Abstract
Organization of proteins into complexes is crucial for many cellular functions. However, most proteomic approaches primarily detect protein interactions for soluble proteins but are less suitable for membrane-associated complexes. Here we describe a mating-based split ubiquitin system (mbSUS) for systematic identification of interactions between membrane proteins as well as between membrane and soluble proteins. mbSUS allows in vivo cloning of PCR products into a vector set, detection of interactions via mating, regulated expression of baits, and improved selection of interacting proteins. Cloning is simplified by introduction of λ attachment sites for GATEWAY. Homo- and heteromeric interactions between Arabidopsis K+ channels KAT1, AKT1, and AKT2 were identified. Tests with deletion mutants demonstrate that the C terminus of KAT1 and AKT1 is necessary for physical assembly of complexes. Screening of a sorted collection of 84 plant proteins with K+ channels as bait revealed differences in oligomerization between KAT1, AKT1, and AtKC1, and allowed detection of putative interacting partners of KAT1 and AtKC1. These results show that mbSUS is suited for systematic analysis of membrane protein interactions.
Keywords: split ubiquitin, proteomics, KAT1, Arabidopsis, GATEWAY
An important part of proteomics is associomics, i.e., the systematic analysis of protein interactions, which may be important for the regulation of cellular processes (1). Exploring protein interactions not only extends information about known proteins but can also help to identify functions of unknown proteins. The yeast two-hybrid system was successfully adapted to study the yeast proteome by in vivo cloning of the ORFeome and subsequent testing of interactions via the mating approach (2).
To study membrane protein interactions in yeast, the split ubiquitin system (SUS) was developed (3). SUS is based on the ability of the N-terminal “Nub” and C-terminal “Cub” ubiquitin halves to reconstitute a functional protein. NubG is a mutant with severely decreased affinity for Cub; functional ubiquitin can only be reconstituted when NubG and Cub are in close vicinity by fusion with proteins that interact. Functional ubiquitin is then recognized by ubiquitin-specific proteases (USPs). In the system developed by Stagljar et al. (4), USPs release an artificial transcription factor, protein A-LexA-VP16 (PLV), at the C terminus of Cub, leading to activation of lexA-driven reporter genes HIS3 and lacZ in the nucleus. This selectable SUS was successfully applied to detect pairs of interactions among membrane proteins from yeast (4), plants (5), and animals (6). However, this system is not suitable for systematic analysis of large protein collections as performed with the classic yeast two-hybrid system (2).
As a prerequisite for the development of a tool suitable for systematic analysis of membrane protein interactions, it is important to choose well characterized membrane proteins as controls. KAT1, AKT1, and AtKC1 are α-subunits of shaker-like K+ channels from Arabidopsis and belong to the best-characterized membrane proteins in plants. Similar to their homologues from animals, they consist of six membrane-spanning domains with a pore-forming region between transmembrane domains 5 and 6. The functional pore is formed within tetrameric complexes formed by four subunits of similar structure (7). Analyses using the classical yeast two-hybrid system as well as functional studies in Xenopus oocytes demonstrated that the cytosolic C terminus of plant K+ channels is important for complex assembly (8, 9). KAT1 plays a role in K+ influx in guard cells during stomatal opening, whereas AKT1 and AtKC1 are important for K+ uptake from soil (10, 11). Recently, heteromers between AtKC1 and other K+ channels were detected, which may potentially be relevant for regulation of in planta activity (12).
To enable systematic analyses of membrane protein interactions a mating-based SUS (mbSUS) was developed. Interaction analysis of a set of Arabidopsis membrane proteins revealed physical interactions between KAT1, AKT1, and AKT2. Screening of a sorted cDNA collection of 84 proteins revealed differences regarding the oligomerization of KAT1, AKT1, and AtKC1 and detected the putative interacting partners of KAT1 and AtKC1. Because the vectors are compatible with GATEWAY cloning (13), mbSUS can be used for large-scale studies of membrane and membrane-associated proteins.
Materials and Methods
Strains and Vectors. Construction of yeast strains for the mating approach THY.AP4 [MATa ura3 leu2 lexA::lacZ::trp1 lexA::HIS3 lexA::ADE2] and THY.AP5 [MATα URA3 leu2 trp1 his3 loxP::ade2] and of pSUgate vectors pMetYCgate, pNXgate, pXN-gate, and pNubWT-2 is described in Supporting Text, which is published as supporting information on the PNAS web site.
Cloning into pSUgate Vectors and Interaction Screens. Arabidopsis ORFs (Table 1, which is published as supporting information on the PNAS web site) were amplified from the first-strand DNA with TripleMaster DNA polymerase (Eppendorf) by using gene-specific primers (acaagtttgtacaaaaaagcaggctctccaaccaccATGX19–25-5′ ORF and tccgccaccaccaaccactttgtacaagaaagctgggtaX19–25-3′ strand ORF without stop). Purified PCR products were cloned in vivo (14). For NubG fusions, pNXgate and pXNgate were cleaved with EcoRI/SmaI and used together with PCR products to transform THY.AP5. Transformants were selected on synthetic complete (SC) media lacking tryptophan (T) and uracil (U). For CubPLV fusions, pMetYCgate was cleaved with PstI/HindIII and used together with PCR products to transform THY.AP4. Transformants were selected on SC lacking leucine (L). Several clones from each THY.AP5 and THY.AP4 transformation were incubated in appropriate SC media with and without G418. Stationary cultures without G418 were harvested, and plasmids were isolated and amplified in Escherichia coli. DNA sequence of constructs was determined, and constructs were used for interaction assays. For interaction screening full-length ORFs and partial cDNAs KAT1-ΔR5-6 (KAT1 from 1–1,506) and AKT1ΔR5-6 (AKT1 from 1–1,488) were cloned into pSUgate vectors as described above. Thirty clones from each THY.AP5 and THY.AP4 transformation were mixed, and these pools were incubated in appropriate SC media with and without G418. Stationary cultures without G418 were used for subsequent interaction assays.
Interaction Assays. Stationary cultures (see above) were harvested and resuspended in yeast extract/peptone/dextrose (YPD). The THY.AP4 (Cub) and THY.AP5 (Nub) suspensions were mixed and plated on YPD. After 6–8 h at 28°C, cells were selected for diploids by replica plating on SC media -TUL, and incubated at 28°C for 2–3 days. For growth assays, diploid cells were either replica-plated or streaked out on synthetic dextrose minimal media (SD) with different concentrations of methionine (Met) as indicated. Growth was monitored for 2–9 days. For β-galactosidase assays and Western blot, see supporting information.
Construction of a Sorted cDNA Collection and 96-Well Array Screening. Construction of a sorted NubG collection, the list of selected Arabidopsis ORFs, and the screening procedure in 96-well plate format, as well as a table summarizing all results, are described in the supporting information.
Results
Yeast Strains THY.AP4 and THY.AP5. To improve selection of interacting proteins and to enable mating-based large-scale screening, two yeast strains were constructed. The reporter yeast strain THY.AP4 contains reporter genes HIS3, ADE2, and lacZ, all under control of lexA-driven promoters. Compared with the SUS-reporter strain L40 (4), THY.AP4 grows significantly faster and contains the additional lexA::ADE2 reporter. THY.AP4 permits detection of interacting proteins by (i) selection on media lacking adenine and histidine, (ii) monitoring β-galactosidase activity of lacZ with 5-bromo-4-chloro-3-indolyl-β-d-galactosidase or o-nitrophenylglucoside as substrates, and (iii) monitoring ADE2 enzymatic activity by visual analysis of accumulation of red pigment (15). THY.AP5 was designed for transformation with Nub fusions and testing for interactions between Nub and Cub (in THY.AP4) fusions by mating.
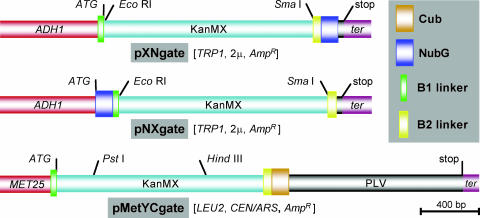
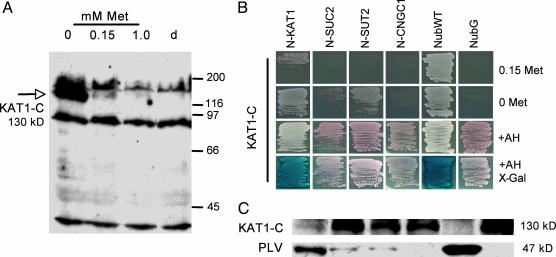
Linkers and Vectors. Linkers B1 and B2 were fused into pSUgate plasmids (Fig. 1). B1 and B2 contain attB1 and attB2 sites of the GATEWAY cloning system (13) and allow in vivo cloning (14) of a single PCR product of each ORF into the different pSUgate vectors (Fig. 7, which is published as supporting information on the PNAS web site). Because efficiency of in vivo cloning depends on efficient cleavage of vectors, a cassette containing the kanamycin resistance gene and a set of two noncompatible restriction sites were inserted between the linkers, permitting counterselection on G418. pNXgate and pXNgate are multiple-copy plasmids containing TRP1 for selection in yeast, suitable for NubG-X and X-NubG fusions of prey polypeptides X, respectively (Fig. 1). pMetYCgate is a low-copy plasmid with the selection marker LEU2, comprised of the Met-repressible MET25 promoter, B1-KanMX-B2 cassette, and CubPLV. pMetYCgate is suited for Y-CubPLV fusions of bait peptides Y. To verify that the MET25 promoter can be used to modulate bait levels, Arabidopsis KAT1 was cloned into pMetYCgate. Regulation of MET25-KAT1-CubPLV by Met was analyzed with a PLV-specific antibody (Fig. 2A). The antibody detected an ≈130-kDa polypeptide corresponding approximately to the sum of the calculated molecular masses of KAT1 (78 kDa), B2-Cub (6.4 kDa), and PLV (47 kDa). KAT1-CubPLV levels were lower at 0.15 mM Met and repressed at higher concentrations.
Fig. 1.
Expression cassettes of pSUgate vectors. pNXgate and pXNgate vectors are suitable for “NubG-X” and “X-NubG” fusions of prey polypeptides “X,” and pMetYCgate is suited for “Y-CubPLV” fusions of bait peptides “Y.” B1-KanMX-B2 is identical in all vectors. Promoters are red, terminators are purple; NubG, Cub, and the linkers are shown on the right. Marked restriction sites are used to produce linear vectors for in vivo cloning. “ATG” and “stop” mark the start and the stop codons in the expression cassettes, respectively. AmpR refers to ampicillin resistance cassette. 2μ refers to a high-copy, whereas CEN/ARS refers to a low-copy, yeast origin of replication. The scale gives length in base pairs.
Fig. 2.
Interaction of KAT1-CubPLV with different NubG-X fusions (A, adenine; H, histidine; N, NubG; C, CubPLV). (A) Expression of met25-KAT1-CubPLV is regulated by Met. Diploid cells carrying KAT1-CubPLV and pNXgate plasmid were grown on synthetic complete -TLU (tryptophan, leucine, uracil) media and different concentrations of Met. Western analysis of cell extracts with anti-PLV antibody was performed; lane d indicates diploids with empty pMetYCgate and pNXgate. Size of proteins is given in kDa (130 kDa corresponds to KAT1-CubPLV fusion; other polypeptides are unspecific because they are present in control). (B) Growth on minimal medium (lacking amino acids) reporting KAT1-CubPLV interactions. Interactions were selected on media with and without 0.15 mM Met. +AH shows the growth of diploids on nonselective minimal medium. X-Gal shows detection of lacZ activity with 5-bromo-4-chloro-3-indolyl β-d-galactosidase (X-Gal) as a substrate. Soluble NubWT and NubG were positive and negative controls, respectively. (C) Western analysis of interaction-dependent PLV cleavage with anti-PLV antibody. Extracts were prepared from diploid cells grown on minimal medium +AH. Order of lanes corresponds to B.
Detection of KAT1 Homomers. mbSUS was tested by analyzing interactions between the K+ channel KAT1 and a set of Arabidopsis prey: KAT1, the putative cyclic nucleotide-gated channel CNGC1, and the sucrose transporters SUC2 and SUT2. pNX-gate and pNubWT-2, vectors expressing soluble NubG and wild-type Nub (NubWT), were used as controls. Growth of diploid cells under selective conditions revealed interaction of KAT1-CubPLV with NubG-KAT1 and NubWT on 0.15 mM Met (Fig. 2B). In the absence of Met, cells coexpressing KAT1-CubPLV with NubG-KAT1 or NubWT grew faster than cells coexpressing KAT1-CubPLV with the sucrose transporters. Interaction of KAT1 with itself was confirmed by ADE2 and lacZ assays. Interactions were further verified with anti-PLV antibodies. Significant levels of the released PLV (47 kDa) were detected only in cells expressing KAT1-CubPLV with NubG-KAT1 or NubWT (Fig. 2C). The increased amount of released PLV correlated with a decrease of KAT1-CubPLV polypeptide.
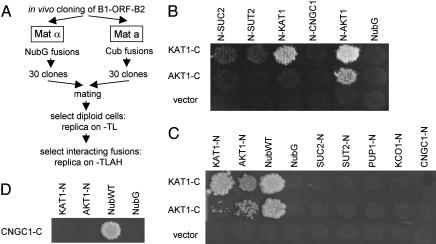
Screens Reveal Specific Heteromerization of KAT1 with AKT1. To test the suitability of mbSUS for systematic approaches, screens using K+ channels KAT1 and AKT1 as baits and SUC2, SUT2, KAT1, CNGC1, AKT1, vacuolar K+ channel KCO1, and purine transporter PUP1 as prey were carried out. For this purpose, THY.AP4 was cotransformed with linear pMetYC-gate and bait PCR products, and THY.AP5 was cotransformed with linear pNXgate or pXNgate and prey PCR products. To avoid negative effects of PCR-induced mutations (16), 30 clones from THY.AP4 and THY.AP5 transformations were pooled. The respective pools were mated, and diploids were selected under prototrophic conditions (Fig. 3A). Diploids were replica-plated, and interactions were monitored regarding HIS3 and ADE2 reporters by incubating plates at 28°C and analyzing growth after different incubation periods. The NubG-X screen revealed interactions of KAT1-CubPLV with NubG-KAT1 and NubG-AKT1, and of AKT1-CubPLV with NubG-AKT1 after 2 days of incubation (Fig. 3B). Interaction of AKT1-CubPLV with NubG-KAT1 was observed after 6 days of incubation (Fig. 8, which is published as supporting information on the PNAS web site). Screens with the inverse X-NubG fusions confirmed homo- and heteromeric interactions for KAT1-CubPLV with KAT1-NubG and AKT1-NubG, and of AKT1-CubPLV with AKT1-NubG and KAT1-NubG (Fig. 3C). Both CubPLV fusions interacted with soluble NubWT, but not with soluble NubG. In general, X-NubG interactions seemed weak compared with NubG-X interactions. Therefore, longer incubation times were needed (2 versus 9 days for comparable growth). Neither KAT1 nor AKT1 showed significant interaction with KCO1, PUP1, or with SUC2 and SUT2, which was consistent with previous tests using the original SUS (5). Moreover, KAT1 and AKT1 did not interact with the structurally closely related shaker-like CNGC1. To verify that CNGC1 was expressed, THY.AP4 was cotransformed with pMetYCgate and a CNGC1 PCR product. After mating with yeast expressing Nub baits, CNGC1-CubPLV interacted with NubWT, but not with NubG, KAT1-NubG, or AKT1-NubG (Fig. 3D).
Fig. 3.
Screening reveals oligomerization of plant K+ channels KAT1 and AKT1. (A) Flow chart of the applied screening strategy (abbreviations are as in Fig. 2). (B and C) Interaction screen with NubG-X and X-NubG fusion proteins. Replica plates showing diploid cells selected on -TLAH media without Met after 2 (NubG-X) and 9 (X-NubG) days of incubation. NubWT and NubG were positive and negative controls, respectively. (D) Results obtained with CNGC1-CubPLV as bait.
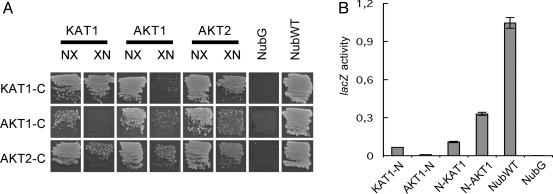
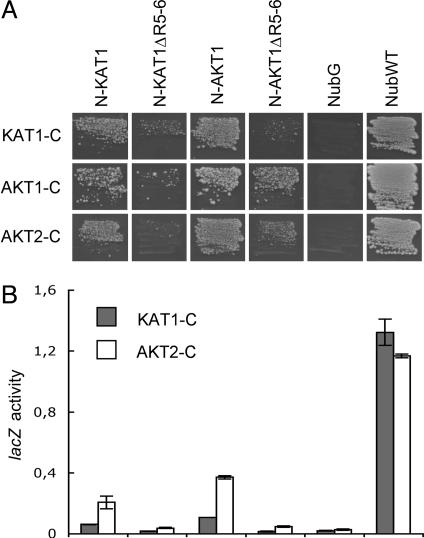
Homo- and Heteromers of K+ Channels. Heterooligomers of KAT1/AKT1 were detected by using mbSUS; however, the existence of KAT1/AKT1 heteromers in plants is controversial (17, 18). To study heteromerization in more detail, a second shaker-like K+ channel AKT2 known to interact with both AKT1 and KAT1 was tested (12, 17). The screening approach involves replica plating, which seems to lead to increased background growth. To increase signal-to-noise ratio, the following assays were performed by using a screening protocol in which cells are directly plated on minimal medium. KAT1-CubPLV and AKT2-CubPLV interacted with NubG fusions of KAT1, AKT1, and AKT2 (Fig. 4A). AKT1-CubPLV interacted with NubG fusions of AKT1 and AKT2, and with NubG-KAT1; however, no significant interaction was detected with KAT1-NubG. The inability to detect interactions between KAT1-NubG and AKT1-CubPLV may reflect low expression levels of AKT1-CubPLV (Fig. 9, which is published as supporting information on the PNAS web site), which translate into reduced growth rates of cells coexpressing AKT1-CubPLV bait and prey compared to cells expressing KAT1-CubPLV and AKT2-CubPLV bait and prey, respectively (6 versus 4 days of incubation for comparable colony sizes, Fig. 4A). Interactions between KAT1 and AKT1 were further analyzed by quantitative lacZ assays (Fig. 4B). LacZ activity was higher in cells coexpressing KAT1-CubPLV with NubG-AKT1 as compared with cells coexpressing KAT1-CubPLV with NubG-KAT1 and KAT1-NubG, suggesting that the interaction of KAT1-CubPLV with NubG-AKT1 may be stronger. Levels of AKT1-CubPLV interactions were too low for quantitation.
Fig. 4.
Interactions among different KAT1, AKT1, and AKT2 fusions. (A) Diploid cells on minimal media after 4 (KAT1-C and AKT2-C) and 6 (AKT1-C) days. NX are NubG-X fusions, and XN are X-NubG fusions. Abbreviations are as in Fig. 2. (B) Liquid β-galactosidase assay using o-nitrophenylglucoside (ONPG) as a substrate. LacZ activity is given in μmol ONPG × min-1 × mg of protein-1. For extracts, cells were grown on -TLAH media without Met. The experiment was repeated twice independently with comparable results.
Involvement of the C Terminus in K+ Channel Interactions. C termini of plant shaker-like K+ channels are known to interact and thought to contribute to channel oligomerization (9, 12). To analyze the role of the C termini, KAT1-ΔR5-6 and AKT1ΔR5-6 deletion mutants lacking the C-terminal domains R5 and R6 were fused to NubG and tested against KAT1-, AKT1-, and AKT2-CubPLV fusions (19). The interaction-dependent growth of diploids expressing NubG-KAT1-ΔR5-6 and NubG-AKT1ΔR5-6 was significantly reduced when compared with corresponding cells expressing NubG-KAT1 and NubG-AKT1 (Fig. 5A). AKT1ΔR5-6-CubPLV and KAT1-ΔR5-6-CubPLV interacted with NubWT, demonstrating that the deletion mutants are expressed (data not shown). NubG fusions of deletion mutants were further analyzed by quantitative lacZ assays. Consistent with growth assays, NubG-KAT1-ΔR5-6 and NubG-AKT1ΔR5-6 revealed significantly lower lacZ activities with KAT1- and AKT2-CubPLV when compared with NubG-KAT1 and NubG-AKT1 (Fig. 5B). Interactions with AKT1-CubPLV were too weak to allow quantitation.
Fig. 5.
Role of C-terminal R5 and R6 domains in AKT1 and KAT1 oligomerization. (A) Diploid cells on 0.15 mM Met after 3 days (KAT1-C and AKT2-C) and without Met after 5 days (AKT1-C). Abbreviations are as in Fig. 2. (B) Liquid β-galactosidase assays were performed as in Fig. 4B. Order of lanes corresponds to A.
Screening of a Sorted X-NubG Collection. It was important to verify that mbSUS is indeed suitable for large-scale approaches, because technical problems such as a high background/signal ratio may appear only when testing large numbers in parallel. A sorted collection of 84 Arabidopsis ORF-NubG fusions was constructed in THY.AP5 as prey and screened with nine ORF-CubPLV fusions in THY.AP4 as bait. To allow high-throughput screening, a liquid–media interaction assay in 96-well format was established. Collection construction, a complete list of ORFs, and the screening procedure are described in Table 1.
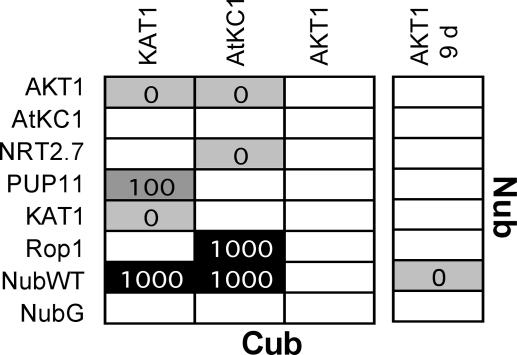
The collection was screened with CubPLV fusions of K+ channel KAT1, AKT1, AtKC1 and six nonrelated transporters as controls (Table 2, which is published as supporting information on the PNAS web site). Screenings were repeated at least twice, and interactions detected in both experiments were scored (2). Fifteen interactions were detected by testing 84 × 9 individual pairs on media containing 0, 0.1, 0.4, and 1 mM Met (Fig. 6 and Table 2). In general, 5 days of incubation were sufficient to detect interactions of CubPLV bait with NubWT or X-NubG prey. After longer incubation, no additional interactions were observed (Fig. 10, which is published as supporting information on the PNAS web site). However, longer incubation times were required for detection of AKT1-CubPLV interactions, consistent with the finding that cells expressing AKT1-CubPLV bait with prey have reduced growth rates because of lower AKT1-CubPLV abundance (Figs. 4, 5, and 9). KAT1-CubPLV bait interacted with AKT1 and KAT1 but not with AtKC1 prey. AtKC1-CubPLV interacted with AKT1 but not with KAT1 and AtKC1. The interactions were specific because neither AKT1 nor KAT1 prey interacted with other CubPLV fusions (Table 2). AKT1-CubPLV interacted with NubWT, but not with X-NubG fusions, a finding different from the previous small-scale interaction screens (Fig. 3C). The inability to detect AKT1-CubPLV interactions with AKT1-NubG and KAT1-NubG suggests that the large-scale procedure may be less sensitive than the small-scale X-NubG tests. In addition to interactions between the K+ channels, other interacting partners of KAT1 and AtKC1 were detected: a purine transporter homolog PUP11 interacted with KAT1-CubPLV, whereas AtKC1-CubPLV interacted with a nitrate transporter homolog NRT2.7 and with a GTPase Rop1 (20–22). The interactions were specific, because neither PUP11 nor NRT2.7 and Rop1 prey interacted with other tested CubPLV fusions (Table 2).
Fig. 6.
Interactions of K+ channels detected by screening of a sorted X-NubG collection. Diploids with interacting X-NubG and CubPLV fusions were selected in liquid media with different concentrations of Met. Growth was monitored after 5 days or, for “AKT1 9 d,” after 9 days. The shade of gray and the numbers give maximum in Met concentration (in μM), in which the corresponding diploids were grown.
Discussion
Evidence is accumulating that oligomerization of membrane proteins is the rule rather than the exception (23). However, because of the difficult biochemistry of membrane proteins, progress in the systematic analysis of interactions is slow. To facilitate rapid and systematic membrane protein interaction screens, an improved split ubiquitin system (mbSUS) was established. One-by-one tests, as well as screening of a sorted Arabidopsis collection, revealed homo- and heteromeric interactions between K+ channels and allowed detection of previously undetected putative of K+ channel interactors. Together with the detection of AMT1 ammonium transport oligomers (32), these results demonstrate that mbSUS is suitable for systematic, large-scale analysis of protein interactions at membranes, such as had been carried out for soluble proteins (2).
Advantages of Using a Regulated Promoter in mbSUS. Efficient detection of specific interactions depends on optimal expression levels of the two fusion proteins. Highly expressed CubPLV bait fusions may accumulate in intracellular membranes (24), leading to nonspecific interaction with Nub fusions, whereas low expression of bait may prevent detection of weak interactions. The use of a regulated promoter, i.e., MET25, provides tight control of CubPLV fusion levels and allows comparison of results with different stringency. In mbSUS one-by-one tests as well as in screens, Met suppressed the expression of KAT1-CubPLV and helped to distinguish between specific and nonspecific NubG-X interactions (Fig. 2 and Fig. 11, which is published as supporting information on the PNAS web site). Thus, Met-controlled expression of bait allows screening for NubG-X interactions under different conditions, helps to distinguish between specific and unspecific interactions and to eliminate false positives. By using different Cub and Nub fusions, it had been demonstrated that Cub has higher affinity to NubG fused to the N termini of proteins (NubG-X) when compared with the corresponding C-terminal X-NubG fusion peptides (25). Consistent with this finding, mbSUS screens of X-NubG fusions did not reveal unspecific growth on interaction-selective media lacking Met even after extended incubation times (Figs. 3C, 10, and 11). Thus, Met-controlled expression of bait helps to distinguish between specific and unspecific interactions in NubG-X screens, but may not be necessary for X-NubG screens in future systematic studies.
Interactions Among K+ Channels. Shaker-like K+ channels can only function as tetramers because each of the four monomers contributes to the single pore formed. K+ channels may be formed by homooligomers; however, a wide spectrum of properties can be generated by heterooligomerization. The Arabidopsis genome contains nine shaker-like K+ channels, three of which were analyzed with respect to their potential to form oligomers: KAT1, AKT1, and AKT2. mbSUS demonstrates that KAT1 and AKT1 interact with themselves, with each other, and with AKT2. Detection of KAT1 and AKT1 homomers and of KAT1/AKT2 and AKT1/AKT2 heteromers is in agreement with previous reports, but contradictory data exist regarding KAT1/AKT1 complexes (12, 17, 18, 26). No AKT1/KAT1 heteromers were detected in insect cell extracts, and C termini of AKT1 and KAT1 did not interact in yeast two-hybrid assays (17). In contrast, the fact that KAT1 and AKT1 interact in Xenopus oocytes and that both KAT1 and AKT1 are expressed in stomata suggests the existence of KAT1/AKT1 heteromers (18, 26). In mbSUS, KAT1-CubPLV and AKT1-CubPLV did not interact with NubG fusions of unrelated plasma membrane proteins such as sucrose transporters, nor with the structurally but not functionally related putative cyclic nucleotide gated channel CNGC1 and the structurally unrelated vacuolar K+ channel KCO1, supporting the notion that interactions between KAT1 and AKT1 fusions were specific (5, 20, 27, 28). Thus, mbSUS provides further evidence for the existence of KAT1/AKT1 complexes.
Yeast two-hybrid studies had suggested that the C-terminal domains R5 and R6 in AKT1 are important for K+ channel assembly (9, 19). Functional analysis of deletion mutants revealed that R5 and R6 are involved in heteromeric assembly of the AKT1 and KAT1 homologues SKT1 and KST1 from potato (8). mbSUS demonstrates that R5 and R6 domains are essential for hetero- as well as homomeric interactions between intact AKT1 and KAT1. Complex assembly may be mediated via physical interaction of two or more R5–R6 regions or via interaction of R5–R6 with other domains, e.g., the putative cyclic nucleotide-binding domain localized between the sixth transmembrane region and the R5 of K+ channels, as had been suggested for AKT1 (9). Further mbSUS experiments may clarify the importance of different regions in physical formation of K+ channel complexes.
K+ Channel Interactions Detected with the Sorted X-NubG Collection. Yeast two-hybrid studies suggest that AtKC1 interacts with AKT1 and with itself, but not with KAT1 (12). mbSUS screens support these observations and show that AtKC1 cannot form homomers in this system. The absence of AtKC1 homomers suggests that AtKC1 is a modulator subunit forming K+ channel heteromers with altered properties with other functional K+ channel subunits (11, 29). Further mbSUS screening may help to elucidate the significance of differential oligomerization within the shaker-like K+ channel family.
Besides oligomers of K+ channel subunits, previously undescribed putative interactions were detected. Although the significance of the interaction between KAT1 and the purine transporter homolog PUP11 is unclear (20), interactions of AtKC1 with NRT2.7 and Rop1 may relate to more obvious physiological functions. K+ is known to increase nitrate (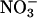 ) uptake from soil, probably by stimulating metabolic activity and ATP production (30). The interaction of AtKC1 with the nitrate transporter homolog NRT2.7 may indicate a more direct coupling. Because NRT2.7 is mainly expressed in leaves, whereas AtKC1 is primarily expressed in roots and only partially in leaves (21), it will be interesting to test whether root-expressed nitrate transporters can also interact with AtKC1 or other K+ channels.
) uptake from soil, probably by stimulating metabolic activity and ATP production (30). The interaction of AtKC1 with the nitrate transporter homolog NRT2.7 may indicate a more direct coupling. Because NRT2.7 is mainly expressed in leaves, whereas AtKC1 is primarily expressed in roots and only partially in leaves (21), it will be interesting to test whether root-expressed nitrate transporters can also interact with AtKC1 or other K+ channels.
Rop proteins are plant orthologues of animal RHO GTPases, involved in regulation of cell morphology and polarity via actin filament organization (22). Interestingly, actin filament reorganization affects K+ channel activities in stomata (31). Members of the Rop family are highly similar but serve different functions: Rop2 is involved in root hair elongation whereas Rop1 regulates pollen tip growth (22). Because AKT1 and AtKC1 are expressed in root hairs, and because AtKC1 interacts with AKT1 and Rop1, it will be interesting to see whether other Rops may also interact with K+ channels and whether similar interactions exist in planta and how they may affect channel function.
Conclusions
mbSUS provides a feasible, easy-to-use, and cost-efficient cloning strategy for large-scale interaction analysis of proteins at membranes. The addition of the third reporter ADE2 and the introduction of the stringently regulated MET25 promoter significantly improve the selection of interacting proteins. The vectors and yeast strains enable in vivo cloning and mating-based analysis of large protein sets. A common pair of linkers in the vectors allows constructing fusions with a single PCR product, further reducing costs of large-scale approaches. The attB1 and attB2 sites in the linkers make mbSUS compatible with GATEWAY cloning (13), so that either the B1/B2-flanked PCRs or the mbSUS fusions can be used to construct different DNA fusions. Thus, the system can serve as an entry point for highly versatile analysis of membrane proteins, as shown for K+ channels in the first mbSUS tests.
Supplementary Material
Acknowledgments
We thank Jens Riexinger for technical support, Daniel Van Belle for designing primers, and H. P. Schmitz (University of Basel, Basel) for the vector LexAox5LacZ. We are grateful for financial support of the consortium by the European Union Project ASSOCIOPORT (QLG2-CT-2001-01297).
Abbreviations: SUS, split ubiquitin system; mbSUS, mating-based SUS; SC, synthetic complete; Met, methionine; PLV, protein A-LexA-VP16.
References
- 1.Phizicky, E., Bastiaens, P. I., Zhu, H., Snyder, M. & Fields, S. (2003) Nature 422, 208-215. [DOI] [PubMed] [Google Scholar]
- 2.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochrat, P., et al. (2000) Nature 403, 623-627. [DOI] [PubMed] [Google Scholar]
- 3.Johnsson, N. & Varshavsky, A. (1994) Proc. Natl. Acad. Sci. USA 91, 10340-10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stagljar, I., Korostensky, C., Johnsson, N. & te Heesen, S. (1998) Proc. Natl. Acad. Sci. USA 95, 5187-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinders, A., Schulze, W., Kühn, C., Barker, L., Schulz, A., Ward, J. M. & Frommer, W. B. (2002) Plant Cell 14, 1567-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, B., Nguyen, M., Breckenridge, D. G., Stojanovic, M., Clemons, P. A., Kuppig, S. & Shore, G. C. (2003) J. Biol. Chem. 278, 14461-14468. [DOI] [PubMed] [Google Scholar]
- 7.Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., Talke, I. N., Amtmann, A., Maathuis, F. J. M., Sanders, D., et al. (2001) Plant Phys. 126, 1646-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmermann, S., Hartje, S., Ehrhardt, T., Plesch, G. & Müller-Röber, B. (2001) Plant J. 28, 517-527. [DOI] [PubMed] [Google Scholar]
- 9.Daram, P., Urbach, S., Gaymard, F., Sentenac, H. & Chérel, I. (1997) EMBO J. 16, 3455-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak, J. M., Murata, Y., Baizabal-Aguirre, V. M., Merrill, J., Wang, M., Kemper, A., Hawke, S. D., Tallman, G. & Schroeder, J. I. (2001) Plant Phys. 127, 473-485. [PMC free article] [PubMed] [Google Scholar]
- 11.Reintanz, B., Szyroki, A., Ivashikina, N., Ache, P., Godde, M., Becker, D., Palme, K. & Hedrich, R. (2002) Proc. Natl. Acad. Sci. USA 99, 4079-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilot, G., Gaymard, F., Mouline, K., Chérel, I. & Sentenac, H. (2003) Plant Mol. Biol. 51, 773-787. [DOI] [PubMed] [Google Scholar]
- 13.Hartley, J. L., Temple, G. F. & Brasch, M. A. (2000) Genome Res. 10, 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fusco, C., Guidotti, E. & Zervos, A. S. (1999) Yeast 15, 715-720. [DOI] [PubMed] [Google Scholar]
- 15.Ugolini, S. & Bruschi, C. V. (1996) Curr. Genet. 30, 485-492. [DOI] [PubMed] [Google Scholar]
- 16.Hudson, J. R., Jr., Dawson, E. P., Rushing, K. L., Jackson, C. H., Lockshon, D., Conover, D., Lanciault, C., Harris, J. R., Simmons, S. J., Rothstein, R., et al. (1997) Genome Res. 7, 1169-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urbach, S., Chérel, I., Sentenac, H. & Gaymard, F. (2000) Plant J. 23, 527-538. [DOI] [PubMed] [Google Scholar]
- 18.Dreyer, I., Antunes, S., Hoshi, T., Müller-Röber, B., Palme, K., Pongos, O., Reintanz, B. & Hedrich, R. (1997) Biophys. J. 72, 2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilot, G., Pratelli, R., Gaymard, F., Meyer, Y. & Sentenac, H. (2003) J. Mol. Evol. 56, 418-434. [DOI] [PubMed] [Google Scholar]
- 20.Bürkle, L., Cedzich, A., Döpke, C., Stransky, H., Okumoto, S., Gillissen, B., Kühn, C. & Frommer, W. B. (2003) Plant J. 34, 13-26. [DOI] [PubMed] [Google Scholar]
- 21.Orsel, M., Krapp, A. & Daniel-Vedele, F. (2002) Plant Phys. 129, 886-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valster, A. H., Hepler, P. K. & Chernoff, J. (2000) Trends Cell Biol. 10, 141-146. [DOI] [PubMed] [Google Scholar]
- 23.Veenhoff, L. M., Heuberger, E. H. & Poolman, B. (2002) Trends Biochem. Sci. 27, 242-249. [DOI] [PubMed] [Google Scholar]
- 24.Supply, P., Wach, A., Thinès-Sempoux, D. & Goffeau, A. (1993) J. Biol. Chem. 268, 19744-19752. [PubMed] [Google Scholar]
- 25.Raquet, X., Eckert, J. H., Müller, S. & Johnsson, N. (2001) J. Mol. Biol. 305, 927-938. [DOI] [PubMed] [Google Scholar]
- 26.Szyroki, A., Ivashikina, N., Dietrich, P., Roelfsema, M. R. G., Ache, P., Reintanz, B., Deeken, R., Godde, M., Felle, H., Steinmeyer, R., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 2917-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czempinski, K., Frachisse, J. M., Maurel, C., Barbier-Brygoo, H. & Mueller-Roeber, B. (2002) Plant J. 29, 809-820. [DOI] [PubMed] [Google Scholar]
- 28.Talke, I. N., Blaudez, D., Maathuis, F. J. M. & Sanders, D. (2003) Trends Plant Sci. 8, 286-293. [DOI] [PubMed] [Google Scholar]
- 29.Véry, A.-A. & Sentenac, H. (2003) Ann. Rev. Plant Biol. 54, 575-603. [DOI] [PubMed] [Google Scholar]
- 30.Wyn Jones, R. G. & Pollard, A. (1983) in Encyclopedia of Plant Physiology, eds. Pirson, A. & Zimmermann, M. H. (Springer, New York), Vol. 15B, pp. 528-562. [Google Scholar]
- 31.Hwang, J.-U., Suh, S., Yi, H., Kim, J. & Lee, Y. (1997) Plant Phys. 115, 335-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludewig, U., Wilken, S., Wu, B.-H., Jost, W., Obralik, P., El-Bakkouri, M., Marini, A.-M., André, B., Hamacher, T., Boles, E., et al. (2003) J. Biol. Chem. 278, 45603-45610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.