Abstract
Angiogenesis is a major therapeutic target. Ideal drug targets are genes expressed only in endothelial cells (ECs) or only during the angiogenic process. Here, we describe a gene, p73RhoGAP (p73), that has both of these properties. By using a PCR-based subtraction–hybridization approach to clone cDNAs from ECs undergoing capillary-tube formation, we identified a RhoGAP member, p73. p73 displays GTPase activity to Rho but not to Rac or Cdc42. Knockdown of p73 protein, achieved by adenovirus delivery of p73 antisense and by small interfering RNA into ECs, demonstrated the importance of this protein in EC function. Under such conditions, EC migration, proliferation, and capillary-tube formation were inhibited. Furthermore, angiogenesis in vivo was also inhibited by antisense p73. A mutant R82A alteration achieved a similar phenotype in vitro to the antisense, demonstrating the importance of the GTPase-activating protein activity to p73 function. Expression profiling of p73 shows that it is vascular cell-selective, being highly expressed in ECs and smooth-muscle cells but not in other cell types. Finally, we show that the mRNA of p73 is up-regulated in an angiogenic milieu with little or no regulation seen under nonangiogenic conditions. p73, a vascular cell-specific GTPase-activating protein, is an important modulator of angiogenesis and displays many of features that make it worthy of being a drug target.
Angiogenesis, the formation of new blood vessels from pre-existing vessels, plays a critical role in many physiological and pathologic processes, including embryogenesis, wound healing, tumor growth, and metastasis (1, 2). Thus, the angiogenic process is considered to be an excellent target for therapeutic intervention. Identification of key regulatory molecules has principally been done by using in vitro models in which endothelial cells (ECs) are cultured on extracellular-matrix components, such as collagen, fibrinogen, or fibronectin, with identification of targets that are involved in events such as migration or proliferation, which are elements of angiogenesis but are not specific for it. Another shortfall in these investigations is the fact that the assays are generally performed on a flat or 2D environment, whereas EC morphogenesis to form capillary tubes requires a 3D matrix, allowing the establishment of important polarity cues. The use of such 3D assays, which include matrices of collagen type 1, fibrin, or Matrigel, recapitulate many of the events in angiogenesis and has allowed dissection of the cellular and molecular events in angiogenesis (3–6). Data using these assays to define genes altered during angiogenesis has supported the idea that there are major fundamental differences between cells responding on 3D versus 2D matrices and that genes specific for angiogenesis might exist.
The mammalian Rho family of small GTPases has been implicated in diverse cellular functions, including reorganization of the actin cytoskeleton, cell-growth control, transcription regulation, and membrane trafficking (7). The Rho family of small GTPases consists of at least the following 20 members: Rho (A, B, and C), Rac (1, 2, and 3), Cdc42, TC10, TCL, Chp (1 and 2), RhoG, Rnd (1, 2, and 3), RhoBTB (1 and 2), RhoD, Rif, and TTF (8). Like other members of the Ras superfamily, Rho proteins act as molecular switches to control cellular processes by cycling between active GTP-bound and inactive GDP-bound states. Regulation of these GTPases occurs by means of three major classes of regulatory proteins. The guanine nucleotide-exchange factors regulate activation through GDP–GTP exchange; GTPase-activating proteins (GAPs), which promote hydrolysis of the GTP to GDP-bound form because the Rho proteins display little if any basal GTPase activity; and guanine nucleotide-dissociation inhibitors, which stabilize the inactive GDP-bound form of the protein (9). At least 134 of these regulatory proteins have now been defined (8).
The function of the Rho family in endothelial morphogenesis is only now being elucidated, and it would appear that different Rho family members play specific roles. Rho and Rac are important for regulation of permeability and cell migration (10, 11). Rho is also important for EC attachment and apoptosis (12, 13), whereas Cdc42 and Rac1 are implicated in vacuole formation and subsequent lumen formation (5). Given the limited number of RhoGTPases and the seeming overabundance of RhoGAPs, it is likely that the RhoGAPs may partly provide the specificity in control of function of the RhoGTPases.
We have cloned and characterized a RhoGAP, p73RhoGAP (p73), which is essential for angiogenesis. It displays characteristics that make it suitable as a potential drug target, namely, a restricted expression profile in primary vascular cells and regulation in the angiogenic milieu with little if any regulation under nonangiogenic conditions.
Materials and Methods
Cells and Cell Culture. Primary human umbilical-vein ECs (HUVECs) were isolated and cultured as described (3), and they were used at passage 4 or less. ECs were plated into fibronectin-coated flasks before infection. HEK293 (293) cells were grown in DMEM (CSL, Melbourne) with 10% FCS/penicillin/streptomycin.
Generation of Recombinant p73 Clones. HUVEC RNA was isolated, and cDNA was generated by reverse transcription. Primers were designed to amplify the p73 5′ and 3′ cDNA fragments (see Table 1, which is published as supporting information on the PNAS web site). Both PCR products were cloned into pGEM-Teasy (Promega), confirmed by sequencing, and ligated together by means of a common internal BamHI site. To generate the p73 mutant (R82A), the arginine codon (CGA) at position 82 was changed to alanine (GCA) by using a PCR-mutagenesis approach. The p73 or R82A mutant cDNA were excised from pGEM-Teasy with NotI and subcloned (in both orientations for p73) into the shuttle vector pAdTrack-CMV (Qbiogene, Carlsbad, CA). We generated 5′-Flag-tagged-p73 by PCR (Table 1) and cloned it into the EcoRV site of pAdTrack-CMV. Recombinant adenovirus was made by using the pAdEasy system (Qbiogene). Viral titres were determined by using the TCID50 method, and viral-particle numbers were quantified at OD at 600 nm.
For gene-transfer experiments, cells were grown to 80% confluence and infected with an amount of pAdEasy empty vector (EV), pAdEasy-p73 antisense (p73R) or pAdEasy-p73R82A (R82A) mutant virus particles, which yielded a similar level of GFP expression.
Generation of a Retroviral Vector Expressing a Small Interfering RNA (siRNA) Against p73. pMSCVpuro (BD Biosciences) was modified to create a short-hairpin RNA-generating retroviral vector. To do this, the 3′ LTR of pMSCVpuro was inactivated by removal of a XbaI/NheI fragment. A H1-RNA polymerase III promoter cassette was then inserted into the MCS of the vector. The p73 specific sequence 5′-GTAGTCGTGCCACCAGTAG-3′ was cloned into the modified pMSCVpuro vector in a short-hairpin sequence format. The sequence 5′-AGGCATCAGCGGACCTCAT-3′ was used as a control siRNA because this sequence contained similar GC content to that specific for p73.
Retroviral Particle Production and HUVEC Infection. We cotransfected 293 T cells with 1.5 μg each of pVPack-VSV-G and pVPack-GP (Stratagene) and 2 μg of siRNA vector DNA by using Lipofectamine 2000 (Invitrogen), and viral particles were harvested according to the manufacturer's instructions (Stratagene). HUVECs were transduced by using an adaptation of the methodology described in ref. 14. Transduced cells were then selected with puromycin (0.35 μg/ml).
Northern Blot Analysis and Virtual Northern Blot Analysis. Total RNA was isolated by Trizol extraction (Invitrogen). For virtual Northern blotting, mRNA was converted into full-length cDNA by using the Smart PCR cDNA-synthesis procedure (Clontech). The cDNA (1 μg) or total RNA (5 μg) was separated by gel electrophoresis, transferred to nylon membranes, and probed with a 32P-labeled p73 RsaI fragment (591 bp; isolated fragment) or cyclophilin A.
Detection of Gene Silencing: Quantitative RT-PCR (Q-RT-PCR) Assay. Total RNA was extracted from HUVECs 4 days after infection by using an RNeasy Mini kit (Qiagen, Valencia, CA) including on-column DNase treatment and reverse transcribed by using an oligo(dT) primer and M-MLV (Promega) reverse transcriptase. Q-RT-PCR was performed by using the appropriate primer pairs (Table 1) on the RotorGene 2000 (Corbett Research, Sydney). All data were normalized to the housekeeping gene POLR2K (RNA polymerase II).
Western Blot Analysis. Primary Abs were incubated at 4°C for 20 h, and secondary Abs were incubated at 20°C for 1 h. Reactive bands were detected by chemiluminescence (ECL Western blotting detection reagents, Amersham Biosciences) and analyzed by densitometry, and results were expressed as arbitrary units.
Rho-, Rac-, and Cdc42-GTP Activity Assays. Cells were serum-depleted (EBM, Clonetics, San Diego) overnight. Rho, Rac, and Cdc42 activity was measured by using the EZ-Detect Rho, Rac, and Cdc42 activation kits (Pierce), respectively. Active protein was detected by Western blotting by using mAbs against Rho, Rac, or Cdc42.
Migration Assay. EC migration was measured essentially as described (15). Cell migration was measured after a 20-h incubation at 37°C.
F-Actin Staining. ECs were plated on fibronectin-coated Lab-Tek chamber slides (Nalge). The cells were grown with or without C3 exoenzyme (30 μg/ml) for 32 h and then serum-starved overnight also in the presence and absence of C3 exoenzyme. The cells were stained with Rho–phalloidin (Molecular Probes). The actin filaments were observed by using an epifluorescence microscope.
Proliferation Assay. HUVECs were plated at 3 × 103 cells per well in 96-well plates. We performed 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2μ-tetrazolium assays (Promega) for cell viability on days 0 (plating) and 3. The absorbance at 490 nm reflects the number of viable cells.
A Matrigel (Becton Dickinson) capillary-tube-formation assay was performed as described (4). ECs were resuspended in complete medium with or without 10 μM Rho-associated kinase (ROCK) inhibitor Y27632 (Santa Cruz Biotechnology) for 30 min, added to the Matrigel-coated wells (4 × 104 cells per well), and incubated at 37°C. Photographs were taken at regular intervals over a 24-h period.
Gel-Foam Assay for Angiogenesis. This assay was performed as described in ref. 16. Essentially, gel-foam sponges (5 mm3) were saturated with vascular endothelial growth factor (VEGF) at a final concentration of 600 ng/ml and either EV or p73R adenovirus at a final concentration of 5 × 105 plaque-forming units per sponge and sealed with 0.4% agarose in PBS. CBA female mice, 6–8 weeks of age, were anesthetized, and an incision (large enough to enable delivery of the sponge to the sides between the first and second mammary glands) was made down the back. The incision was clamped, and the mice were monitored for the next 11 days. At this time, the animals were killed, and the gel-foam sponges were removed and snap-frozen in liquid nitrogen. At least four frozen sections (11-μm thick) were analyzed, representing different levels throughout the sponge. Blood vessels were visualized by using anti-mouse CD31 (PECAM-1) staining (Pharmingen). For each section, the number of blood vessels per high-power field was counted.
C3 Exoenzyme Purification. pGEX2T-C3, expressing GST-C3 exoenzyme fusion protein, was a gift from Shaun Jackson (Monash University Department of Medicine, Australian Centre for Blood Diseases, Box Hill, Australia).
Results
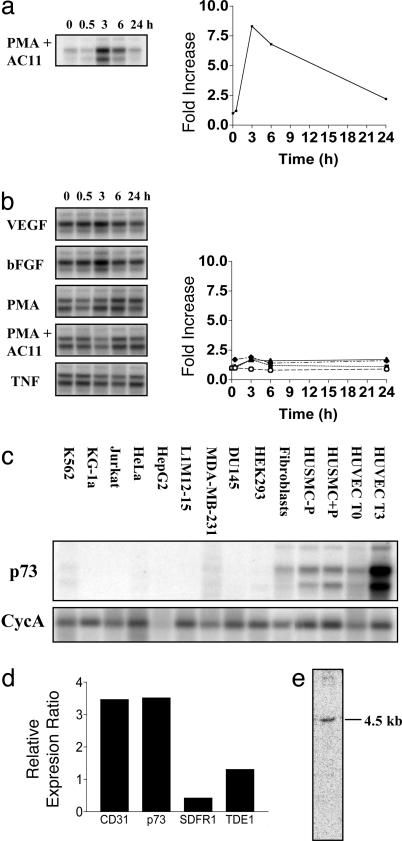
Isolation of p73. Identification of regulated genes during angiogenesis may result in characterization of novel targets for therapeutic drug development. To this end, we have utilized our model of in vitro angiogenesis, in which the morphological events and the time course of changes have been well characterized (4, 6). HUVECs, plated onto a 3D collagen gel in the presence of growth factors, phorbol 12-myristate 13-acetate (PMA), and an Ab to the integrin α2β1, were induced to make capillary tubes over a 24-h period. Isolating these cells at critical time points (namely 0, 0.5, 3, 6, and 24 h) and utilizing a PCR-based suppression subtractive-hybridization approach allowed the isolation of regulated genes. From DNA filters of these regulated cDNAs probed with RNA taken from the different time points, we identified a cDNA, which by virtual Northern blot analysis was highly regulated in the 3D collagen-gel assay (Fig. 1a) at 3 and 6 h, returning to basal levels by 24 h. Although the gene was highly regulated in the 3D collagen system, there was little or no regulation when the cells were plated onto a 2D matrix of collagen in the presence of a panel of stimulators that are utilized in the angiogenesis model (VEGF, fibroblast growth factor, PMA, or PMA plus anti-α2β1) or another known activator of EC, tumor necrosis factor α (Fig. 1b).
Fig. 1.
Expression of p73 during capillary-tube formation and its tissue distribution. (a and b) Virtual Northern blot analysis of p73 expression in capillary-tube formation. (a) HUVECs were plated onto a 3D collagen gel in the presence of PMA and anti-α2β1 to stimulate in vitro capillary-tube formation. Total RNA was harvested at the time points shown, and virtual Northern blots were probed with our isolated cDNA fragment. Blots were quantified by using a phosphorimager and standardized to cyclophilin A (CycA), and the fold increase was determined with respect to time given in the corresponding graph. (b) HUVECs, plated on a 2D collagen matrix, were treated with various stimuli, and the virtual Northern blots were probed as described for a. p73 levels were normalized to CycA, and the fold increase was plotted as follows: VEGF, ○; basic fibroblast growth factor (bFGF), ▪ ; PMA, ▴; PMA plus anti-α2β1 (PMA + AC11), ♦. TNF, tumor necrosis factor α. (c) Tissue distribution of p73 mRNA by virtual Northern blot analysis. Virtual Northern blots were generated from human cell lines and primary cells representing different cell types. HUVECs T0 and T3 refer to ECs at 0 and 3 h on 3D collagen gel. Human umbilical-vein smooth-muscle cells (HUSMCs) treated without and with PMA for 3 h are shown. (d) Expression of p73 in placenta, heart, lung, breast, colon, and HUVECs by using Q-RT-PCR. The ratio of expression in HUVECs over the median of the expression in the other tissues was calculated. This ratio was compared with the ratio obtained with the endothelial-selective marker CD31 (PECAM). (e) Northern blot analysis of p73 expression. Total HUVEC RNA (5 μg) was analyzed by Northern blot analysis by probing with the p73 3′ UTR-specific probe.
National Center for Biotechnology Information (NCBI) database searches (February 2004) of our isolated cDNA identified a 4,567-bp cDNA (Unigene accession no. DKFZp564B1162, GenBank accession no. AL136646) that encoded a hypothetical protein of 655 aa (73 kDa) containing a putative RhoGAP domain spanning amino acids 57–233. Therefore, we called this gene p73.
We next examined the cell expression pattern of this cDNA by using virtual Northern blots made from various cell lines and primary cell cultures. p73 expression is highly restricted to primary vascular cells, namely smooth-muscle cells and ECs. All other tested cells showed little or no expression (Fig. 1c). By using Q-RT-PCR, we investigated the expression of p73 in a number of tissues. Expression analysis of EC markers in primary tissues is skewed by the fact that all tissues contain ECs. To account for this phenomenon in our analysis of the p73 expression profile, we measured the mRNA expression in six primary tissues, including placenta, heart, lung, breast, colon, and HUVECs, and we calculated the ratio of expression in HUVECs over the median of the expression in the other tissues. This ratio was compared with the ratio obtained with the endothelial-selective marker CD31. The expression ratio for p73 was very similar to that of CD31 (Fig. 1d), suggesting that p73 exhibits endothelial-specific expression. In comparison, two other genes, SDFR1 and TDE1, shown in refs. 17 and 18 to display wide tissue distribution, exhibited the expected lower ratios. Northern blot analysis, probing with a p73 3′ UTR-specific probe, revealed a single 4.5-kb transcript (Fig. 1e).
The 3D specificity in terms of regulation and its restricted expression profile suggested that this gene may play a role in angiogenesis.
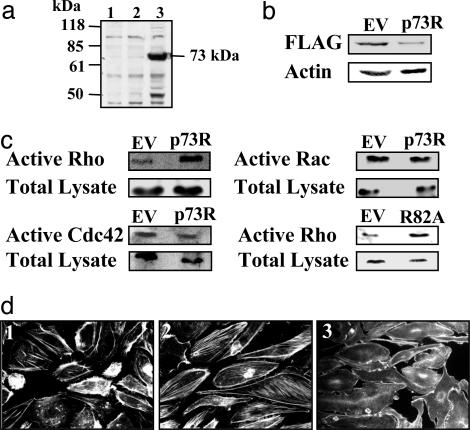
Identification of p73 as a Potential RhoGAP. National Center for Biotechnology Information (NCBI) database comparisons of the RhoGAP domain of p73 revealed considerable identity to other known RhoGAP-containing proteins, such as Bcr, N-chimerin, p50RhoGAP, and p190RhoGAP. This region of homology comprises ≈160 amino acid residues, with 10 residues that are critical to the structural integrity of the GAP domain, 3 residues (Arg-85, Asn-188, and Lys-172, with respect to the p50RhoGAP domain) that are catalytically crucial to GAP activity, and 5 residues (Gly-82, Leu-132, Leu-178, Met-190, and Asn-194) that promote GTP hydrolysis (19–21) (see Fig. 6, which is published as supporting information on the PNAS web site). The hypothetical protein has 9 of the 10 residues that are involved in the structure, with one conservative change from Leu to Ile. All three catalytic amino acids and four of the five GTP hydrolysis-promoting residues are identical, with one conservative change of Leu to Val. The highly conserved identity suggests that this gene may have GAP activity. The full-length cDNA was cloned from an EC library by using PCR, and expression of a Flag-tagged construct in 293 cells gave the predicted protein of 73 kDa (Fig. 2a).
Fig. 2.
Characteristics and substrate specificity of p73. (a) p73 overexpression in 293 cells. EV and pAdtrack-Flag-p73 were transfected into 293 cells, and cell lysates were collected after 48 h. A Western blot was probed with anti-Flag M2 Ab (Sigma). Lanes: 1, cells alone; 2, EV; 3, Flag-p73. (b) p73 protein knockdown by p73R. EV and p73R were transfected into 293 cells and cotransfected with pAdtrack-Flag-p73 7 h later. Cell lysate was collected after 48 h. A Western blot was probed with anti-Flag M2 Ab, and anti-actin was used as a loading control. (c) p73 has activity for Rho but not Rac and Cdc42. HUVECs were adenovirally infected with EV, p73R, and p73 mutant (R82A), and they were then incubated without serum for 20 h. The cells were lysed and assayed for detection of active Rho-GTP, Rac-GTP, and Cdc42-GTP, and the average fold increase compared with EV is shown. Each result represents at least four independent experiments. (d) p73 regulates stress-fiber formation. HUVECs were adenovirally infected with EV (image 1) or p73R (images 2 and 3) 24 h before plating on a fibronectin. p73R-infected HUVECs were treated without (image 2) or with (image 3) C3 exoenzyme (30 μg/ml) for 32 h and stained with Rho–phalloidin.
Analysis of the protein sequence showed, in addition to the putative GAP domain, a potential coiled-coil domain, which are known to mediate protein–protein interactions. Other functional motifs, such as SH2, SH3, pleckstrin homology, or cysteine-rich domains or the catalytic domains of protein kinases, which have been described in many other RhoGAP family members (7), are not present in p73.
To determine whether p73 has functional activity, we utilized antisense constructs as a mechanism for regulating the levels of p73. First, to confirm that such constructs can knockdown gene expression and affect protein levels of p73, we transfected 293 cells with p73R or EV as control. After 6 h, cells were transfected with Flag-tagged p73. Levels of p73 protein were assessed by Western blot analysis 18 h later. The results shown in Fig. 2b indicate that p73R reduced the protein levels by 61 ± 8.6% in three separate experiments. As a mechanism for gene delivery of this antisense construct to EC, we utilized adenovirus and then determined effects on the activity of Rho, Rac, and Cdc42. The results shown in Fig. 2c indicate that p73R increased Rho activity in ECs compared with EV-infected control cells. In the five experiments that were performed, there was a 3.1 ± 0.5-fold increase in Rho activity with p73R. No change in active Rac or Cdc42 (1.1 ± 0.3-fold increase in both cases) with p73R was seen, confirming that the protein contains an active RhoGAP domain that has specificity in ECs for Rho but not Rac or Cdc42.
From structural analysis, the highly conserved Arg residue corresponding to Arg-85 in p50RhoGAP is critical for the binding to Rho target proteins and increasing hydrolysis of Rho-bound GTP (19–21). This amino acid residue corresponds to Arg-82 in p73, which we mutated to Ala (R82A). Expression of this mutant in EC showed a 2.6 ± 0.7-fold increase (n = 4 experiments) in Rho activity (Fig. 2c), consistent with this mutation both eliminating the GAP activity of p73 and generating a dominant-negative form, further confirming that p73 is indeed a RhoGAP family member.
Consistent with Rho activity, p73R-infected ECs showed increased stress-fiber formation compared with EV control cells, which displayed the classic cortical actin-type morphology with minimal stress fibers, characteristic of unstimulated EC (Fig. 2d, image 1). In contrast, p73R-infected ECs displayed prominent, thick F-actin bundles (stress fibers), aligned in parallel arrays (Fig. 2d, image 2). To confirm that the effects of stress-fiber formation were mediated by Rho, p73R-infected HUVECs were treated with the Rho-specific inhibitor C3 transferase and then stained for F-actin. In the presence of C3 transferase, stress-fiber formation was abolished (Fig. 2d, image 3), suggesting that the stress fibers are induced in the p73R-infected cells through an alteration in Rho.
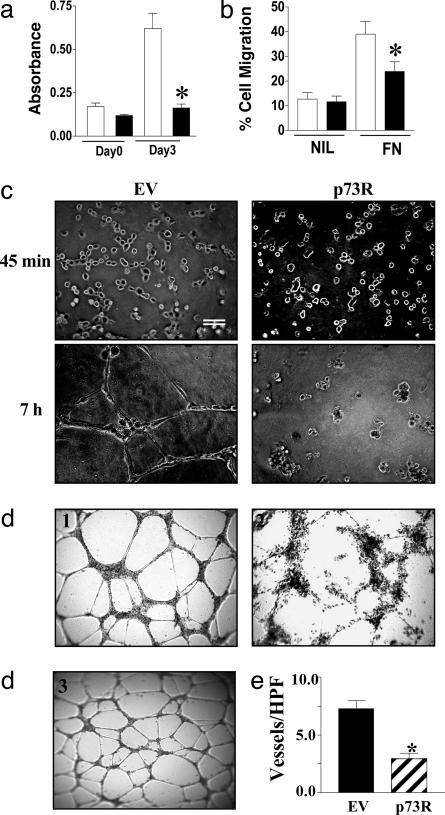
p73 in Angiogenesis. ECs were infected with EV or p73R and analyzed for effects on EC function. The results shown in Fig. 3 indicate that p73R-containing cells were inhibited in their capacity to proliferate (Fig. 3a) and migrate (Fig. 3b), although there was no major change in their capacity to adhere to fibronectin, collagen, or gelatin matrices (data not shown). Importantly, p73R cells also failed to form capillary tubes in the complex matrix Matrigel (Fig. 3c). The changes in the capacity to form tubes was evident as early as 45 min after plating. Although both the EV and p73R-infected cells moved rapidly from single cells to aggregates of small numbers of cells, the p73R cells failed to align or form more extended cellular structures. Video microscopy (Z.-J.S., C.N.H., and J.R.G., un-published data) also demonstrated that p73R cells failed to form cellular protrusions that are normally evident within minutes of plating onto Matrigel. The use of siRNA to inhibit p73 gave a similar phenotype to that seen with antisense adenovirus (Fig. 3d) in comparison either with the EV or an irrelevant siRNA. Silencing of p73 gene by siRNA, as detected by Q-RT-PCR and expressed relative to control, gave a 51 ± 7.5% (n = 3) inhibition. With the siRNA-treated cells, there was a 61 ± 4% inhibition in cell proliferation (n = 4), 44 ± 2% inhibition in cell migration to fibronectin (n = 3) compared with vector-only control, and an inhibition of capillary tubes on Matrigel (Fig. 3d). Finally, to determine whether p73 is essential for angiogenesis in vivo, we used a recently described assay for angiogenesis in the mouse (16). Human p73R adenovirus was used because the human cDNA is 85% identical to the mouse containing long stretches of homology. Mice were implanted on one side with a gel foam bathed in EV and on the other side with a p73R-bathed gel foam. At 10 days later, the gels were removed and vessels were visualized by staining for CD31. The results shown in Fig. 3e show that p73R caused a significant inhibition of blood vessel migration into the gel foam, thus confirming the importance of p73 in angiogenesis.
Fig. 3.
p73 regulates angiogenesis. (a) p73R inhibits HUVEC proliferation. HUVECs were infected with EV (open bars) or p73R (filled bars) adenovirus, and proliferation assays were performed. Each bar is the mean ± SEM (n = 4) of three independent experiments. (b) p73R decreases HUVEC migration. HUVECs infected with EV (open bars) or p73R (filled bars) adenovirus were allowed to migrate toward either no chemotactic factor (NIL) or to fibronectin (FN). The number of migrated cells after 24 h was expressed as a percentage of the total cells added. Data are given as mean ± SEM (n = 3) of at least three independent experiments. (c) p73R inhibits tube formation. HUVECs infected with EV or p73R adenovirus were plated onto Matrigel, and capillary-tube formation was observed over a 12-h time course. Photographs were taken at 45 min and 7 h after plating. (Scale bar, 200 μm.) (d) Silencing of p73 by siRNA inhibits in vitro capillary-tube formation. HUVECs were transduced with either vector control (image 1), p73 siRNA (image 2), or control siRNA (image 3), selected for puromycin resistance, and after 4 days plated onto Matrigel. Tube formation after 24 h is shown and is representative of three experiments. (e) p73R inhibits blood-vessel invasion in vivo. Assessment of the number of blood vessels per high-power field (vessels/HPF) was made from three mice implanted with a VEGF with EV control adenovirus (black) and VEGF with p73R adenovirus (striped) impregnated gel-foam sponge. Results are given as mean ± SEM. Comparisons were made between each group and tested for significance by using a three-way nested ANOVA. *, P < 0.001.
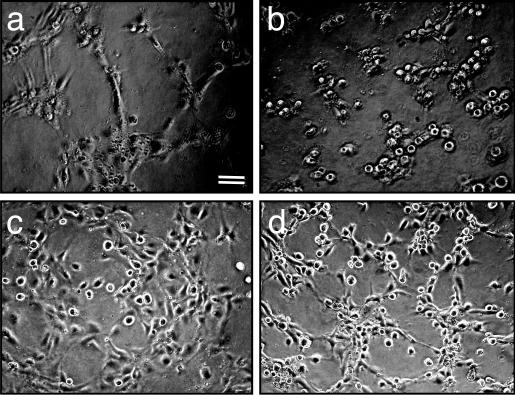
Visual assessment of the block in tube formation in p73R-infected cells showed that these cells failed to reorganize into large aggregates and subsequent sprouts and tubes, suggesting an inability to reorganize the actin cytoskeleton. To test this possibility, the EV and p73R adenovirus-infected cells were treated with the ROCK inhibitor Y27632. ROCK is the downstream effector of Rho. Consistent with published results showing a dependence on Rho activity for tube formation (22, 23), EV control cells treated with Y27632 were inhibited from forming tubes (Fig. 4c). p73R-infected cells did not undergo realignment (Fig. 4b). However, when these cells were treated with Y27632 they were able to initiate reorganization to form tubular structures (Fig. 4d) more like that seen in control cells (Fig. 4a). Similar results were obtained with the use of another ROCK inhibitor, HA1077 (see Fig. 7, which is published as supporting information on the PNAS web site). These results show that a ROCK inhibitor is able to reverse the phenotype induced as a result of the enhanced Rho activity in p73R cells.
Fig. 4.
The ROCK inhibitor, Y27632, reverses the p73 antisense phenotype. HUVECs were infected with either EV control (a and c) or p73 antisense (b and d) adenovirus. Cells were plated onto Matrigel in the absence (a and b) or presence (c and d) of the ROCK inhibitor Y27632 at a final concentration of 10μM. Formation of capillary tubes was assessed over the next 12 h. Photographs were taken at 4 h after plating.
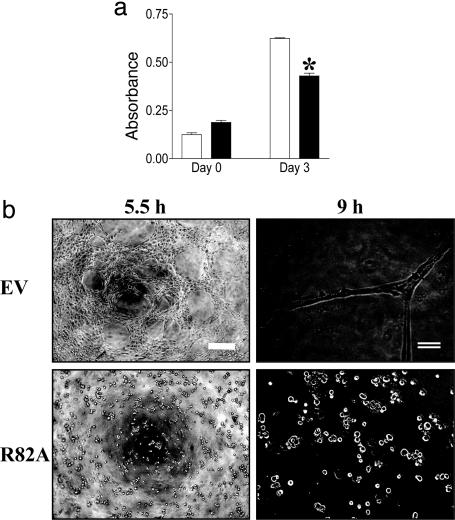
To determine whether the GAP activity is responsible for the changes in biological activity induced in the p73R cells, the functional capacity of mutant R82A-infected cells was investigated. R82A cells were inhibited in their ability to proliferate (Fig. 5a), and they failed to form capillary tubes in Matrigel (Fig. 5b), demonstrating that the regulation of the GAP activity is indeed responsible for the functional effects.
Fig. 5.
p73 mutant inhibits EC function. (a) p73 (R82A) inhibits proliferation. EV-infected (open bars) and R82A-infected (filled bars) HUVECs were plated on day 0, and proliferation was measured 3 days later. Results are expressed as absorbance. Each bar represents the mean ± SEM (n = 4) of three independent experiments. (b) R82A inhibits tube formation. HUVECs infected with either EV or R82A were plated onto Matrigel, and capillary-tube formation was observed over 12 h. Photographs were taken at 5 and 9 h after plating. Scale bars are as follows: single line, 50 μm; double line, 200 μm. *, P < 0.05.
Discussion
In this study, we have identified a RhoGAP, p73, which has specificity to Rho but not Rac or Cdc42, and its function is essential for angiogenesis. Further, it is highly expressed in vascular cells, and its regulation pattern is specific for a milieu that is permissive to angiogenesis.
To our knowledge, the identification of p73 is the first description of a vascular-specific RhoGAP. Other functionally specific or tissue-specific RhoGAPs that have been identified include the brain-specific p200RhoGAP, which controls neurite outgrowth and is important in neuronal and oligodendrocyte function (24), and p190-B RhoGAP, which regulates the insulin/insulin-like growth factor signaling pathway, having major effects on cell and animal size regulation and on the switch between adipogenesis and myogenesis (25, 26).
Sequence analysis of p73 shows that its GAP domain, although quite similar to known functional RhoGAP proteins, is most highly related to two hypothetical proteins, RhoGAP2 and KIAA0053, showing 81% and 63% amino acid identity, respectively. All three of these RhoGAPs have coiled-coil domains at the very C-terminal end, which are also highly homologous (54% and 40% amino acid identity to p73, respectively). A database search showed the domain in p73 is also very similar to two other coiled-coil domains contained within the proteins, cortexillin I and myosin light chain 2, both of which interact with actin. Thus, it is tempting to speculate that p73 may also show actin-binding properties, which could account for its functional effects. Furthermore, these three RhoGAP proteins also have potential alternative splice forms, incorporating a pleckstrin homology domain at the N terminus. The role of such p73 isoforms is intriguing.
The arginine at position 82 within the GAP domain is essential for function because crystallography studies show that this amino acid is inserted within the catalytic domain of the RhoGTPases and stabilizes the GTPase activity (19–21). In p73, mutation of Arg-82 to Ala eliminated its GAP activity (Fig. 2c). Furthermore, this mutant brought about similar changes in cell function to the p73R (Fig. 5), confirming that the GAP activity of p73 is essential for its function.
EC morphogenesis in vitro, as a reflection of what takes place in vivo, involves a coordinated set of events, including migration, proliferation, vacuole formation, and coalescence (6). Vacuole and lumen formation occurs by means of integrin-mediated pinocytosis involving vesicle and membrane trafficking events, and these morphogenic processes depend on Rac and Cdc42 activity (5). Thus, it is unlikely that p73 is implicated in the formation of vacuoles and lumen, given its specificity to Rho. Although our profiles showed that p73 is up-regulated at 3–6 h during tube formation, the antisense-containing cells suggest that p73 regulates an early event in tube formation. When plated onto Matrigel, the p73R cells have only a limited ability to undergo reorganization. Within the first 15–30 min, they are able to form small clumps of EC but fail to form protrusions or undergo more extensive reorganization at later times (Fig. 3). These features suggest that p73 may regulate attachment and/or cytoskeletal reorganization involved in cell movement because these events are known to depend on Rho activity (27, 28). However, measurement of cell attachment to different matrices showed no significant changes (data not shown), suggesting that initial integrin-mediated attachment is not altered. p73R cells did show high levels of basal actin stress-fiber assembly, even when the cells are first attached to matrix. Recent studies by Connolly et al. (29) showed that soon after Matrigel contact by EC, there is a generalized reduction in actin stress fibers and the formation of cellular protrusions with actin-rich lamellipodia. They suggest that this loss of actin stress fibers allows the subsequent cell movement and capillary assembly. Our results support this possibility, and we further postulate that p73 plays such a role to disassemble the actin cytoskeleton by means of the down-regulation of active Rho. Thus, inhibition of p73 function essentially locks the cell into a static nonmotile configuration, and indeed, p73R cells were inhibited in their capacity to migrate. The requirement for actin disassembly in tube formation and the potential role of p73 in this process is strengthened by the finding that p73R transfected cells treated with ROCK inhibitors adopted a more normal phenotype in terms of cellular alignment on Matrigel (Figs. 4d and 7) and clearly displayed the capacity to migrate and reorganize into the beginnings of capillary sprouts.
The most notable feature of p73, besides its cell specificity, is its regulation profile. Within the angiogenic microenvironment of collagen gels, p73 is up-regulated at 3 and 6 h but declines to basal levels by 24 h. However, given similar exogenous cues on 2D collagen with the addition of growth factors PMA and anti-α2β1 (RMAC11), little or no up-regulation was observed. Equally, no change was seen with the powerful inflammatory activator of EC, tumor necrosis factor α. Thus, alteration in mRNA levels for p73, which is postulated to reflect a regulatory cycle of p73, is restricted to the angiogenic milieu, suggesting that p73 activity may be “angiogenic-specific,” a possibility that supports the use of 3D cell-based assays as more appropriate to our understanding of gene regulation during morphogenesis. These results also highlight the possibility that p73 may be an excellent target for control of angiogenesis in pathologies such as tumor growth while sparing action on the mature vasculature.
Supplementary Material
Acknowledgments
We thank Chris Drogemuller, Michelle Parsons, Peter Brautigan, Jenny Drew, and Anna Sapa for expert technical assistance; Sue Lester from the Arthritis Laboratory (Hanson Institute) for helping with the statistics; and staff at the Adelaide Women's and Children's Hospital and Burnside War Memorial Hospital for collection of umbilical cords. We also thank Drs. Xiao-Chun Li, Pu Xia, and Yeesim Khew-Goodall for their expert advice. This work was supported by a Program Grant from the National Health and Medical Research Council of Australia, grants from the Anti-Cancer Foundation of South Australia, the National Heart Foundation of Australia, and Bionomics, Ltd.
Abbreviations: EC, endothelial cell; HUVEC, human umbilical-vein EC; siRNA, small interfering RNA; GAP, GTPase-activating protein; Q-RT-PCR, quantitative RT-PCR; p73, p73RhoGAP; p73R, pAdEasy-p73 antisense; R82A, pAdEasy-p73R82A; EV, empty vector; 293, HEK293; ROCK, Rho-associated kinase; VEGF, vascular endothelial growth factor; PMA, phorbol 12-myristate 13-acetate.
References
- 1.Augustin, H. G. (1998) Trends Pharmacol. Sci. 19, 216-222. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan, D. (1997) Science 277, 48-50. [DOI] [PubMed] [Google Scholar]
- 3.Gamble, J. R., Matthias, L. J., Meyer, G., Kaur, P., Russ, G., Faull, R., Berndt, M. C. & Vadas, M. A. (1993) J. Cell Biol. 121, 931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamble, J., Meyer, G., Noack, L., Furze, J., Matthias, L., Kovach, N., Harlant, J. & Vadas, M. (1999) Endothelium 7, 23-34. [DOI] [PubMed] [Google Scholar]
- 5.Bayless, K. J. & Davis, G. E. (2002) J. Cell Sci. 115, 1123-1136. [DOI] [PubMed] [Google Scholar]
- 6.Meyer, G. T., Matthias, L. J., Noack, L., Vadas, M. A. & Gamble, J. R. (1997) Anat. Rec. 249, 327-340. [DOI] [PubMed] [Google Scholar]
- 7.Van Aelst, L. & D'Souza-Schorey, C. (1997) Genes Dev. 11, 2295-2322. [DOI] [PubMed] [Google Scholar]
- 8.Etienne-Manneville, S. & Hall, A. (2002) Nature 420, 629-635. [DOI] [PubMed] [Google Scholar]
- 9.Mackay, D. J. & Hall, A. (1998) J. Biol. Chem. 273, 20685-20688. [DOI] [PubMed] [Google Scholar]
- 10.Wojciak-Stothard, B., Potempa, S., Eichholtz, T. & Ridley, A. J. (2001) J. Cell Sci. 114, 1343-1355. [DOI] [PubMed] [Google Scholar]
- 11.Nobes, C. D. & Hall, A. (1999) J. Cell Biol. 144, 1235-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chrzanowska-Wodnicka, M. & Burridge, K. (1996) J. Cell Biol. 133, 1403-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hippenstiel, S., Schmeck, B., N′Guessan, P. D., Seybold, J., Krull, M., Preissner, K., Eichel-Streiber, C. V. & Suttorp, N. (2002) Am. J. Physiol. 283, L830-L838. [DOI] [PubMed] [Google Scholar]
- 14.Denk, A., Goebeler, M., Schmid, S., Berberich, I., Ritz, O., Lindemann, D., Ludwig, S. & Wirth, T. (2001) J. Biol. Chem. 276, 28451-28458. [DOI] [PubMed] [Google Scholar]
- 15.Leavesley, D. I., Schwartz, M. A., Rosenfeld, M. & Cheresh, D. A. (1993) J. Cell Biol. 121, 163-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarty, M. F., Baker, C. H., Bucana, C. D. & Fidler, I. J. (2002) Int. J. Oncol. 21, 5-10. [PubMed] [Google Scholar]
- 17.Bossolasco, M., Lebel, M., Lemieux, N. & Mes-Masson, A. M. (1999) Mol. Carcinog. 26, 189-200. [PubMed] [Google Scholar]
- 18.Langnaese, K., Mummery, R., Gundelfinger, E. D. & Beesley, P. W. (1998) FEBS Lett. 429, 284-288. [DOI] [PubMed] [Google Scholar]
- 19.Rittinger, K., Walker, P. A., Eccleston, J. F., Smerdon, S. J. & Gamblin, S. J. (1997) Nature 389, 758-762. [DOI] [PubMed] [Google Scholar]
- 20.Barrett, T., Xiao, B., Dodson, E. J., Dodson, G., Ludbrook, S. B., Nurmahomed, K., Gamblin, S. J., Musacchio, A., Smerdon, S. J. & Eccleston, J. F. (1997) Nature 385, 458-461. [DOI] [PubMed] [Google Scholar]
- 21.Musacchio, A., Cantley, L. C. & Harrison, S. C. (1996) Proc. Natl. Acad. Sci. USA 93, 14373-14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Somlyo, A. V., Phelps, C., Dipierro, C., Eto, M., Read, P., Barrett, M., Gibson, J. J., Burnitz, M. C., Myers, C. & Somlyo, A. P. (2003) FASEB J. 17, 223-234. [DOI] [PubMed] [Google Scholar]
- 23.Uchida, S., Watanabe, G., Shimada, Y., Maeda, M., Kawabe, A., Mori, A., Arii, S., Uehata, M., Kishimoto, T., Oikawa, T. & Imamura, M. (2000) Biochem. Biophys. Res. Commun. 269, 633-640. [DOI] [PubMed] [Google Scholar]
- 24.Moon, S. Y., Zang, H. & Zheng, Y. (2003) J. Biol. Chem. 278, 4151-4159. [DOI] [PubMed] [Google Scholar]
- 25.Sordella, R., Classon, M., Hu, K. Q., Matheson, S. F., Brouns, M. R., Fine, B., Zhang, L., Takami, H., Yamada, Y. & Settleman, J. (2002) Dev. Cell 2, 553-565. [DOI] [PubMed] [Google Scholar]
- 26.Sordella, R., Jiang, W., Chen, G. C., Curto, M. & Settleman, J. (2003) Cell 113, 147-158. [DOI] [PubMed] [Google Scholar]
- 27.Hall, A. (1998) Science 279, 509-514. [DOI] [PubMed] [Google Scholar]
- 28.Burridge, K. & Chrzanowska-Wodnicka, M. (1996) Annu. Rev. Cell Dev. Biol. 12, 463-518. [DOI] [PubMed] [Google Scholar]
- 29.Connolly, J. O., Simpson, N., Hewlett, L. & Hall, A. (2002) Mol. Biol. Cell 13, 2474-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.