Abstract
Gene disruption and overexpression play central roles in the analysis of gene function. Homologous recombination is, in principle, the most efficient method of disrupting, modifying, or replacing a target gene. Although homologous integration of exogenous DNA into the genome occurs readily in Saccharomyces cerevisiae, it is rare in many other organisms. We identified and disrupted Neurospora crassa genes homologous to human KU70 and KU80, which encode proteins that function in nonhomologous end-joining of double-stranded DNA breaks. The resulting mutants, named mus-51 and mus-52, showed higher sensitivity to methyl methanesulfonate, ethyl methanesulfonate, and bleomycin than wild type, but not to UV, 4-nitroquinoline 1-oxide, camptothecin, or hydroxyurea. Vegetative growth, conidiation, and ascospore production in homozygous crosses were normal. The frequency of integration of exogenous DNA into homologous sequences of the genome in the KU disruption strains of N. crassa was compared with that in wild type, mei-3, and mus-11. In mei-3 and mus-11, which are defective in homologous recombination, none or few homologous integration events were observed under any conditions. When mtr target DNA with ≈2-kb 5′ and 3′ flanking regions was used for transformation of the KU disruption strains, 100% of transformants exhibited integration at the homologous site, compared to 10 to 30% for a wild-type recipient. Similar results were obtained when the ad-3A gene was targeted for disruption. These results indicate that KU disruption strains are efficient recipients for gene targeting.
Keywords: Ku70, Ku80, gene targeting, Neurospora crassa
In eukaryotes, two main recombination pathways have been identified, differing as to whether double-strand break (DSB) repair depends on DNA sequence homology. The first, homologous recombination (HR), involves interaction between homologous sequences, whereas the second, nonhomologous end-joining (NHEJ), involves direct ligation of the strand ends independent of DNA homology. Saccharomyces cerevisiae mainly uses an HR system of DSB repair, in which exogenous DNA fragments are integrated at homologous sites in the genome if the DNA has a short region of homology at both ends (1, 2). The RAD52 gene of S. cerevisiae is essential for this integration event (3). In contrast, many other organisms, including humans, plants, and insects, seem to use NHEJ preferentially in DSB repair. As a result, exogenous DNA can be integrated anywhere in the genome, even if it carries a long stretch of homologous sequence. The NHEJ process is mediated by the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), the Ku70–Ku80 heterodimer, and the DNA ligase IV-Xrcc4 complex (4, 5). The Ku protein was originally found as an autoantigen in patients with polymyositis-scleroderma overlap syndrome (6). Human Ku is a heterodimer of 69- and 83-kDa polypeptides, Ku70 and Ku80, which bind tightly to the DNA ends. This is a key step in NHEJ repair. Homologs of Ku70 and Ku80 have been identified in vertebrates, insects, the worm Caenorhabditis elegans, yeasts, and plants (7, 8). However, NHEJ repair has not been reported in Neurospora crassa.
The mei-3, mus-11, and mus-25 genes in N. crassa are homologs of S. cerevisiae RAD51, RAD52, and RAD54, which are specific for HR repair (9–11). The rate of homologous integration of plasmid pMTR (12) into the chromosomal mtr locus was assayed in the wild type and in mutants defective in these HR repair genes (11). In the wild type, 3–5% of transformants showed homologous integration, whereas <0.3% of transformants did so in mei-3 and mus-25 mutants. Because the homologous integration rate is so low in N. crassa, it is not easy to disrupt a particular gene by gene replacement. Repeat induced point mutation (RIP), which exploits the premeiotic inactivation of duplicated sequences (13), has been used as an alternative. Although this method is useful and effective, it is time consuming.
We simply hypothesized that the rate of homologous integration of exogenous DNA can be increased by blocking NHEJ function, and searched the Neurospora genome database (14) to find N. crassa genes homologous to human KU70 and KU80. We disrupted these genes by a PCR-based method of direct replacement (15) and constructed two strains defective in N. crassa KU70 and KU80 homologs. These mutants showed mild sensitivity to mutagens such as methyl methanesulfonate, ethyl methanesulfonate, and bleomycin. The Neurospora KU-70 and KU-80 homologs were therefore named mus-51 and mus-52, respectively. Targeting of two genes, mtr and ad-3A, on chromosomes IV and I, respectively, was conducted in these strains by a PCR-based method (16). In this study, exogenous DNA was introduced by electroporation (17, 18), which is more efficient and convenient than the spheroplast-polyethylene glycol (PEG) method (19, 20) used previously for transformation of Neurospora.
Materials and Methods
Strains and Plasmids. Table 1 shows N. crassa strains used in these experiments. C1-T10–37A and C1-T10–28a are wild-type strains closely related to the standard Oak Ridge wild type (21). Escherichia coli strains DH1 and XL-1 Blue were used for amplification of plasmids (22). Plasmids pBluescript SK+ (Stratagene) and pGEM (Promega) were generally used for construction of new vectors. Plasmids pBARGEM7–1 (23) and pCSN43 (24) and cosmids G7H3 and G8B12 were obtained from the Fungal Genetics Stock Center, University of Kansas Medical School (Kansas City).
Table 1. N. crassa strains used in this study.
| Strain | Genotype | Source/ref. |
|---|---|---|
| C1-T10–37A | A (wild type) | Laboratory stock |
| C1-T10–28a | a (wild type) | Laboratory stock |
| 74-OR256–14a | a al-2 pan-2 cot-1 | Laboratory stock |
| C-P-1 | a pan-1 arg-2 leu-2 | Laboratory stock |
| C20–1 | A ad-2 ser-1 | Laboratory stock |
| 54yo-728–5 | A mus-51::Hygr | This study |
| 54yo-728–7 | a mus-51::Hygr | This study |
| 54yo-828–3 | A mus-52::Hygr | This study |
| 54yo-828–4 | a mus-52::Hygr | This study |
| FGSC#2764 | A mei-3 | FGSC |
| FGSC#6409 | A mus-11 | FGSC |
| FGSC#8341 | A mus-23 al-2 pan-2 cot-1 | FGSC |
FGSC, Fungal Genetics Stock Center.
Genetic Studies in N. crassa. Genetic analysis was carried out as described by Davis and de Serres (25).
PCR Method. PCR amplification was carried out with the Expand High-Fidelity PCR system (Roche Diagnostics) following the manufacturer's protocol.
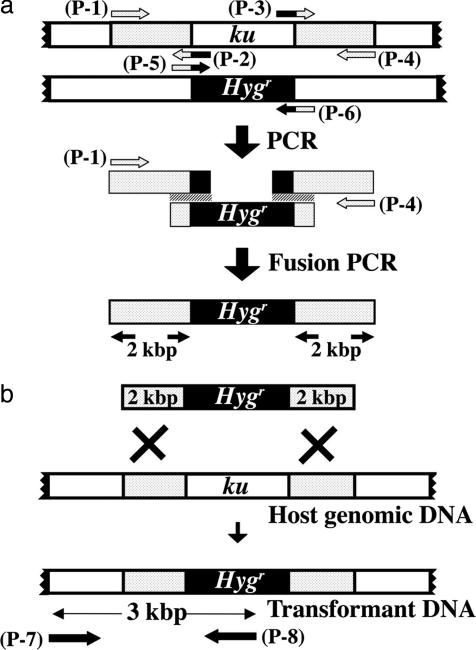
Construction of Plasmids for the Replacement of mus-51 and mus-52 by the Hygr Gene. The strategy for replacing the mus-51 and mus-52 genes by the hygromycin-resistance gene (Hygr) is presented in Fig. 1.
Fig. 1.
Strategy for replacing mus-51 and mus-52 by Hygr. (a) The construction of a ku-ortholog gene-targeting vector. The 5′ region of the ku-ortholog gene was amplified by using primers (p-1) and (p-2), and the 3′ region was amplified with primers (p-3) and (p-4). The size of both products was ≈2 kb. The Hygr gene was amplified with primers (p-5) and (p-6). These three PCR products were used as a template in fusion PCR using primers (p-1) and (p-4), and the fusion product was used for transformation. (b) Homologous integration of a fusion PCR product into a target gene. This product was introduced into recipient cells by electroporation, and homologous replacement of the ku-ortholog gene by the Hygr gene was confirmed by PCR using primers (p-7) and (p-8).
Preparation of DNA for replacement of mus-51. The flanking DNA (2 kb 5′ and 3′) of the mus-51 gene was amplified by PCR using cosmid G7H3 as a template. Primers for amplification of the 5′-flanking DNA were: (p-1) 5′-GTGCTGTAGCCGTTTTGGGTATCGC-3′ and (p-2) 5′-GGCGTAATAGCGAAGAGATAGTTGCTGGAAATAA-3′. Primers for the 3′-flanking DNA were: (p-3) 5′-AAGCATAAAGTGTAAAGGCTTGTTGATGACCGT-3′ and (p-4) 5′-TTGGACGCCGCACACCTCTCGCTCT-3′. The Hygr gene was amplified by PCR using plasmid pCSN43 as a template. The primers used were (p-5) 5′-TTATTTCCAGCAACTATCTCTTCGCTATTACGCC-3′ and (p-6) 5′-CACGGTCATCAACAAGCCTTTACACTTTATGCTT-3′. The three PCR-products were mixed and used as a template in a fusion PCR (15) that was carried out by using primers (p-1) and (p-4) under the following conditions: initial denaturation 94°C for 2 min, subsequent steps at 94°C for 15 sec, annealing at 60°C for 30 sec, extension at 68°C for 5 min, 10 cycles. Then, 94°C for 15 sec, 60°C for 30 sec, 68°C for 5 min (each cycle at 68°C 5 sec added), 20 cycles, final extension at 72°C for 7 min, hold at 4°C. The fusion PCR product was analyzed on an agarose gel (0.7%) and used for transformation of the N. crassa wild type.
DNA preparation for replacement of mus-52. Amplification of the 2 kb 5′ and 3′ flanking DNA of mus-52 was done by using cosmid G8B12 as a template. The method was as described above. Primers were: (p-1) 5′-GCGCCGGGAGGTTGTTCGTAAGCTG-3′,(p-2) 5′-GGCGTAATAGCGAAGAGGCTTTTCGGCTTTGCTG-3′, (p-3) 5′-AAGCATAAAGTGTAAAGCAGGGTTGGAGACAGGT-3′, and (p-4) 5′-AAGGCGGAGTTGTTGGCTGCGAAGG-3′. Primers (p-1) and (p-2) were used for amplification of 2 kb of 5′ flanking DNA of mus-52. Primers (p-3) and (p-4) were used for amplification of 2 kb of 3′ flanking DNA of mus-52. The primers for amplification of the Hygr gene were (p-5) 5′-CAGCAAAGCCGAAAAGCCTCTTCGCTATTACGCC-3′ and (p-6) 5′-ACCTGTCTCCAACCCTGCTTTACACTTTATGCTT-3′. The fusion PCR was carried out with primers (p-1) and (p-4) as described above.
Electroporation. A suspension of conidia (2.0 × 109 per ml) was prepared in 1 M sorbitol. A solution (5 μl) containing 10 μg DNA of the fusion PCR product was mixed with 50 μl of the conidial suspension, incubated on ice for 5 min, and 40 μl of the mixture was transferred to an electroporater cell (BTX Electro Cell Manipulation 600, Genetronics) and shocked by using a charging voltage of 1.5 kV and a resistance of 186 ohm.
After the electroshock, 1 ml of Vogel's minimal medium containing 1.2% sucrose was added, and the conidia were incubated at 30°C for 2 h. The solution (200 μl) was spread on agar medium containing hygromycin B (500 μg/ml). Colonies resistant to hygromycin were isolated and tested by PCR using primers (p-7) and (p-8) to determine whether replacement occurred at the target gene (Fig. 1b). Southern blotting was used to confirm whether the colonies contained extra copies of the Hygr gene. The targeted strain was crossed to wild type to test for normal segregation of the markers.
Mutagen Sensitivity. Sensitivity to UV and various chemicals was tested by spot test (26). Survival to methyl methanesulfonate (MMS) killing was determined as described (27).
Results
Replacement of mus-51 and mus-52. We searched the Neurospora genome database (www.broad.mit.edu/annotation/fungi/neurospora) to find N. crassa genes homologous to human KU70 and KU80. The candidate genes encode proteins of 645 and 661 aa, respectively. Alignment of the amino acid sequences of human Ku70 and Neurospora Ku70 homolog, and human Ku80 and Neurospora Ku80 homolog is shown in Fig. 6, which is published as supporting information on the PNAS web site. Cosmids G7H3 and G8B12 from the Orbach/Sachs libraries (28) contain Neurospora KU70 homolog gene mus-51 and KU80 homolog gene mus-52, respectively. These cosmids were used as templates to amplify mus-51 and mus-52 by PCR. As described in Materials and Methods, DNA fragments carrying Hygr with 2-kb 5′ and 3′ flanking DNAs from the Neurospora KU homolog gene were produced by fusion PCR. The fusion PCR products were introduced into wild type, and hygromycin-resistant colonies were isolated. About 200 transformants were subcultured, and genomic DNA was extracted to determine by PCR whether mus-51 or mus-52 was replaced by Hygr. Primers in this PCR were set so that one is outside the targeted gene and the other is inside. About 10% of hygromycin-resistant colonies contained Hygr in place of the KU ortholog. Strains 54yo-728 carrying mus-51::Hygr and 54yo-828 carrying mus-52::Hygr were then used as mus-51 and mus-52 mutants.
Characterization of mus-51 and mus-52 Mutants. Conventional mapping. The genome database indicates that mus-51 and mus-52 are located in linkage groups IV and III, respectively. To prove genetic linkage, we crossed mus-51 to strain C-P-1 (a pan-1 arg-2 leu-2) and mus-52 to strain C20–1 (A ad-2 ser-1). The mus-gene was scored as hygromycin-resistant. mus-51 was near arg-2 (<1%) and mus-52 was near ser-1 (1.2%), as expected.
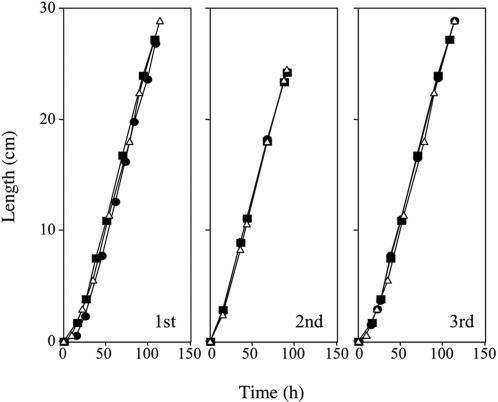
Vegetative growth, conidiation, sporulation, and telomere length. Race tubes were used to measure linear growth rate (Fig. 2). Even after three consecutive transfers, growth rate of mus-51 or mus-52 was identical to wild type. Conidiation was also normal. Homozygous crosses of mus-51 or mus-52 were fertile and produced normal ascospores. There was no apparent elongation or shortening of telomeres (data not shown).
Fig. 2.
Measurement of linear growth at 25°C race tubes (30 cm length × 1.5 cm) containing minimal medium (sucrose 0.5%) were used. Distance from inoculated point to front of elongating tip was measured at various times. 1st, 2nd, and 3rd indicate order of consecutive transfers of mycelia. Open triangles, wild type; filled circles, mus-51; filled squares, mus-52.
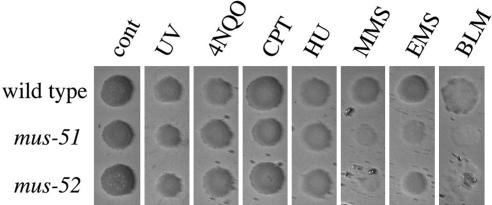
Mutagen sensitivity. Mutagen sensitivity of the mus-51 and mus-52 was determined with spot tests using conidial suspensions. Camptothecin, 4-nitroquinoline 1-oxide, hydroxyurea, MMS, ethyl methanesulfonate (EMS), and bleomycin (BLM) were tested, as was sensitivity to UV. The mutants showed mild sensitivity to MMS, EMS, and BLM, but they were not sensitive to other chemical agents or to UV (Fig. 3).
Fig. 3.
Sensitivity of wild type, mus-51, and mus-52 to UV and chemical agents. A conidial suspension was spotted on the agar surface of plates containing 4-nitroquinoline 1-oxide (4NQO, 75 ng/ml), camptothecin (CPT, 0.4 μg/ml), hydroxyurea (HU, 1.5 mg/ml), MMS (0.175 μl/ml), ethyl methane-sulfonate (EMS, 6 μl/ml), and bleomycin (BLM, 5 μg/ml). UV irradiation was 375 J/m2.
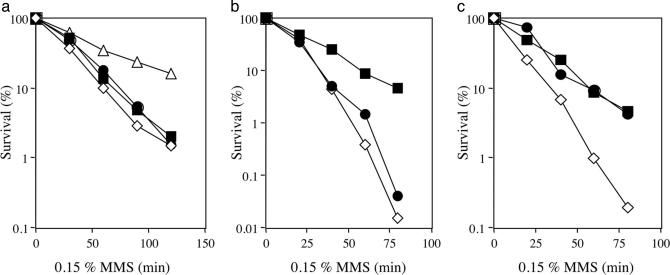
Epistasis relationship. Double mutants were constructed by combining mus-51 and mus-52 with DNA repair mutants representing various epistasis groups. MMS was used to compare sensitivity of each strain. The mus-51 mus-52 double mutant was identical in sensitivity to the single-mutant parents (Fig. 4a). The mus-23 mutation (which is homologous to S. cerevisiae MRE11) (29) was epistatic to mus-52 (Fig. 4b), but the double mutant carrying mei-3 and mus-52 was more sensitive than either parental single mutant (Fig. 4c). Therefore, mus-51 and mus-52 belong to the NHEJ group of recombination repair mutants, not to the HR group.
Fig. 4.
Comparison of sensitivity to MMS in the double mutants and their parental single mutants. A conidial suspension was treated by MMS (1.5 μl/ml) for the indicated time. (a) Wild type (open triangles), mus-51 (filled circles), mus-52 (filled squares), and mus-51 mus-52 (open diamonds). (b) mus-52 (filled squares), mus-23 (filled circles), and mus-52 mus-23 (open diamonds). (c) mus-52 (filled squares), mei-3 (filled circles), and mus-52 mei-3 (open diamonds).
Targeting the mtr and ad-3A Genes by Using Wild Type, mus-51, mus-52, mei-3, and mus-11 as Recipients of Transforming DNA. The mtr and ad-3A genes on chromosome IV and I, respectively, were selected as targets for gene disruption experiments. mtr mutants are resistant to the amino acid analog p-fluorophenylalanine (PFP). ad-3A mutants accumulate purple pigment (30). Targeting vectors were constructed by replacing the mtr or ad-3A ORF's with the blasticidin-resistance gene bar. A 2.7-kb DNA fragment containing the bar gene was cut from pBARGEM7–1 with restriction enzymes ScaI and SmaI.
In the case of mtr, the construction and introduction of targeting vectors was carried out as follows: pMTR, containing the mtr gene, was digested by MscI to delete ≈1 kb containing the promoter and a part of the mtr ORF. A 2.7-kb bar fragment was inserted to make plasmid pGS1 (9.5 kb). This plasmid was digested by NotI, and the resulting 6.7-kb linear fragment carrying 1.8-kb 5′ and 1.9-kb 3′ of the mtr gene on both sites of bar was introduced into host strains of different genetic background by electroporation. Transformants resistant to blasticidin (200 μg/ml) were isolated and tested to determine whether they are resistant to PFP (20 μg/ml). If bar DNA is integrated into the mtr locus, the strain should be resistant to PFP. We further investigated by PCR whether the integration is through homologous replacement. Table 2 shows that 10–30% of the blasticidin-resistant transformants in the wild-type strain resulted from homologous integration. All transformants in the mus-51 and mus-52 strains resulted from homologous integration and replacement. There was little homologous integration in mei-3 and mus-11 strains, consistent with their defect in HR repair.
Table 2. Rate of homologous integration at the mtr locus in the wild type and in mus-51, mus-52, mei-3, and mus-11 mutant strains.
| Strains | Exp. no. | Bla-resist | PFP-resist | Homologous integration, % |
|---|---|---|---|---|
| Wild type | 1 | 9 | 3 | |
| 2 | 22 | 3 | ||
| 3 | 11 | 3 | ||
| 4 | 16 | 3 | ||
| Total | 58 | 12 | 21 | |
| mus-51 | 1 | 41 | 41 | |
| 2 | 18 | 18 | ||
| Total | 59 | 59 | 100 | |
| mus-52 | 1 | 12 | 12 | |
| 2 | 23 | 23 | ||
| 3 | 12 | 12 | ||
| 4 | 26 | 26 | ||
| Total | 73 | 73 | 100 | |
| mei-3 | 1 | 58 | 2 | |
| 2 | 35 | 1 | ||
| Total | 93 | 3 | 3* | |
| mus-11 | 1 | 45 | 0 | |
| 2 | 20 | 0 | ||
| Total | 65 | 0 | 0 |
Bla-resist, blastcitidin resistance, PFP-resist, PFP resistance; homologous integration rate = number of PFP-resist/number of Bla-resist.
Confirmation of homologous integration was not obtained by PCR
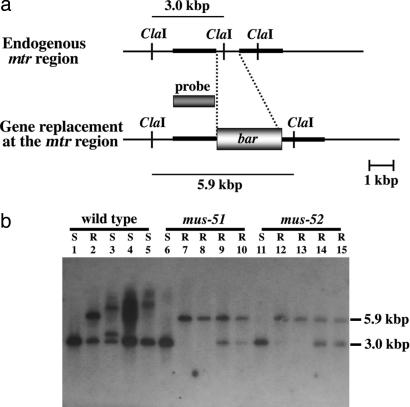
To confirm that transformants showing resistance to both blasticidin and PFP carry the mtr gene integrated homologously into genomic DNA, Southern hybridization analysis was carried out by using mtr as a probe (Fig. 5). Four transformants from each strain, wild type, mus-51, and mus-52, were isolated by random sampling. Genomic DNAs from these were digested by ClaI and electrophoresed for Southern analysis. In the wild type, one transformant (Fig. 5, lane 2) was resistant to both chemicals, indicating homologous integration, whereas the other three were resistant to blasticidin but sensitive to PFP, indicating ectopic integration. In mus-51 and mus-52, all four transformants were resistant to both chemicals, indicating homologous integration.
Fig. 5.
Southern hybridization analysis in the wild type, mus-51, mus-52, and their transformants. (a) Diagram of the relevant region of the endogenous mtr gene and the homologously integrated mtr::bar gene. The thick lines show 2-kb mtr segments attached to the bar gene. The target DNA was introduced into the recipient strain, and blastocidin-resistant transformants were isolated. (b) Results of Southern analysis. Lanes 1, 6, and 11 are non-transformants, showing the 3-kb ClaI fragment of mtr. All other lanes are transformants. Homologous integration produces a 5.9-kb ClaI fragment. Because transformants are frequently heterokaryons carrying both transformed and nontransformed nuclei, both 3- and 5.9-kb bands may be observed. Ectopic integration produces different-sized bands, as shown in lanes 3–5. S, sensitive to PFP; R, resistant to PFP.
Construction of an ad-3A targeting vector carrying 1.6 kb 5′ and 2.0 kb 3′ of the ad-3A gene on both sites of the bar was similar to the mtr construction, and the vector was introduced into host strains. Transformants resistant to blasticidin were isolated and subcultured in minimal medium supplemented with adenine for 10 days to confirm the pigmentation. The numbers of colonies with purple mycelia, indicating loss of ad-3A function, were scored. As shown in Table 3, 40% of blasticidin-resistant transformants from the wild-type strain had purple mycelia, whereas 100% of transformants in the mus-51 and mus-52 backgrounds were purple. PCR was used to confirm that the strains with purple mycelia were disruptants resulting from homologous replacement.
Table 3. Rate of homologous integration at the ad-3A locus in the wild type and in mus-51 and mus-52 mutant strains.
| Strains | Bla-resist | Adenine-requiring | Homologous integration, % |
|---|---|---|---|
| Wild type | 85 | 34 | 40 |
| mus-51 | 46 | 46 | 100 |
| mus-52 | 36 | 36 | 100 |
Bla-resist, blasticitidin resistance.
Relationship Between Targeting Frequency and Length of Homology. To investigate the relationship between the targeting frequency and the length of the homologous sequence, 50, 100, 500, and 1,000 base pairs homologous to 5′ and 3′ flanking DNA's of the mtr gene were amplified by PCR and attached to both sides of the bar gene. Blasticidin-resistant transformants were tested for resistance to PFP (Table 4). Few blasticidin-resistant transformants into which DNA had been introduced with 50- and 100-bp homology showed resistance to PFP, either in the mus-51 and mus-52 strains or in wild type. With 500-bp homology, <10% of blasticidin-resistant transformants in the wild-type strain showed resistance to PFP, whereas ≈90% in the mus-51 and mus-52 strains were resistant to PFP. With 1,000-bp homology, ≈20% of blasticidin-resistant transformants in the wild type were resistant to PFP, and all blasticidin-resistant transformants in the mus-51 and mus-52 strains were resistant. Greater than 1,000-bp homology is apparently required for completely efficient replacement of the target gene when NHEJ is not functional.
Table 4. Relationship between the frequency of integration into a homologous site in the genome and the length of the homologous sequence.
| Strain | Length of homology, bp | Exp. no. | Bla-resist | PFP-resist | Homologous integration, % |
|---|---|---|---|---|---|
| Wild type | 50 | 1 | 22 | 0 | |
| 2 | 29 | 1 | |||
| Total | 51 | 1 | 2 | ||
| mus-51 | 50 | 1 | 30 | 0 | |
| 2 | 29 | 1 | |||
| Total | 59 | 1 | 2 | ||
| mus-52 | 50 | 1 | 29 | 0 | |
| 2 | 30 | 0 | |||
| Total | 59 | 0 | 0 | ||
| Wild type | 100 | 1 | 20 | 0 | |
| 2 | 30 | 1 | |||
| Total | 50 | 1 | 2 | ||
| mus-51 | 100 | 1 | 8 | 2 | |
| 2 | 13 | 0 | |||
| Total | 21 | 2 | 10 | ||
| mus-52 | 100 | 1 | 14 | 0 | |
| 2 | 32 | 2 | |||
| Total | 46 | 2 | 4 | ||
| Wild type | 500 | 1 | 40 | 4 | |
| 2 | 40 | 3 | |||
| Total | 80 | 7 | 9 | ||
| mus-51 | 500 | 1 | 40 | 35 | |
| 2 | 39 | 37 | |||
| Total | 79 | 72 | 91 | ||
| mus-52 | 500 | 1 | 30 | 28 | |
| 2 | 39 | 36 | |||
| Total | 69 | 64 | 93 | ||
| Wild type | 1,000 | 1 | 9 | 3 | |
| 2 | 22 | 7 | |||
| 3 | 40 | 6 | |||
| 4 | 40 | 7 | |||
| Total | 111 | 23 | 21 | ||
| mus-51 | 1,000 | 1 | 4 | 4 | |
| 2 | 8 | 8 | |||
| 3 | 10 | 10 | |||
| 4 | 8 | 8 | |||
| Total | 30 | 30 | 100 | ||
| mus-52 | 1,000 | 1 | 31 | 31 | |
| 2 | 19 | 19 | |||
| Total | 50 | 50 | 100 |
Bla-resist, blasticidin resistance; PFP-resist, PFP resistance.
Discussion
We demonstrated that exogenous DNA is integrated into homologous sequences in the genome of N. crassa at a frequency of 100% in mus-51 and mus-52 strains, indicating that suppression of NHEJ increases the frequency of HR. This finding has strong implications for recombination technology, because it is easy to make genetic alterations in the mus-51 or mus-52 mutants as recipients for transformation, and because these mutants do not show severe defects, but differ from wild type only in being moderately sensitive to mutagens that produce double strand breaks.
Systematic gene disruption is important for determining the functions of target genes and for adding new markers and reporters in studies of gene expression. The virtually complete Neurospora genome sequence is now available for searching (14, 31). Sequences of potential target genes are easily amplified by PCR even if they are >1 kb long. The mus-51 and mus-52 mutations are marked with Hygr, and once a targeted gene has been disrupted, the mus mutation can easily be eliminated by crossing to a wild type strain and isolating hygromycin-sensitive progeny.
Until now, ability to achieve 100% homologous integration of transforming DNA has been a unique advantage of S. cerevisiae. Knocking out targeted genes in other eukaryotes has been far less efficient. Our findings with Neurospora change this situation. They should be widely applicable to other systems.
Supplementary Material
Acknowledgments
We thank George R. Hoffmann for his review of this paper. This work was supported by Rational Evolutionary Design of Advanced Biomolecules, Saitama Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence, Japan Science and Technology Agency.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HR, homologous recombination; NHEJ, nonhomologous end-joining; MMS, methyl methanesulfonate; PFP, p-fluorophenylalanine.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AB177394 (mus-51) and AB177395 (mus-52)].
References
- 1.Takita, Y., Takahara, M., Nogami, S., Anraku, Y. & Ohya, Y. (1997) Yeast 13, 763-768. [DOI] [PubMed] [Google Scholar]
- 2.Wach, A., Brachat, A., Pohlmann, R. & Philippsen, P. (1994) Yeast 10, 1793-1808. [DOI] [PubMed] [Google Scholar]
- 3.Schiestl, R. H., Zhu, J. & Petes, T. D. (1994) Mol. Cell. Biol. 14, 4493-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker, J. R., Corpina, R. A. & Goldberg, J. (2001) Nature 412, 607-614. [DOI] [PubMed] [Google Scholar]
- 5.Critchlow, S. E. & Jackson, S. P. (1998) Trends Biosci. 23, 394-398. [DOI] [PubMed] [Google Scholar]
- 6.Mimori, T., Arizuki, M., Yamagata, H., Inada, S., Yoshida, S. & Homma, M. (1981) J. Clin. Invest. 68, 611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dynan, W. S. & Yoo, S. (1998) Nucleic Acids Res. 26, 1551-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura, K., Adachi, Y., Chiba, K., Oguchi, K. & Takahashi, H. (2002) Plant J. 29, 771-781. [DOI] [PubMed] [Google Scholar]
- 9.Hatakeyama, S., Ishii, C. & Inoue, H. (1995) Mol. Gen. Genet. 249, 439-446. [DOI] [PubMed] [Google Scholar]
- 10.Sakuraba, Y., Schroeder, A. L., Ishii, C. & Inoue, H. (2000) Mol. Gen. Genet. 264, 392-401. [DOI] [PubMed] [Google Scholar]
- 11.Handa, N., Noguchi, Y., Sakuraba, Y., Ballario, P., Macino, G., Fujimoto, N., Ishii, C. & Inoue, H. (2000) Mol. Gen. Genet. 264, 154-163. [DOI] [PubMed] [Google Scholar]
- 12.Schroeder, A. L., Pall, M. L., Lotzgesell, J. & Siino, J. (1995) Fungal Genet. Newslett. 42, 65-68. [Google Scholar]
- 13.Selker, E. U. (1990) Annu. Rev. Genet. 24, 579-613. [DOI] [PubMed] [Google Scholar]
- 14.Galagan, J. E., Calvo, S. E., Borkovich, K. A., Selker, E. U., Read, N. D., Jaffe, D., FitzHugh, W., Ma, L. J., Smirnov, S., Purcell, S., et al. (2003) Nature 422, 859-868. [DOI] [PubMed] [Google Scholar]
- 15.Kuwayama, H., Obara, S., Mario, T., Katoh, M., Urushihara, H. & Tanaka, Y. (2002) Nucleic Acids Res. 30, E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wendland, J. (2003) Curr. Genet. 44, 115-123. [DOI] [PubMed] [Google Scholar]
- 17.Vann, D. C. (1995) Fungal Genet. Newslett. 42A, 53. [Google Scholar]
- 18.Margolin, B. S., Freitag, M. & Selker, E. U. (1997) Fungal Genet. Newslett. 44, 34-35. [Google Scholar]
- 19.Vollmer, S. J. & Yanofsky, C. (1986) Proc. Natl. Acad. Sci. USA 83, 4869-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomita, H., Soshi, T. & Inoue, H. (1993) Mol. Gen. Genet. 238, 225-233. [DOI] [PubMed] [Google Scholar]
- 21.Tamaru, H. & Inoue, H. (1989) J. Bacteriol. 171, 6288-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., Fritsch, E. F. & Maniatis, T. M. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 23.Pall, M. L. & Brunelli, J. P. (1993) Fungal Genet. Newslett. 40, 59-62. [Google Scholar]
- 24.Staben, C., Jensen, B., Singer, M., Pollock, J., Schechtman, M., Kinsey, J. & Selker, E. (1989) Fungal Genet. Newslett. 36, 79-81. [Google Scholar]
- 25.Davis, R. H. & de Serres, F. J. (1970) Methods Enzymol. 17, 79-143. [Google Scholar]
- 26.Kato, A., Akamatsu, Y., Sakuraba, Y. & Inoue, H. (2004) Curr. Genet. 45, 37-44. [DOI] [PubMed] [Google Scholar]
- 27.Inoue, H. & Ishii, C. (1984) Mutat. Res. 125, 185-194. [DOI] [PubMed] [Google Scholar]
- 28.Orbach, M. J. (1994) Gene 150, 159-162. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe, K., Sakuraba, Y. & Inoue, H. (1997) Mol. Gen. Genet. 256, 436-445. [DOI] [PubMed] [Google Scholar]
- 30.Perkins, D. D., Radford, A. & Sachs, M. S. (2001) The Neurospora Compendium (Academic, New York).
- 31.Borkovich, K. A., Alex, L. A., Yarden, O., Freitag, M., Turner, G. E., Read, N. D., Seiler, S., Bell-Pedersen, D., Paietta, J., Plesofsky, N., et al. (2004) Mol. Biol. Rev. 68, 1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.