Abstract
Prions are thought to replicate in an autocatalytic process that converts cellular prion protein (PrPC) to the disease-associated misfolded PrP isoform (PrPSc). Our study scrutinizes this hypothesis by in vitro protein misfolding cyclic amplification (PMCA). In serial transmission PMCA experiments, PrPSc was inoculated into healthy hamster brain homogenate containing PrPC. Misfolded PrP was amplified by rounds of sonication and incubation and reinoculated into fresh brain homogenate every 10 PMCA rounds. The amplification depended on PrPC substrate and could be inhibited by recombinant hamster PrP. In serial dilution experiments, newly formed misfolded and proteinase K-resistant PrP (PrPres) catalyzed the structural conversion of PrPC as efficiently as PrPSc from brain of scrapie (263K)-infected hamsters, yielding an ≈300-fold total amplification of PrPres after 100 rounds, which confirms an autocatalytic PrP-misfolding cascade as postulated by the prion hypothesis. PrPres formation was not paralleled by replication of biological infectivity, which appears to require factors additional to PrP-misfolding autocatalysis.
Transmissible spongiform encephalopathies such as Creutzfeldt–Jakob disease, bovine spongiform encephalopathy, and scrapie are caused by unique transmissible agents consisting of proteinaceous infectious particles, termed prions (1). Prions are considered to consist mainly, if not solely, of a misfolded scrapie-associated, aggregated, and proteinase K-resistant isoform (PrPSc) of the cellular prion protein (PrPC) (2). The infectious process is believed to follow an autocatalytic mechanism in which PrPC is converted into the misfolded “infectious” state by PrPSc. PrPC recruited and misfolded by PrPSc subsequently catalyzes the conversion of further PrPC, eventually leading to massive replication of infectivity and fatal disease. To distinguish between PrPSc, which is isolated from infectious tissue and is by definition associated with the transmissible spongiform encephalopathy agent, on the one hand and structurally altered PrP, which has been converted into a misfolded proteinase K-resistant (PrPres) form in vitro but which does not necessarily correlate with infectivity, on the other, we defined the latter as PrPres.
For a long time, PrPC has been shown to acquire proteinase K resistance in vitro under the impact of PrPSc (3). Saborio et al. (4) reported a new experimental approach, protein misfolding cyclic amplification (PMCA), for the generation of PrPres in vitro by using brain homogenates taken from healthy Syrian hamsters (SHas) mixed with PrPSc. In PMCA, the amount of PrPres increases in a cyclic process of alternating incubation and sonication steps. The amplification is assumed to follow a mechanism of seeded aggregation in which the ultrasonic treatment breaks the PrP aggregates into smaller units. These fragments in turn then provide additional seeds for further aggregate growth (5, 6). Amplification has also been observed without sonication (7) or with a single sonication step (8) and seems to depend on the presence of small RNA fragments (9).
In particular, the “protein-only” model of the prion hypothesis implies that not only the original PrPSc but also newly formed PrPres aggregates exert a converting activity on PrPC. According to the data available so far (4, 7, 9), the generation of misfolded PrP by PMCA would be consistent with such autocatalysis but, alternatively, could also be explained by a simple catalytic conversion of PrPC by the initial PrPSc present in the sample. Thus, so far, whether PrPres generated in a PMCA reaction is infectious (i.e., able to transmit disease in vivo), or, indeed, possesses any catalytic activity to structurally convert PrPC at all has not been demonstrated.
Therefore, the present study intends to further investigate whether the PMCA conversion of PrPC to PrPres is mediated by an autocatalytic mechanism, and models the replication of infectivity according to the prion hypothesis. For this purpose, we scrutinized the replication mechanism implied by the prion hypothesis by addressing the following questions on PMCA. (i) Does PrPres generation depend on the presence of a specific PrP substrate? (ii) Can PrPres generation be specifically inhibited? (iii) Does PrPres generated by PMCA serve as a template for conversion of further PrPC to PrPres? (iv) Is PrPres generated by PMCA infectious in animal experiments?
Methods
PMCA of PrPres. Normal SHa brain homogenate (10% wt/vol) was prepared as described by Saborio et al. (4) and was spiked with brain homogenate prepared from terminally ill hamsters intracerebrally (i.c.) infected with 263K scrapie. Samples of 200 μl were subjected to 23 amplification rounds consisting of 5 × 1 s sonication with a ultrasonic microtip probe at 40% power setting (Sonopuls 2070, Bandelin, Germany) followed by 1 h of incubation at 37°C. The probe tip was washed between samples with NaOH (2 M) and water to remove prions adherent to the metal surface (10). Control samples were frozen immediately or were incubated for the duration of the experiment at 37°C. After 10 rounds and 23 PMCA rounds, aliquots of 20 μl were frozen and stored at -80°C.
PrP0/0 brain homogenate was prepared as above from Prnp0/0 mice (11). For inhibition experiments, hamster recombinant PrP (rPrP) (23–231) (Prionics, Schlieren, Switzerland), mouse rPrP (23–231) prepared from Escherichia coli according to ref. 12, or BSA (Sigma) was added before the PMCA reaction at final concentrations of 7.5–250 nM. Concentrations of rPrP were determined by UV absorption measurement, and relative levels of rPrP and PrPC were determined by comparing Western blot band intensities of a rPrP dilution series with the following band intensity of PrPC from hamster brain homogenate after deglycosylation with PNGase F (New England Biolabs) according to the manufacturer's instructions (data not shown).
In PMCA with serially diluted PrPres, the amplification procedure was carried out as above. After each amplification cycle consisting of 10 rounds of alternating sonication and incubation, the reaction mixture was diluted 1.5- 2-, or 2.5-fold into normal hamster brain homogenate. In total, 10 amplification cycles (I–X in Fig. 3) with nine dilution steps were performed. Samples of 20 μl were taken before and after each round and archived at -80°C. At the end of the experiment, samples were thawed for Western blot analysis and bioassay (compare Fig. 4). The sample obtained after ten passages with 2-fold dilution was used in bioassay titration (sample B). Control samples were frozen immediately (sample A) or after 29-fold (sample C) dilution in 10% (wt/vol) normal hamster brain homogenate. All samples used for bioassay titrations and Western blotting were subjected to not more than one freezing and thawing process to avoid distortion of results by loss of PrPres or infectivity due to freezing.
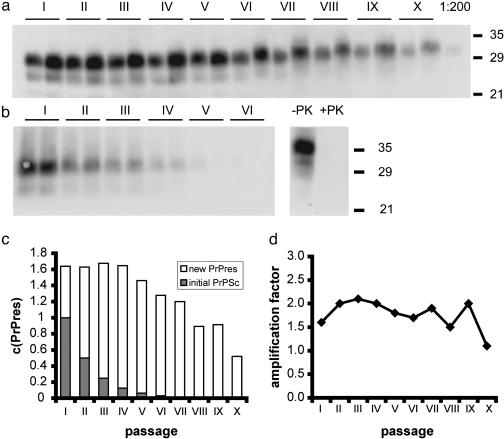
Fig. 3.
PMCA with serially passaged PrPres. (a) 263K-homogenate was diluted 1:20 in SHa-homogenate for PMCA. One cycle consisting of 10 rounds of amplification was performed and the reaction mixture was diluted 2-fold in normal hamster brain homogenate after the cycle. The process was repeated to a total of 10 amplification/dilution cycles. Samples of 20 μl were taken before and after each cycle, frozen for storage, digested with proteinase K, and subjected to Western blot analysis by using 3F4 antibody and quantified densitometrically. Lane X (1:200): a 10-fold dilution of the starting homogenate was included as a PrP concentration reference. (b) Sonication/dilution cycles were carried out as in a by diluting into brain homogenate (10% wt/vol) from PrP0/0 mice. Uninfected hamster brain homogenate was treated with proteinase K under identical conditions to the PMCA samples to document complete proteinase K (PK) digestion of PrPC (lanes -PK and +PK). (c) Quantification of PrPres in the serial transmission PMCA experiment shown in a; relative amounts of initial PrPSc (filled bars) and newly formed PrPres (open bars). (d) Amplification factors derived from c. The catalytic activity is independent of the ratio of PrPSc derived from the initial scrapie homogenate and newly formed PrPres. The average amplification factor was 1.75 ± 0.3 with a 270-fold total amplification. Serial transmission PMCA was repeated several times for 5–10 consecutive cycles with different dilution factors (data not shown), yielding ≈20% variation in amplification factors.
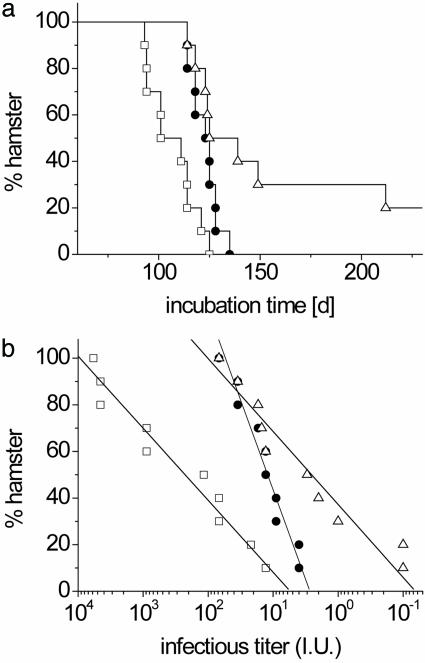
Fig. 4.
Incubation times and corresponding infectivity titers in hamster bioassay experiments. The following samples were analyzed: sample A, initial reaction mixture taken before PMCA and containing 5 × 10-3 g of scrapie brain tissue per ml (□); sample B, PMCA reaction mixture after 10 serial passages (•); and sample C, initial reaction mixture after 512-fold dilution without PMCA (▵). Samples B and C each contained 1 × 10-5 g of the initially added scrapie brain tissue per ml. The 50-μl aliquots of the samples were inoculated i.c. into 10 SHas after 103-fold dilution in PBS. (a) Mean incubation times were 107 ± 11 and 125 ± 6 days for samples A and B, respectively, before and after serial PMCA. Only 8 of 10 animals challenged with sample C succumbed to scrapie until 250 days after infection with a mean incubation time of 138 days for the diseased animals. (b) Infectivity titers were calculated from incubation times by dose–incubation curves as in ref. 15 and were plotted for individual animals on a logarithmic scale. The distribution of titers could be described by a linear fit with a slope of 30 ± 3 for samples A and C and a mean logarithmic titer of 2.4 (250 LD50i.c.) and ≤0.5 (3 LD50i.c.), respectively. The infectivity distribution of sample B showed a slope of 73 ± 5 with a mean logarithmic titer of 1.1 (13 LD50i.c.). Titers refer to the mean infectivity in 50-μl inocula of samples A–C.
Proteinase K Digestion, Western Blotting, and Quantification of PrPres. For PrPres quantification, samples were thawed, digested with proteinase K (100 μg/ml) for 1 h at 37°C, and analyzed by Western blotting with 3F4-antibody at a 1:2,000 dilution (13). PrP was visualized by enhanced chemiluminescence reaction (Amersham Pharmacia, Freiburg, Germany) according to the instructions of the manufacturer and was immediately evaluated densitometrically by using the Diana III luminescence imaging system (Raytest, Straubenhardt, Germany) and the aida software package (Raytest). Protein sizes were determined by using Bio-Rad high molecular weight marker.
Amplification Factors/Normalization. Amplification (Amp) factors (AmpF) were determined in serial dilution experiments by comparing the intensities (I) of diglycosylated PrP bands in the molecular mass range between 27 and 30 kDa after amplification to the corresponding PrP signal after the previous amplification round (e.g., AmpF(II) = IafterAmpII/IafterAmpI × dilution factor), except for cycle I where the AmpF was determined directly from Amp(I) = IafterAmpI/IbeforeAmpI as was completed for amplification experiments without serial dilution.
In inhibition experiments with rPrP, all PrPres signals were normalized to the amplification product obtained without the addition of rPrP for each blot. Normalized mean values and SD of 4–6 independent experiments are shown in each column (Fig. 2). P values were calculated by using Student's paired t test between the data sets of two samples as indicated. P values <0.05 were considered to indicate statistically significant differences.
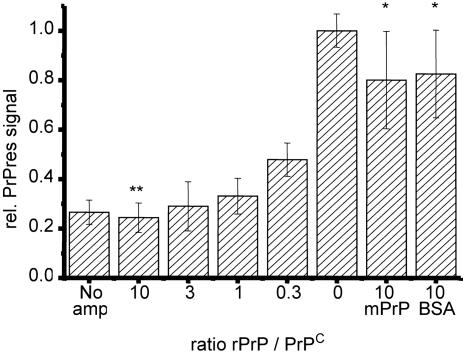
Fig. 2.
Inhibition of PrPres formation by rPrP during PMCA. Dilutions of 263K homogenate in SHa-homogenate(1:100) containing a 0- to 10-fold excess of SHa rPrP (23–231) (Prionics) over PrPC were subjected to eight amplification cycles. After proteinase K digestion and analytical Western blotting, PrPres was quantified densitometrically. PrPres signals of 4–6 independent experiments were normalized by setting the densitometric signal after PMCA to a relative intensity of one. Mean values and SD are shown. P values <0.05 indicate statistical significant differences between two mean values. A 10-fold molar excess of mouse rPrP and BSA was given into additional samples as an unspecific reference. Hamster rPrP significantly inhibits PrPres amplification when compared with amplification without SHa rPrP (**, P < 0.003) and when compared with amplification in the presence of excess murine rPrP or BSA (*, P < 0.05).
Bioassay for Titration of Infectivity. Archived samples were thawed and diluted 1:1,000 in PBS, and 50-μl aliquots were inoculated i.c. into SHas as described (14). The amount of infectivity [50% i.c. lethal doses (LD50i.c.)] in the inoculated samples (50 μl) was assayed as described by Kimberlin and Walker (14) by the observed incubation times (t, in days) until terminal scrapie, using dose–incubation curves (15). For the calculation of infectivity from incubation times, the following empirical equation was used: log(LD50i.c.) = 0.0008 t2 - 0.2575t + 20.7929 [mean error of assay: ± 0.4 log(LD50i.c.)]. For the purpose of fitting logarithmic titers, infectivity in animals without scrapie symptoms at 250 days after inoculation was set to 0.1 LD50i.c. and the percentage of SHas was plotted against log(LD50i.c.).
Results and Discussion
Substrate-Dependent Generation of Protease-Resistant PrP. Analogous to the approach used by Saborio et al. (4), we spiked brain homogenate from normal SHas with brain homogenate from terminally ill scrapie hamsters infected with strain 263K and subjected them to sonication and incubation cycles at different dilutions. The samples were then digested by proteinase K. The amount of PrPres was quantified by Western blotting, using 3F4 antibody. Densitometric quantification of Western blot bands showed a linear response to the amount of PrPres present in the concentration range used in PMCA experiments. Sonicating samples before loading onto the gel did not change the signal (see Fig. 5, which is published as supporting information on the PNAS web site). Variations in densitometric signal of parallel samples were found to be 10–20% between Western blots and <10% within the same blot.
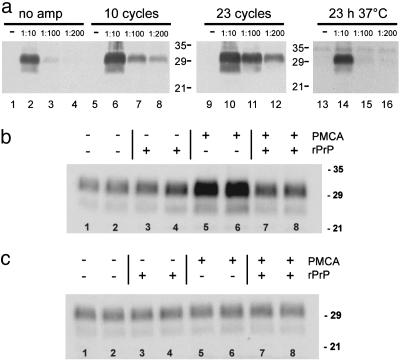
Fig. 1a shows an increase in PrPres signals after amplification by a factor of 5 (10 rounds) and 10 (23 rounds), respectively. No significant increase in PrPres signals was observed after incubation (23 h at 37°C) without sonication.
Fig. 1.
(a) Amplification of PrPres. Brain homogenate (10% wt/vol) prepared from 263K-infected hamsters in the terminal stage of scrapie (263K-homogenate) was diluted 1:10, 1:100 and 1:200 in brain homogenate (10% wt/vol) from healthy SHas. Brain homogenates were subjected to 10 (lanes 5–8) and 23 (lanes 9–12) amplification cycles, and control samples were frozen immediately (lanes 1–4) or were incubated for 23 h at 37°C without sonication (lanes 14–16). Samples were analyzed by Western blotting, using 3F4 antibody after digestion with proteinase K. The amount of PrPres increased at least 10-fold after 23 amplification cycles (lane 1 vs. lane 8). (b and c) Substrate-dependent amplification. 263K homogenate was diluted 1:100 into brain homogenate of healthy SHas (b) or PrP0/0 knockout mice (c). A 10-fold excess of SHa rPrP (23–231) (Prionics) was added where indicated and the reaction mixture was subjected to eight PMCA amplification rounds. All experiments were performed in duplicate and the mean deviation between two independent samples after amplification was found to be ≈30%. Samples subjected to PMCA in hamster brain homogenate (b, lanes 5 and 6) showed an increase in PrPres (3.2- to 3.4-fold), when compared with control samples (b, lanes 1 and 2), and amplification of PrPres could be blocked by excess SHa rPrP (b, lanes 7 and 8). In brain homogenates devoid of PrPC, no amplification occurred with or without the presence of hamster rPrP (c, lanes 1–8).
Next, we tested whether the observed increase in immunoreactive PrP is due to generation of new PrPres. Alternatively, it would be conceivable that PrPSc trapped in cellular membrane fragments, which has not been detected originally, could have been released by repeated sonication of the sample leading to an apparent increase in PrPres. To address this concern, scrapie hamster brain homogenate (263K) was diluted in brain homogenate of PrP0/0 mice (11) lacking PrPC for the conversion reaction. In this reaction mixture, no increase of the immunostaining of hamster PrPres occurred (Fig. 1c). However, similarly treated 263K brain homogenates diluted in brain homogenate of healthy SHa showed an increase in PrPres immunostaining of 3.2- to 3.4-fold after eight PMCA rounds (Fig. 1b). Therefore, the increased intensity of the Western blot signals for hamster PrPres observed after PMCA in Fig. 1a cannot be simply attributed to a release of previously undetected material but must be due to PrPres newly generated from PrPC substrate.
Inhibition of PrPres Formation by rPrP. No detectable PrPres was generated in PMCA samples containing SHa rPrP (23–231) at a 10-fold excess to PrPC (Fig. 1b). To test whether the inhibition of PrPres formation was specific, we added SHa rPrP at molar rPrP/PrPC ratios of 0.3:10 to the amplification reaction (Fig. 2). Even substoichiometric quantities of SHa rPrP significantly inhibited PrPres generation. Half-maximal PrPres formation occurred at a molar ratio of 0.3 and SHa rPrP at a molar ratio of ≥1 almost completely inhibited the conversion reaction (P < 0.003). Murine rPrP (23–230), however, as well as albumin (BSA), did not significantly decrease the amount of PrPres generated by PMCA, even when added at a 10-fold excess over PrPC (Fig. 2).
These findings show that hamster rPrP (23–231) inhibits the PMCA conversion reaction specifically. Inhibition of PrP conversion even by substoichiometric amounts of homologous rPrP fits well into the model of prion replication. For example, the inhibitor could stick to the active site in PrPSc, thus blocking further PrPC conversion or compete with PrPC in binding a cofactor necessary for conversion. A specific catalytic cofactor, such as the proposed protein X (16), would be expected to have different binding affinities to PrP from different species, making it a likely target for the specific inhibition observed in PMCA.
Alternatively, Masel et al. (17, 18) have modeled in detail how PrPC conversion is efficiently inhibited by PrP-substrate analogs, which bind to the active center of conversion, e.g., the sticky ends of PrPSc fibrils, without being converted properly into a catalytically active conformation. The observed inhibition of amplification indicates high specificity of the PMCA reaction at the molecular level.
PMCA with Serially Passaged PrPres. To test for an autocatalytic molecular conversion process, we generated PrPres by PMCA and serially passaged the reaction mixture into fresh brain homogenate.
In our experiment, each cycle of the amplification reaction consisted of 10 rounds of alternating sonication and subsequent incubation at 37°C. After 10 rounds, the reaction mixture was passaged by diluting aliquots 1.5-, 2-, or 2.5-fold into fresh brain homogenate from normal SHas or from PrP0/0 mice. By performing nine sequential passages, the initial material was diluted by a factor of 38, 512, or 3,815, respectively. Samples were taken before and after the amplification reaction in each passage and analyzed as above (Fig. 3 a and b and see Fig 6, which is published as supporting information on the PNAS web site, for 1.5× and 2.5× dilutions). The PrPres signal was ≈50% of the initial signal in the 2× dilution series after 10 amplification cycles and 9 subsequent passages (Fig. 3c), which corresponds to an average amplification factor of 1.8 ± 0.3 per cycle. We found the amplification factor to be constant during the passage series ranging from 1.5 to 2.0 (Fig. 3d). The average amplification factor of all dilution experiments (1.8 ± 0.4) was independent of the dilution factor within experimental error, with a total amplification factor of 270 and 280 for 2- and 2.5-fold dilutions, respectively (Table 1).
Table 1. Amplification factors in serial transmission PMCA.
| Dilution factor per cycle | 1.5 | 2.0 | 2.5 |
| Amplification per cycle | 1.7 ± 0.4 | 1.75 ± 0.3 | 1.8 ± 0.3 |
| Total amplification factor | 50* | 270 | 280 |
Eight amplification cycles
In additional experiments with five serial passages and various dilution factors, a higher apparent amplification factor was sometimes observed during the initial round of amplification, possibly resulting from a superimposed process of recruitment of PrPSc from previously insoluble high molecular weight debris (data not shown). However, in subsequent passages, we always found amplification factors consistent with the results shown above. Note also that amplification factors depended on the ratio of PrPSc to PrPC, a 1:100 starting dilution of PrPSc, producing 4- to 5-fold amplification after 10 rounds, whereas increasing PrPSc concentration to 1:20 lowered the amplification factor to the value observed in serial dilution experiments.
By serially amplifying PrPres and diluting it into fresh brain homogenate, the reaction is not limited by the supply of PrPC substrate. If the dilution factor exactly matches the growth rate, the system, in principle, allows indefinite autocatalytic propagation of an infectious agent (19). The observed slow decrease in PrPres signal in the course of the amplification/dilution cycles is consistent with an exponential growth process in which the dilution factor slightly exceeds the growth factor.
If the initial PrPSc merely acted as a catalytic seed for the formation of PrPres, however, amplification would only be observed in the initial amplification cycles and the reaction would quickly come to a halt as the original PrPSc is diluted out exponentially. Likewise, a hypothetical mechanism that exposes new catalytic surfaces in the initial PrPSc with each sonication step would not result in maintained amplification, unless the increase in exposed catalytic surfaces exactly matches the dilution factor. Whereas an exponential increase in catalytic sites by fragmenting the initial PrPSc is, in principle, possible over nine dilution cycles, resulting in 512-fold smaller PrPSc particles, fragmentation of PrPSc and thus amplification efficiency should then increase monotonously with sonication time. In PMCA reactions, however, the dependency of amplification efficiency on sonication time shows a distinct maximum at a sonication of 5–20 s per round (data not shown).
Taken together, our findings outlined so far demonstrate that PrPres does serve as a catalytically active template for structural PrPC conversion in PMCA. This finding provides proof for autocatalytic replication of misfolded PrPres in an ex vivo conversion system. The PrPres catalyzes conversion of PrPC to PrPres apparently as efficiently as observed for the PrPSc initially present in the sample and furthermore exhibits a resistance to proteinase K digestion indistinguishable from that of PrPSc. Therefore, at the molecular level, PrPres formed in the PMCA reaction possesses the key properties attributed to infectious PrPSc by the prion hypothesis, i.e., proteinase K resistance and autocatalytic protein misfolding activity.
Infectivity of Material Generated by PMCA with Serially Passaged PrPres. The catalytic conversion activity of PrPres created in the serial PMCA reaction suggested that, according to the prion model, it might exhibit biological infectivity in an appropriate bioassay. To test for a gain in infectivity during serial PMCA, we archived samples of the initial infectious brain homogenate before amplification (sample A), of the PMCA reaction mixture after 10 serial passages (sample B), and of the starting material after a 29-fold dilution without PMCA (sample C) at -80°C. Of each of the samples, 50-μl aliquots were inoculated i.c. into 10 hamsters after a 103-fold dilution in PBS. The resulting incubation times until the occurrence of terminal scrapie are shown in Fig. 4a. With 107 ± 11 days, sample A showed the shortest incubation time, corresponding to a mean infectious titer in the diluted sample of 250 LD50i.c. Only 8 of 10 hamsters challenged with sample C developed disease until 250 days after inoculation, with a mean incubation time of 138 days for the diseased animals. If the inocula administered to the yet surviving animal in this group contained no infectivity, the mean titer for sample C would be ≈2 LD50i.c per 50 μl. The mean incubation time for sample B was 125 ± 6 days, corresponding to 13 LD50i.c. in 50 μl of the diluted sample, and lay between the two other groups.
Apparent infectivity titers in individual animals showed a narrower distribution in the PMCA sample B than in control samples A and C, suggesting a more homogeneous distribution of the infectious agent, which is most plausibly due to the sonication process repeatedly performed during PMCA (Fig. 4b). Up to 10-fold increased apparent infectivity values generated by ultrasonication of purified PrPSc in the presence of liposomes have been reported (20, 21) and predicted by kinetic modeling (22). However, control experiments comparing infectivity titers in sonicated and untreated samples by using limited dilution series of Rocky Mountain Laboratory mouse scrapie strain (RML) mouse prions in PrP-overexpressing mice (Tga20) showed no significant difference in titers (data not shown).
Thus, the titration experiments do not provide conclusive evidence whether PrPres formed by PMCA is associated with “new” infectivity because the moderate increase in infectivity observed might well be due to fragmentation of PrPSc. Clearly, the rate of PrPres generation is not matched by a corresponding increase of infectivity during the PMCA reaction. Possibly, this specific form of misfolded PrP needs additional structural features for infectivity or it might require a specific cofactor that may not be incorporated into the prion during PMCA such as highly complex polysaccharides, which have been found associated with purified PrPSc (23). Short RNA molecules have been implicated as essential components of the in vitro amplification of PrPres and thus possibly for infectivity (9). A drop in amplification efficiency is observed in the last reaction cycle (Fig. 3d). Even though preparation of brain homogenates and sonication of PMCA samples were carried out under sterile conditions, 100 h of incubation at 37°C might still have led to microbial growth and elevated proteolytic or RNase activity, destroying potential cofactors for prion infectivity.
Conclusion
We serially passaged misfolded protease-resistant PrPres in an in vitro amplification reaction. Amplification depends on cellular PrPC as a substrate for conversion. Hamster rPrP acts as an inhibitory substrate analogue, which specifically blocks PrPC conversion at stoichiometric concentrations. Therefore, the PMCA assay promises to be a valuable tool to screen compounds for their therapeutic and prophylactic potential in fighting transmissible spongiform encephalopathies.
PrPres generated by serial in vitro PMCA is indistinguishable from PrPSc in terms of proteinase K resistance and catalyzes PrP misfolding as efficiently as PrPSc. However, if PrPres generated by the PMCA reaction is infective, its titer is at least 10-fold lower than PrPSc derived from the brains of Scrapie hamsters, which suggests that the final make-up of PrPSc in terms of conformation and aggregation state or an essential cofactor for efficient infection of animals is lacking from the PrPres produced in vitro.
On the other hand, serially passaging misfolded PrPres in the PMCA procedure has produced misfolded PrP in an in vitro reaction that meets all of the requirements of the biochemical PrP misfolding process as postulated by the prion hypothesis, providing formal proof for the autocatalytic propagation of PrP misfolding.
Supplementary Material
Acknowledgments
We thank U. Bertsch (Center of Neuropathology and Prion Research, Ludwig Maximilians University of Munich) for rPrP, and Marion Joncic (Robert Koch-Institute) and Salah Soliman (Center of Neuropathology and Prion Research, Ludwig Maximilians University of Munich) for technical assistance. This work was supported by European Union Grant QLK3-CT-2001-02345, State of Bavaria Grants 830302-6 and 822796-9, and Ludwig Maximilians University of Munich FöFoLe Grant 822784-5.
Abbreviations: PMCA, protein misfolding cyclic amplification; PrP, prion protein; PrpC, cellular PrP; PrPSc, scrapie-associated PrP isoform; PrPres, proteinase K-resistant PrP; LD50i.c., 50% intracerebral lethal dose; rPrP, recombinant PrP; SHa, Syrian hamster.
References
- 1.Prusiner, S. B. (1982) Science 216, 136-144. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kocisko, D. A., Priola, S. A., Raymond, G. J., Chesebro, B., Lansbury, P. T., Jr., & Caughey, B. (1994) Nature 370, 471-474. [DOI] [PubMed] [Google Scholar]
- 4.Saborio, G. P., Permanne, B. & Soto, C. (2001) Nature 411, 810-813. [DOI] [PubMed] [Google Scholar]
- 5.Jarrett, J. T. & Lansbury, P. T., Jr. (1993) Cell 73, 1055-1058. [DOI] [PubMed] [Google Scholar]
- 6.Eigen, M. (1996) Biophys. Chem. 63, A1-A18. [DOI] [PubMed] [Google Scholar]
- 7.Lucassen, R., Nishina, K. & Supattapone, S. (2003) Biochemistry 42, 4127-4135. [DOI] [PubMed] [Google Scholar]
- 8.Vorberg, I. & Priola, S. A. (2002) J. Biol. Chem. 277, 36775-36781. [DOI] [PubMed] [Google Scholar]
- 9.Deleault, N. R., Lucassen, R. W. & Supattapone, S. (2003) Nature 425, 717-720. [DOI] [PubMed] [Google Scholar]
- 10.Zobeley, E., Flechsig, E., Cozzio, A., Enari, M. & Weissmann, C. (1999) Mol. Med. 5, 242-243. [PMC free article] [PubMed] [Google Scholar]
- 11.Büeler, H., Fischer, M., Lang, Y., Bluethmann, H., Lipp, H. P., DeArmond, S. J., Prusiner, S. B., Aguet, M. & Weissmann, C. (1992) Nature 356, 577-582. [DOI] [PubMed] [Google Scholar]
- 12.Liemann, S. & Glockshuber, R. (1999) Biochemistry 38, 3258-3267. [DOI] [PubMed] [Google Scholar]
- 13.Kascsak, R. J., Rubenstein, R., Merz, P. A., Tonna-DeMasi, M., Fersko, R., Carp, R. I., Wisniewski, H. M. & Diringer, H. (1987) J. Virol. 61, 3688-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimberlin, R. H. & Walker, C. A. (1986) J. Gen. Virol. 67, 255-263. [DOI] [PubMed] [Google Scholar]
- 15.Kimberlin, R. H. & Walker, C. A. (1977) J. Gen. Virol. 34, 295-304. [DOI] [PubMed] [Google Scholar]
- 16.Prusiner, S. B., Scott, M. R., DeArmond, S. J. & Cohen, F.E. (1998) Cell 93, 337-348. [DOI] [PubMed] [Google Scholar]
- 17.Masel, J. & Jansen, V. A. (2000) Biophys. Chem. 88, 47-59. [DOI] [PubMed] [Google Scholar]
- 18.Masel, J., Jansen, V. A. & Nowak, M. A. (1999) Biophys. Chem. 77, 139-152. [DOI] [PubMed] [Google Scholar]
- 19.Spiegelman, S., Haruna, I., Holland, I. B., Beaudreau, G. & Mills, D. (1965) Proc. Natl. Acad. Sci. USA 54, 919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabizon, R., McKinley, M. P. & Prusiner, S. P. (1987) Proc. Natl. Acad. Sci. USA 84, 4017-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabizon, R. & Prusiner, S. B. (1990) Biochem. J. 266, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masel, J. & Jansen, V. A. (1999) Proc. R. Soc. London Ser. B 266, 1927-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Appel, T. R., Dumpitak, C., Matthiesen, U. & Riesner, D. (1999) Biol Chem. 380, 1295-1306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.