated SNPs showed no significantly enriched contribution for any of the eight brain phenotypes we examined.
Figure 3.
Enrichment analysis results for various disorder-associated SNP-set-based heritability. We examined seven disorders: (1) SCZ: schizophrenia; (2) ADHD: attention deficit hyperactivity disorder; (3) AUT: autism spectrum disorder; (4) AN: anorexia nervosa; (5) BIP: bipolar disorder; (6) MDD: major depressive disorder; and (7) CD: Crohn’s disease. For each disorder, the results of publicly available genome-wide case/control association study were obtained from the Psychiatric Genomics Consortium and used to rank genotype SNPs based on the significance of disease-association. The x-axis represents five SNP set selection thresholds: top 10%, 20%, 30%, 40%, and 50%. For each of the SNP selection thresholds, we divided genotype SNPs into two sets: (1) one set of top ranked disorder-associated SNPs within the selection threshold; and (2) the other set of the remaining genotype SNPs. GCTA was used to learn the genome-wide SNP heritability of a brain phenotype using a joint analysis of the two SNP set-based genetic components. The y-axis represents estimated SNP heritability contributable to the set of disorder-associated SNPs. Error bars represent the analytic standard errors of the estimates. Heritability estimates for seven disorders are displayed using solid navy-color bars, while the null expectations (i.e., estimates calculated based on the proportion of SNPs with respect to the genome-wide SNP heritability of the corresponding brain phenotype) are marked by dashed gray-line bars. Brain phenotypes with enriched disorder-set-based heritability at FDR q ≤ 0.1 are highlighted with asterisk (*** uncorrected p<0.001; ** p <0.01; * p<0.05).
Discussion
Using an unbiased investigation of 139 brain-wide cortical ROIs in 1,750 healthy young adults, we identified eight brain structural phenotypes for which individual differences are significantly modulated by schizophrenia-associated genetic variants en masse. We specifically focused on cortical measures as they are related to normal development, aging, cognitive functioning, as well as neurodevelopmental disorders(43). In particular, cortical thickness reflects progressive changes of developmental processes that last from birth to adulthood and to aging, which are driven by axonal and dendrite remodeling, myelination, and synaptic pruning that contribute to active modification of brain functional circuitry(44–46). Cortical surface areas, on the other hand, have been suggested to develop independently from cortical thickness(37). Variability in cortical surface areas and ICV is thought to reflect exceptionally dynamic early childhood developmental processes, including neural stem cell proliferation and migration(43, 44). Genetic modulation of these biological processes may thus underlie a common genetic basis between brain anatomy and the etiology of neurodevelopmental disorders, such as schizophrenia.
Our study highlights several brain cortical regions as potential endophenotypes of schizophrenia(9, 47). The identified regions include right temporal pole, left superior frontal, superior temporal, inferior parietal, lateral occipital, and entorhinal cortices, and the cuneus. While structural changes in a broad range of brain regions have been associated with schizophrenia in prior research, it has often been difficult to determine whether such correlations reflect shared etiologic factors or secondary effects of schizophrenia or its treatment on brain structure. Our findings support the hypothesis that these brain anatomical correlates share a common genetic basis with schizophrenia. While these regions are anatomically distributed throughout the brain, most reside within the cortical association networks, which are disrupted in patients with psychotic disorders(48). Our findings thus suggest that distributed cortical changes in these regions, which arise due to a shared genetic basis with schizophrenia, may lead to changes in large-scale anatomical and functional systems affected in schizophrenia.
Among the eight cortical phenotypes, we found intracranial volume (ICV) and superior frontal cortical thickness as most robustly related to schizophrenia genetic risk. In particular, cortical thickness in the left superior frontal gyrus (SFG) showed a very specific relationship with schizophrenia-associated genetic variants. This finding is consistent with a large body of literature implicating volumetric reductions in SFG in psychotic illness, including in (1) both unmedicated and medicated schizophrenia patients(49), (2) child onset schizophrenia patients (who are believed to have increased genetic loading; see Ordóñez et al. 2015(50)), and (3) clinical high risk patients who ultimately convert to psychosis(51). Reductions in cortical thickness in the superior frontal gyrus have also been demonstrated recently to be mediated by cognitive control deficits(52), a core feature of schizophrenia that is associated with poor functional outcome. The SFG includes the dorsolateral prefrontal cortex and, as defined for the present analysis, is situated near the intersection of three large-scale cortical systems subserving higher cognitive functions, the fronto-parietal control network, the default network, and the dorsal attention network (53). In particular, the left SFG is functionally connected to most of the default mode network and suggested to play a key role in information processing between the default mode network and a number of distributed fronto-parietal control networks(54, 55). A series of studies have reported that patients with schizophrenia have exhibited distinct connectivity patterns compared to controls within(56, 57) and between(58, 59) these two networks, each tied to key domains of cognition. The superior frontal gyrus is also among the cortical territories that have vastly expanded, as a proportion of overall cortical tissue, in humans versus other primates(60). This expansion presumably provides adaptive advantages for human behavior and survival. Consistent with their recent origins, these territories also develop late during post-natal development, with cortical thinning – presumably reflecting synaptic pruning among other circuit refinement processes – occurring into late adolescence and early adulthood(61). As this is the time window in which schizophrenia and related psychotic disorders typically present, it is intriguing to speculate that superior frontal gyrus thickness - one of the last structures to undergo developmental thinning – is a key neurogenetic correlate of schizophrenia that reflects either delayed or incomplete circuit refinements during the critical developmental period. This may lay the foundation for cognitive decline(62) and risk for psychotic experiences(51, 63, 64).
We also found that ICV, the phenotype most strongly associated with schizophrenia risk variants, is also associated with SNP sets contributing to other neurodevelopmental disorders, particularly ADHD, and a little lesser extent to ASD. Prior evidence suggests a genetic effect of disease liability on ICV in these three disorders(2, 65, 66). In particular, reduced total brain gray matter volume was observed among unaffected relatives of individuals with schizophrenia(67–69) and ADHD(65). ICV is a global measure of brain structure, and it thus may reflect the integration of multiple processes that have been implicated in diverse neurodevelopmental disorders, including altered neuronal migration and synaptic pruning.
In this work, we demonstrated the utility of partitioning-based SNP heritability analysis for investigating the shared genetic basis of two complex traits. Several other approaches aim to achieve the same goal. Polygenic scoring analysis(11) has been widely used in recent neuroimaging genetics work(14, 15, 17, 33, 70). As opposed to standard single-SNP-based association analyses, polygenic scoring analysis uses genome-wide SNP data to relate aggregated disease risk scores to phenotypic differences. Estimated polygenic risk scores have shown impressive power for predicting case versus control status in independent disorder samples(40). However, the polygenic scoring strategy has a limited utility if two complex traits do not preserve consistent directional effects at a genome-wide level. This may have contributed to several recent studies that have discovered limited evidence for association between polygenic risk scores of schizophrenia and cortical(71) or subcortical neuroimaging phenotypes(72). Bivariate analysis of the covariance between two traits(73), as implemented in bivariate GCTA, is another popular approach. Using a multivariate linear mixed-effects model, this method estimates genetic correlation between two complex traits using SNP-derived genomic relationships. Bivariate GCTA however requires raw genotype data for both traits, which may not be readily available. Recently, another powerful SNP co-heritability analysis tool(74) has been developed that employs LD score regression(29). This method estimates genetic correlation between two traits using only genome-wide summary statistics, and offers great utility for genetic studies of complex traits. Nevertheless, it requires a large sample size - typically tens of thousands of samples - to achieve reasonable statistical power(30) .
Our study results should be interpreted in the context of several limitations. First, linear mixed effects modeling used in this study only consider the contribution of additive genetic effects. A recent meta-analysis of 17,804 human traits reports a predominant contribution of additive genetic effects(75), but non-additive genetic effects may play an important role in brain structure and function(76). Similarly, our analysis does not include sex chromosome data, which may harbour relevant genetic contributions to disorder-related neuroanatomy. Secondly, SNP heritability analysis using mixed linear effects modeling is a useful tool to estimate what fraction of the phenotypic variance can be explained by a set of target variants en masse. However, it does not imply that the selected disease associated SNPs carry the same direction of effects for the studied brain phenotype, nor does it identify the most significantly associated variants. Thirdly, we did not have statistical power to dissect the partitioned heritability attributed to by disease-associated SNPs into more finely defined categories (e.g., specific chromosomes, functional categories or biological pathways). As neuroimaging sample sizes increase, we anticipate more fine-scaled partitioning of variance by annotation and genomic region will become feasible and thus facilitate the dissection of genetic architectures between disease and brain structural phenotypes. Finally, recent studies have noted nicotine usage could affect the brain structure and function(77, 78), including the measures of cortical thickness and volume. The MGH/Harvard GSP samples include about 106 smokers (7.9%). We investigated whether there are statistically significant differences in major brain anatomical measures between a group of smokers and a demographically matched group of non-smokers in our sample (conditioning on age, sex, handedness, principal component analysis factors for ethnicity, and ICV for cortical volumetric measures). After regressing out covariates, the Wilcoxon rank sum test was used to compare the differences. In brief, we did not find statistically significant differences after performing multiple testing correction (at FDR q = 0.1; Supplementary Table 5). This null result is likely because, among a small proportion of smokers in our sample (N=106; 7.9%), most were light daily smokers who use zero or one cigarette per day (N=76; 6%). Considering a small proportion of daily smokers in our sample (<8%), and non-significant differences in the brain morphometric measures between the groups of smokers and non-smokers, we decided not to include nicotine usage as a covariate in the present study. It is though possible that the long duration of smoking or the effects of the second-hand smoking exposure may lead to more extensive structural changes, which could not be tested in our sample due to lack of data.
In summary, our study demonstrates that certain brain anatomical phenotypes are influenced by genetic variants that also contribute to schizophrenia disease risk. While the effect of common genetic variants is only modest individually, aggregate genomic effects account for a substantial proportion of the variance in brain measures(25, 41, 76, 79). Further studies are warranted to elucidate the role of these neuroanatomical phenotypes in the pathogenesis of schizophrenia. Along with anatomical measures, brain connectivity and neural integrity are also key markers of neuropsychiatric disorders. Our future work will investigate the genetic overlap between brain functional connectivity and schizophrenia disease risk. For example, healthy individuals with heightened genetic risk and signs of structural changes may or may not exhibit concomitant changes in functional architecture. These changes might reflect compensatory processes in these ostensibly healthy individuals. In addition, studies of clinical samples may offer crucial information about how variation in cortical regions highlighted in our study are associated with genetic liability, symptom severity, and treatment response.
Supplementary Material
Acknowledgments
Neuroimaging genetics data were provided by the Brain Genomics Superstruct Project (GSP) of Harvard University and MGH, with support from the Center for Brain Science
Neuroinformatics Research Group, Athinoula A. Martinos Center for Biomedical Imaging, Center for Human Genetic Research, and Stanley Center for Psychiatric Research. Twenty individual investigators at Harvard and MGH generously contributed data to the overall project. The QTIM study is supported by grants from NIH (R01 HD050735) and the NHMRC (389875, 486682, 1009064). We thank the twins and siblings for their participation, Marlene Grace and Ann Eldridge for twin recruitment, Aiman Al Najjar and other radiographers for scanning, Kerrie McAloney and Daniel Park for research support, and Anjali Henders and staff for DNA sample processing and preparation. P.M.T., N.J., and M.J.W. were supported in part by a Consortium grant (U54 EB020403 to P.M.T.) from the NIH Institutes contributing to the Big Data to Knowledge (BD2K) Initiative. This research was also funded in part by NIH Grants K99MH101367 (to P.H.L.); K23MH104515 (to J.T.B), K01MH099232 (to A.J.H.); K24MH094614 and R01 MH101486 (to J.W.S.); J.W.S. is a Tepper Family MGH Research Scholar.
Footnotes
Supplementary information is available at Molecular Psychiatry’s website.
Author Contribution
Project conception and experiment design: P.H.L.
Statistical analysis and interpretation of findings: P.H.L., J.T.B., J-Y.J., J.W.S., D.O., D.S.M
Imaging and genetics data generation and processing: A.J.H., P.H.L., R.B., J.R., J.W.S., T.G., N.J, D.P.H, J.F, K.L.M, G.I.Z., N.G.M, M.J.W., P.M.T.
Writing of the manuscript: P.H.L., J.T.B., J.W.S., T.G., Y.C.
Revision of the manuscript: A.J.H., J.T.B., J.W.S, T.G., N.J., P.M.T.
Conflicts of Interest
Authors declare no conflicts of interest.
Web Resources
FREESURFER: http://surfer.nmr.mgh.harvard.edu
PLINK (ver. 1.9): https://www.cog-genomics.org/plink2/
EIGENSOFT: http://www.hsph.harvard.edu/alkes-price/software/
The GCTA-GREML Power Calculator: http://cnsgenomics.com/shiny/gctaPower/
Genome-wide Complex Trait Analysis (GCTA ver. 1.24.4): http://www.complextraitgenomics.com/software/gcta/
Psychiatric Genomic Consortium (PGC) SCZ, BIP, ASD, ADHD, MDD summary statistics: http://www.med.unc.edu/pgc/downloads
Crohn’s disease summary statistics: http://www.ibdgenetics.org/downloads.html
References
- 1.Sullivan P, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187–92. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.van Erp T, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA, et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry. 2016;21(4):547–53. doi: 10.1038/mp.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhojraj T, Francis AN, Montrose DM, Keshavan MS. Grey matter and cognitive deficits in young relatives of schizophrenia patients. Neuroimage. 2011;(Suppl 1):S287–92. doi: 10.1016/j.neuroimage.2010.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modinos G, Vercammen A, Mechelli A, Knegtering H, McGuire PK, Aleman A. Structural covariance in the hallucinating brain: a voxel-based morphometry study. J Psychiatry Neurosci. 2009;34(6):465–9. [PMC free article] [PubMed] [Google Scholar]
- 5.Modinos G, Costafreda SG, van Tol MJ, McGuire PK, Aleman A, Allen P. Neuroanatomy of auditory verbal hallucinations in schizophrenia: a quantitative meta-analysis of voxel-based morphometry studies. Cortex. 2013;49(4):1046–55. doi: 10.1016/j.cortex.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 6.van Tol M, van der Meer L, Bruggeman R, Modinos G, Knegtering H, Aleman A. Voxel-based gray and white matter morphometry correlates of hallucinations in schizophrenia: The superior temporal gyrus does not stand alone. Neuroimage Clin. 2013;4:249–57. doi: 10.1016/j.nicl.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Lutterveld R, van den Heuvel MP, Diederen KM, de Weijer AD, Begemann MJ, Brouwer RM, et al. Cortical thickness in individuals with non-clinical and clinical psychotic symptoms. Brain. 2014;137(Pt 10):2664–9. doi: 10.1093/brain/awu167. [DOI] [PubMed] [Google Scholar]
- 8.Wible C, Anderson J, Shenton ME, Kricun A, Hirayasu Y, Tanaka S, et al. Prefrontal cortex, negative symptoms, and schizophrenia: an MRI study. Psychiatry Res. 2001;108(2):65–78. doi: 10.1016/s0925-4927(01)00109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman I, Gould TD. The Endophenotype Concept in Psychiatry: Etymology and Strategic Intentions. Am J Psychiatry. 2003;160(4):636–45. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 10.Mattai A, Chavez A, Greenstein D, Clasen L, Bakalar J, Stidd R, et al. Effects of clozapine and olanzapine on cortical thickness in childhood-- –onset schizophrenia. Schizophr Res. 2010;116(1):44–8. doi: 10.1016/j.schres.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell S, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tesli M, Espeseth T, Bettella F, Mattingsdal M, Aas M, Melle I, et al. Polygenic risk score and the psychosis continuum model. Acta Psychiatr Scand. 2014;130(4):311–7. doi: 10.1111/acps.12307. [DOI] [PubMed] [Google Scholar]
- 13.Meyer-Lindenberg A. Imaging genetics of schizophrenia. Dialogues Clin Neurosci. 2010;12(4):449–56. doi: 10.31887/DCNS.2010.12.4/amlindenberg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walton E, Geisler D, Lee PH, Hass J, Turner JA, Liu J, et al. Prefrontal inefficiency is associated with polygenic risk for schizophrenia. Schizophr Bull. 2014;40(6):1263–71. doi: 10.1093/schbul/sbt174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C, et al. Neural mechanisms of a genomewide supported psychosis variant. Science. 2009;324(5927):605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- 16.Kauppi K, Westlye LT, Tesli M, Bettella F, Brandt CL, Mattingsdal M, et al. Polygenic risk for schizophrenia associated with working memory-related prefrontal brain activation in patients with schizophrenia and healthy controls. Schizophr Bull. 2015;41(3):736–43. doi: 10.1093/schbul/sbu152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walton E, Turner J, Gollub RL, Manoach DS, Yendiki A, Ho BC, et al. Cumulative genetic risk and prefrontal activity in patients with schizophrenia. Schizophr Bull. 2013;39(3):703–11. doi: 10.1093/schbul/sbr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Erp T, Guella I, Vawter MP, Turner J, Brown GG, McCarthy G, et al. Schizophrenia miR-137 locus risk genotype is associated with dorsolateral prefrontal cortex hyperactivation. Biol Psychiatry. 2014;75(5):398–405. doi: 10.1016/j.biopsych.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oertel-Knöchel V, Lancaster TM, Knöchel C, Stäblein M, Storchak H, Reinke B, et al. Schizophrenia risk variants modulate white matter volume across the psychosis spectrum: evidence from two independent cohorts. Neuroimage Clin. 2015;7:764–70. doi: 10.1016/j.nicl.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terwisscha vSA, Bakker SC, van Haren NE, Derks EM, Buizer-Voskamp JE, Boos HB, et al. Genetic schizophrenia risk variants jointly modulate total brain and white matter volume. biol psychiatry. 2013;73(6):525–31. doi: 10.1016/j.biopsych.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Lee H, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–94. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis L, Yu D, Keenan CL, Gamazon ER, Konkashbaev AI, Derks EM, et al. Partitioning the heritability of Tourette syndrome and obsessive compulsive disorder reveals differences in genetic architecture. PLoS Genet. 2013;9(10):e1003864. doi: 10.1371/journal.pgen.1003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S, DeCandia TR, Ripke S, Yang J, et al. Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ), International Schizophrenia Consortium (ISC) Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–50. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge T, Nichols T, Lee PH, Holmes AJ, Roffman JL, Buckner RL, et al. Massively expedited genome-wide heritability analysis (MEGHA) Proc Natl Acad Sci U S A. 2015;112(8):2479–84. doi: 10.1073/pnas.1415603112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryant C, Giovanello KS, Ibrahim JG, Chang J, Shen D, Peterson BS, et al. Mapping the genetic variation of regional brain volumes as explained by all common SNPs from the ADNI study. PLoS One. 2013;8(8):e71723. doi: 10.1371/journal.pone.0071723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gusev A, Bhatia G, Zaitlen N, Vilhjalmsson BJ, Diogo D, Stahl EA, et al. Quantifying missing heritability at known GWAS loci. PLoS Genet. 2013;9(12):e1003993. doi: 10.1371/journal.pgen.1003993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gusev A, Lee SH, Trynka G, Finucane H, Vilhjálmsson BJ, Xu H, et al. Regulatory and Cell-Type-Specific Variants across 11 Common Diseases. Am J Hum Genet. 2014;95(5):535–52. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulik-Sullivan B, Loh PR, Finucane HK, Ripke S, Yang J, et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finucane H, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015 doi: 10.1038/ng.3404. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Zubicaray G, Chiang MC, McMahon KL, Shattuck DW, Toga AW, Martin NG, et al. Meeting the Challenges of Neuroimaging Genetics. Brain Imaging Behav. 2008;2(4):258–63. doi: 10.1007/s11682-008-9029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmes A, Hollinshead MO, O’Keefe TM, Petrov VI, Fariello GR, Wald LL, et al. Brain Genomics Superstruct Project initial data release with structural, functional, and behavioral measures. Sci Data. 2015;2:150031. doi: 10.1038/sdata.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes A, Lee PH, Hollinshead M, Bakst L, Roffman JL, Smoller JW, et al. Individual differences in amygdala-medial prefrontal anatomy link negative affect, impaired social functioning, and polygenic depression risk. J Neurosci. 2012;32(50):18087–100. doi: 10.1523/JNEUROSCI.2531-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rentería M, Hansell NK, Strike LT, McMahon KL, de Zubicaray GI, Hickie IB, et al. Genetic architecture of subcortical brain regions: common and region-specific genetic contributions. Genes Brain Behav. 2014;13(8):821–30. doi: 10.1111/gbb.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–81. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desikan R, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–80. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 37.Winkler A, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, et al. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53(3):1135–46. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein J, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44(5):552–61. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnes J, Ridgway GR, Bartlett J, Henley SM, Lehmann M, Hobbs N, et al. Head size, age and gender adjustment in MRI studies: a necessary nuisance? Neuroimage. 2010;53:1244–55. doi: 10.1016/j.neuroimage.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 40.Ripke S, Neale BM, Corvin A, Walters JT, Farh KH, Holmans PA, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toro R, Poline JB, Huguet G, Loth E, Frouin V, Banaschewski T, et al. Genomic architecture of human neuroanatomical diversity. Mol Psychiatry. 2015;20(8):1011–6. doi: 10.1038/mp.2014.99. [DOI] [PubMed] [Google Scholar]
- 42.Smoller JW, Ripke S, Lee PH, Neale B, Nurnberger JI, Santangelo S, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyall A, Shi F, Geng X, Woolson S, Li G, Wang L, et al. Dynamic Development of Regional Cortical Thickness and Surface Area in Early Childhood. Cereb Cortex. 2015;25(8):2204–12. doi: 10.1093/cercor/bhu027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baribeau D, Anagnostou E. A Comparison of Neuroimaging Findings in Childhood Onset Schizophrenia and Autism Spectrum Disorder: A Review of the Literature. Front Psychiatry. 2013;4(175) doi: 10.3389/fpsyt.2013.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Nedivi E. Neuronal structural remodeling: Is it all about access? Curr Opin Neurobiol. 2010;20(5):557–62. doi: 10.1016/j.conb.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moyer C, Shelton MA, Sweet RA. Dendritic spine alterations in schizophrenia. Neurosci Lett. 2015;601:46–53. doi: 10.1016/j.neulet.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roffman J, Weiss AP, Goff DC, Rauch SL, Weinberger DR. Neuroimaging-genetic paradigms: a new approach to investigate the pathophysiology and treatment of cognitive deficits in schizophrenia. Harv Rev Psychiatry. 2006;14(2):78–91. doi: 10.1080/10673220600642945. [DOI] [PubMed] [Google Scholar]
- 48.Baker J, Holmes AJ, Masters GA, Yeo BT, Krienen F, Buckner RL, et al. Disruption of cortical association networks in schizophrenia and psychotic bipolar disorder. JAMA Psychiatry. 2014;71(2):109–18. doi: 10.1001/jamapsychiatry.2013.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong Q, Lui S, Sweeney JA. A selective review of cerebral abnormalities in patients with first-episode schizophrenia before and after treatment. Am J Psychiatry. 2015 doi: 10.1176/appi.ajp.2015.15050641. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 50.Ordóñez A, Luscher ZI, Gogtay N. Neuroimaging findings from childhood onset schizophrenia patients and their non-psychotic siblings. Schizophr Res. 2015;(15):132–2. doi: 10.1016/j.schres.2015.03.003. S0920-9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cannon T, Chung Y, He G, Sun D, Jacobson A, van Erp TG, et al. Progressive Reduction in Cortical Thickness as Psychosis Develops: A Multisite Longitudinal Neuroimaging Study of Youth at Elevated Clinical Risk. Biol Psychiatry. 2015;77(2):147–57. doi: 10.1016/j.biopsych.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tully L, Lincoln SH, Liyanage-Don N, Hooker CI. Impaired cognitive control mediates the relationship between cortical thickness of the superior frontal gyrus and role functioning in schizophrenia. Schizophr Res. 2014;152(2–3):358–64. doi: 10.1016/j.schres.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 53.Yeo B, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–65. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spreng R, Sepulcre J, Turner GR, Stevens WD, Schacter DL. Intrinsic architecture underlying the relations among the default, dorsal attention, and frontoparietal control networks of the human brain. J Cogn Neurosci. 2013;25(1) doi: 10.1162/jocn_a_00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W, Qin W, Liu H, Fan L, Wang J, Jiang T, et al. Subregions of the human superior frontal gyrus and their connections. Neuroimage. 2013;78:46–58. doi: 10.1016/j.neuroimage.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 56.Du Y, Pearlson GD, Yu Q, He H, Lin D, Sui J, et al. Interaction among subsystems within default mode network diminished in schizophrenia patients: A dynamic connectivity approach. Schizophr Res. 2016;170(1):55–65. doi: 10.1016/j.schres.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pomarol-Clotet E, Salvador R, Sarró S, Gomar J, Vila F, Martínez A, et al. Failure to deactivate in the prefrontal cortex in schizophrenia: dysfunction of the default mode network? Psychol Med. 2008;38(8):1185–93. doi: 10.1017/S0033291708003565. [DOI] [PubMed] [Google Scholar]
- 58.Chang X, Shen H, Wang L, Liu Z, Xin W, Hu D, et al. Altered default mode and fronto-parietal network subsystems in patients with schizophrenia and their unaffected siblings. Brain Res. 2014;1562:87–99. doi: 10.1016/j.brainres.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 59.Nekovarova T, Fajnerova I, Horacek J, Spaniel F. Bridging disparate symptoms of schizophrenia: a triple network dysfunction theory. Front Behav Neurosci. 2014;8:171. doi: 10.3389/fnbeh.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deacon T. Human brain evolution II. Embryology and brain allometry. In: Jerison H, Jerison I, editors. Intelligence and Evolutionary Biology. Berlin: Springer-Verlag; 1988. pp. 383–415. [Google Scholar]
- 61.Thompson P. Cracking the brain’s genetic code. Proc Natl Acad Sci. 2015 doi: 10.1073/pnas.1520702112. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Narr K, Woods RP, Thompson PM, Szeszko P, Robinson D, Dimtcheva T, et al. Relationships between IQ and regional cortical gray matter thickness in healthy adults. Cereb Cortex. 2007;17(9):2163–71. doi: 10.1093/cercor/bhl125. [DOI] [PubMed] [Google Scholar]
- 63.Narr K, Bilder RM, Toga AW, Woods RP, Rex DE, Szeszko PR, et al. Mapping Cortical Thickness and Gray Matter Concentration in First Episode Schizophrenia. Cereb Cortex. 2005;15(6):708–19. doi: 10.1093/cercor/bhh172. [DOI] [PubMed] [Google Scholar]
- 64.Schultz C, Koch K, Wagner G, Roebel M, Schachtzabel C, Gaser C, et al. Reduced cortical thickness in first episode schizophrenia. Schizophr Res. 2010;116(2–3):204–9. doi: 10.1016/j.schres.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 65.Greven CU, Bralten J, Mennes M, O’Dwyer L, van Hulzen KJ, Rommelse N, et al. Developmentally stable whole-brain volume reductions and developmentally sensitive caudate and putamen volume alterations in those with attention-deficit/hyperactivity disorder and their unaffected siblings. JAMA Psychiatry. 2015 May;72(5):490–9. doi: 10.1001/jamapsychiatry.2014.3162. [DOI] [PubMed] [Google Scholar]
- 66.Kucharsky HR, Alter R, Sojoudi S, Ardekani BA, Kuzniecky R, Pardoe HR. Corpus Callosum Area and Brain Volume in Autism Spectrum Disorder: Quantitative Analysis of Structural MRI from the ABIDE Database. J Autism Dev Disord. 2015;45(10):3107–14. doi: 10.1007/s10803-015-2468-8. [DOI] [PubMed] [Google Scholar]
- 67.McIntosh A, Owens DC, Moorhead WJ, Whalley HC, Stanfield AC, Hall J, et al. Longitudinal volume reductions in people at high genetic risk of schizophrenia as they develop psychosis. Biol Psychiatry. 2011;69(10):953–8. doi: 10.1016/j.biopsych.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Lawrie S, McIntosh AM, Hall J, Owens DG, Johnstone EC. Brain structure and function changes during the development of schizophrenia: the evidence from studies of subjects at increased genetic risk. Schizophr Bull. 2008;34(2):330–40. doi: 10.1093/schbul/sbm158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brans R, van Haren NE, van Baal GC, Schnack HG, Kahn RS, Hulshoff Pol HE. Heritability of changes in brain volume over time in twin pairs discordant for schizophrenia. Arch Gen Psychiatry. 2008;65(11):1259–68. doi: 10.1001/archpsyc.65.11.1259. [DOI] [PubMed] [Google Scholar]
- 70.Sabuncu M, Buckner R, Smoller JW, Lee PH, Fischl B, Sperling R. The association between a polygenic alzheimer score and cortical thickness in cognitively normal subjects. Cereb Cortex. 2011;22(11):2653–61. doi: 10.1093/cercor/bhr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voineskos A, Felsky D, Wheeler AL, Rotenberg DJ, Levesque M, Patel S, et al. Limited Evidence for Association of Genome-Wide Schizophrenia Risk Variants on Cortical Neuroimaging Phenotypes. Schizophr Bull. 2015 doi: 10.1093/schbul/sbv180. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franke B, Stein JL, Ripke S, Anttila V, Hibar DP, van Hulzen KJ, et al. Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept. Nat Neurosci. 2016;19(3):420–31. doi: 10.1038/nn.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee S, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using SNP-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28(19):2540–2. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015 doi: 10.1038/ng.3406. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Polderman T, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nature Genet. 2015;47(7):702–9. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- 76.Chen C, Peng Q, Schork AJ, Lo MT, Fan CC, Wang Y, et al. Pediatric Imaging Neurocognition and Genetics Study; Alzheimer’s Disease Neuroimaging Initiative Large-scale genomics unveil polygenic architecture of human cortical surface area. Nat Commun. 2015;6(7549) doi: 10.1038/ncomms8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morales A, Ghahremani D, Kohno M, Hellemann GS, London ED. Cigarette exposure, dependence, and craving are related to insula thickness in young adult smokers. Neuropsychopharmacology. 2014;39(8):1816–22. doi: 10.1038/npp.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, Yuan K, Cai C, Feng D, Yin J, Bi Y, et al. Reduced frontal cortical thickness and increased caudate volume within fronto-striatal circuits in young adult smokers. Drug Alcohol Depend. 2015;151:211–9. doi: 10.1016/j.drugalcdep.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 79.Bryant C, Giovanello KS, Ibrahim JG, Chang J, Shen D, Peterson BS, et al. Mapping the genetic variation of regional brain volumes as explained by all common SNPs from the ADNI study. PLoS One. 2013;8(8):e71723. doi: 10.1371/journal.pone.0071723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.