Abstract
Avian pathogenic Escherichia coli (APEC) can cause significant morbidity in chickens. The thymus provides the essential environment for T cell development; however, the thymus transcriptome has not been examined for gene expression in response to APEC infection. An improved understanding of the host genomic response to APEC infection could inform future breeding programs for disease resistance and APEC control. We therefore analyzed the transcriptome of the thymus of birds challenged with APEC, contrasting susceptible and resistant phenotypes. Thousands of genes were differentially expressed in birds of the 5-day post infection (dpi) challenged-susceptible group vs. 5 dpi non-challenged, in 5 dpi challenged-susceptible vs. 5 dpi challenged-resistant birds, as well as in 5 dpi vs. one dpi challenged-susceptible birds. The Toll-like receptor signaling pathway was the major innate immune response for birds to respond to APEC infection. Moreover, lysosome and cell adhesion molecules pathways were common mechanisms for chicken response to APEC infection. The T-cell receptor signaling pathway, cell cycle, and p53 signaling pathways were significantly activated in resistant birds to resist APEC infection. These results provide a comprehensive assessment of global gene networks and biological functionalities of differentially expressed genes in the thymus under APEC infection. These findings provide novel insights into key molecular genetic mechanisms that differentiate host resistance from susceptibility in this primary lymphoid tissue, the thymus.
Keywords: RNASeq, APEC, thymus, transcriptome, immune response
INTRODUCTION
Colibacillosis, caused by avian pathogenic Escherichia coli (APEC), is an extraintestinal disease that may manifest as septicemia, pericarditis, or airsacculitis in poultry (JanBen et al., 2001; Stordeur et al., 2004). APEC also has been recently identified as a possible cause of human disease (Rodriguez-Siek et al., 2005; Ewers et al., 2007; Johnson et al., 2007; Russo and Johnson, 2003). Studies report that APEC shares similar phylogenic background and certain virulence genes with human extraintestinal pathogenic Escherichia coli (ExPEC), suggesting the potential of zoonotic risk of APEC (Manges and Johnson, 2012). Moreover, contaminated chicken meat and eggs are potential sources of human infections (Vincent et al., 2010; Bergeron et al., 2012).
APEC generally gains entry to the host bird via the respiratory tract (Dho-Moulin and Fairbrother, 1999). From there, bacteria enter the bloodstream and gain access to the viscera resulting in a multisystemic disease. Colibacillosis causes multimillion-dollar annual losses in the US poultry industry due to morbidity, mortality, and condemnation of infected products (Kabir, 2010). In the United Kingdom, a recent longitudinal survey of 4 broiler flocks sampled weekly for 4 wk showed 39% of dead birds resulted from colibacillosis (Kemmett et al., 2013). Also in the U.K., 70% of dead birds were caused by colibacillosis from a separate analysis of causes of mortality 2 to 3 d after placement of broiler chicks (Kemmett et al., 2014). Although antibacterial agents have been used successfully to prevent this disease, restrictions on antibiotic usage in poultry production and APEC's increasing resistance to antimicrobial agents have made colibacillosis control problematic (Lanz et al., 2003; Yang et al., 2004). Thus, control of colibacillosis by means other than antimicrobial agents is highly desirable.
Variation in gene expression can be very useful in studying specimens treated under different conditions at a genome-wide level (Alizadeh et al., 2000; Ross et al., 2000; Bahar et al., 2006). Many types of chicken microarrays have been used in genome-wide gene expression studies, including a macrophage microarray, avian innate immunity microarray, 44 K Agilent microarray, and Affymetrix chicken genome array (Call et al., 2001; Lavric et al., 2008; Li et al., 2008; Kranis et al., 2013). The new technology of RNAseq is an efficient and reliable tool to investigate genetic architecture and sequence variation and to quantify gene expression through whole transcriptome analysis (Ozsolak and Milos, 2011). We have reported its use in previous studies of the transcriptomic response of bone marrow and bursa to systemic APEC infection (Sun et al., 2015a; 2015b). The current study used RNAseq technology to characterize the transcriptomic response of genes involved in the early phases of immune response against APEC by studying the primary lymphoid organ, the thymus.
MATERIALS AND METHODS
Ethics Statement
All animal care and experimental procedures were reviewed and approved by the Iowa State University Institutional Animal Care and Use Committee (#11-07-6460-G).
Avian Pathogenic Escherichia Coli (APEC) Experimental Design
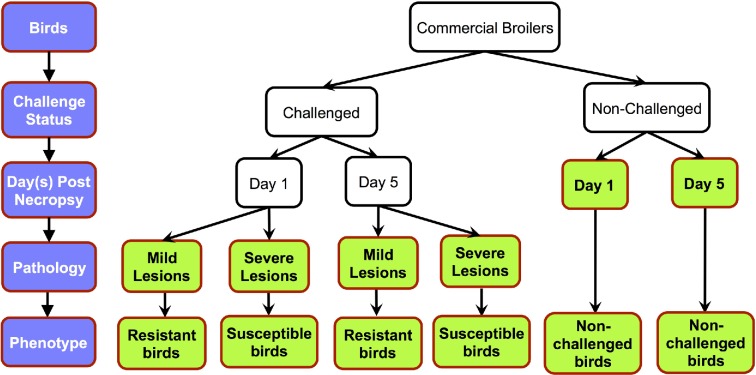
A total of 360 commercial male broilers (meat-type chickens) were used in the pathogen-challenge trial. At 4 wk of age, 288 birds were inoculated with APEC O1 intra-air sac and, for the control group (same type and age), 72 were injected with the same volume of phosphate buffered saline (PBS). The APEC O1 strain and experimental procedures have been previously described in detail (Sandford et al., 2011; Sandford et al., 2012). At necropsy, the lesions on the liver, air sacs, and pericardium were scored. The range of scores for each tissue was: liver, 0 to 2; air sac, 0 to 3; and pericardium, 0 to 2. The sum of its tissue lesion scores was used to assign the level of pathology of the individual bird. If the summed lesion scores were 0 to 3, birds were classified as resistant (mild lesion). If the summed lesion scores were 4 to 7, birds were classified as susceptible (severe lesion). The lesion scores were normally distributed over the 288 infected birds (Sun et al. 2015a). Thymi were collected at one or 5 days post infection (dpi). A total of 6 treatments were thus classified: 1 dpi challenged-resistant birds; 1 dpi challenged-susceptible birds; 1 dpi non-challenged birds; 5 dpi challenged-resistant birds; 5 dpi challenged-susceptible birds; and 5 dpi non-challenged birds (Figure 1). Birds selected for RNA-seq analysis were from the 2 phenotypic extremes for APEC-induced pathology: resistant birds with 0 to 1 lesion scores and susceptible birds with 6 to 7 lesion scores. These were the same birds as the study of Sun et al. (2015a). Four individual bird replicates were used for each treatment group, totaling 24 samples.
Figure 1.
Graphical representation of experimental design. There were 3 levels of experimental variables: challenge status, necropsy d, and pathology level of infected birds. The 6 studied groups are indicated in bold.
mRNA isolation, cDNA Library Preparation, and Sequence Analysis
An Ambion MagMAX-96 Kit (AM1839) (Applied Biosystems, Foster City, CA) was used to isolate RNA from the thymus samples. The quality and quantity of RNA were assessed using the Agilent 2100 Bioanalyzer according to the manufacturer's instructions (Agilent Technologies). RNA Integrity Numbers (RIN) for all the RNA samples selected to construct the cDNA libraries were greater than 8.0. Next, an Illumina TruSeq ® RNA Sample Preparation v2 Kit was utilized to convert 0.1 to 4 μg RNA into cDNA libraries. Twenty-four cDNA libraries, which included 4 biological replicates (birds) for each treatment, were constructed. Briefly, fragment mRNA was purified using oligo-dT beads from the initial RNA and reverse transcribed into a double strand cDNA fragment. End repair, adenylation, adapter ligation, and PCR amplification were then carried out in conformance with the TruSeq® manufacturer's instructions (Protocol: #15026495, May 2012). A Qubit® Quantitation Platform and HS dsDNA kit (Invitrogen, Paisley, UK) were then used to test and quantify the cDNA libraries. Six cDNA libraries, including one for each of the 6 treatments, were sequenced in the same lane of the Illumina® HiSeq 2000 at the Iowa State University DNA facility (4 lanes for the 24 cDNA libraries) with single end 100 bp cycles.
Read Quality Control, Alignment, and Reads Number Calculation
Fastx toolkit software (version 0.0.13) was used to remove the adapter for each read, and quality of RNA-seq reads from all the samples was checked using FastQC software (version 0.10.1) keeping a Phred score of 32. Then the filtered reads from each sample were separately aligned to the Gallus gallus 4.0 reference genome from Ensembl using TopHat2 (version 2.0.9) and Bowtie (version 2.1.0) software with default parameters. The abundance of reads for all annotated genes was counted using the HTseq software package (version 0.5.4p3) in Python.
Statistical and Biological Analysis
To test the samples’ relationship, Qlucore Omics Explorer (version 3.0) was used to conduct principal component analysis (PCA) by using the read count data from the 24 samples. Then, the software package edgeR (version 3.0.8) was run in R software (version 2.15.3) to identify differentially expressed (DE) genes. The generalized linear model (GLM) analysis in edgeR based on the negative binomial distribution was applied. Then relevant linear contrasts were constructed to compare treatment conditions. The Benjamini-Hochberg method was used to control false discovery rate (FDR) (Benjamini and Hochberg, 1995) at 5%. To avoid gene length bias, the GOseq package (version 1.10.0) (Young et al., 2010) was utilized for further gene ontology (GO) and pathway analysis while controlling FDR at 5%. Animal systems biology analysis and modeling center (ASBAMC) was used to generate the significant pathways.
Candidate Genes for qPCR Validation
Quantitative real time PCR (qPCR) was performed to measure the mRNA expression levels of 11 selected genes (IL7, IL7R, LCK, ZAP70, CD3Z, IL18, IL8, IFNGR, NOD1, LIG4, TLR6) using the same 24 RNA samples used for sequencing. The gene selection criteria were involvement in multiple immune response pathways and significance in the RNAseq analysis. An internal control gene (28 S rRNA) was used for normalization of the initial concentration of RNA. Primers were designed for amplifying fragments in the qPCR reaction using sequences from NCBI and Primer 3 (Rozen and Skaletsky, 2000). Primer sequence detail is displayed in Table 1. qPCR was performed in triplicate on individual thymus samples. Reactions of qPCR were carried out using the QuantiTect SYBR Green kit (Qiangen Inc., Valencia, CA) as described by Redmond and co-workers (Redmond et al., 2010). The following equation was used to calculate the adjusted cycle threshold (Ct) values: 40 – [Ct target gene mean + (Ct 28S median – Ct 28S mean)(slope of target gene/slope of 28S)]. The Fit Model procedure in JMP software (SAS Institute Inc., Cary, NC) was used to analyze the Ct value. Relative gene expression values were calculated for different treatment contrasts.
Table 1.
Primers sequence for qPCR validation
| Gene | Forward sequence (5′-3′) | Reverse sequence (5′-3′) |
|---|---|---|
| IL7 | CATCGAAGAGCTGGTAAATATG | GCCATACTCTGTAGTGATCC |
| IL7R | ATGGTGATGGGACCTTTG | CACAGCCAGGGTATAGTTAG |
| LCK | CACCGGAGGCTATCAATTAC | GTTGGTCATCCCTGGATATG |
| ZAP70 | ACCCACGAGGAAGATTAAG | ATGCTGCCATAGTAGAAGG |
| CD3Z | GCCAGGACGATGTGTATAA | TCTGCAGGGAAGAGTAAAC |
| IL18 | AGGTGAAATCTGGCAGTGGAAT | ACCTGGACGCTGAATGCAA |
| IL8 | GCCCTCCTCCTGGTTTCAG | TGGCACCGCAGCTCATT |
| IFNGR | TGGCAGAGAGAAACACTAC | CCCAGTAGGACACATGATAC |
| NOD1 | CTGTGTCCTGCAGAAAGT | CCTGCTAACTGGATCTGTATT |
| LIG4 | CACAGTGCTCTCCATCAA | TCCATACGCCATCCTTTC |
| TLR6 | TGCATAAGAGTGAGAATCTGG | TACTACATAGGCTCCTCACA |
Availability of Supporting Data
The RNAseq data can be obtained from the NCBI Gene Expression Omnibus (GEO) database with the accession number GSE69014.
RESULTS
mRNAseq Read Alignment and Sample Variability
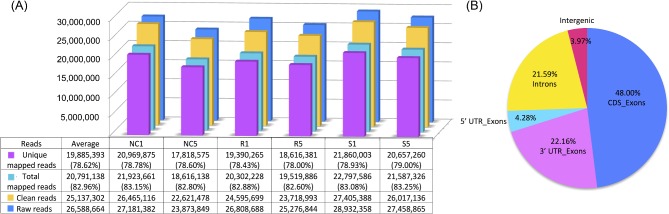
Twenty-four individual thymus samples were analyzed by RNAseq. These included one sample from each of 6 treatment conditions from 4 biological replicates. After sequencing the cDNA libraries, the average total raw reads were 26.59 million. By trimming the adaptor contamination using the Fastx toolkit and FastQC quality control, the average number of clean reads over all samples was 25.14 million. The number of raw and clean reads for each treatment group is displayed in Figure 2A. Using TopHat2, an average of 82.96% of the reads mapped back to the reference genome and the unique mapped reads accounted for an average of 78.62% (Figure 2A). Examination of the total mapped reads distribution is illustrated in Figure 2B. Distribution of reads among the 6 treatment groups was relatively consistent. On average, 74.44% of the reads mapped to exons, including 48.00% CDS_exons, 22.16% 3′ UTR_exons, and 4.28% 5′ UTR_exons (Figure 2B). There were 21.59% and 3.97% of the reads located into introns and intergenic, respectively (Figure 2B).
Figure 2.
Types of reads and total mapped reads distribution. A: The raw, clean, total mapped, and unique mapped reads in each of the 6 treatment groups. NC1, 1 d post infection (dpi) non-challenged birds; NC5, 5 dpi non-challenged birds; R1, 1 dpi resistant birds; R5, 5 dpi resistant birds; S1, 1 dpi susceptible birds; S5, 5 dpi susceptible birds. B: The total mapped reads distribution of the thymus transcriptome.
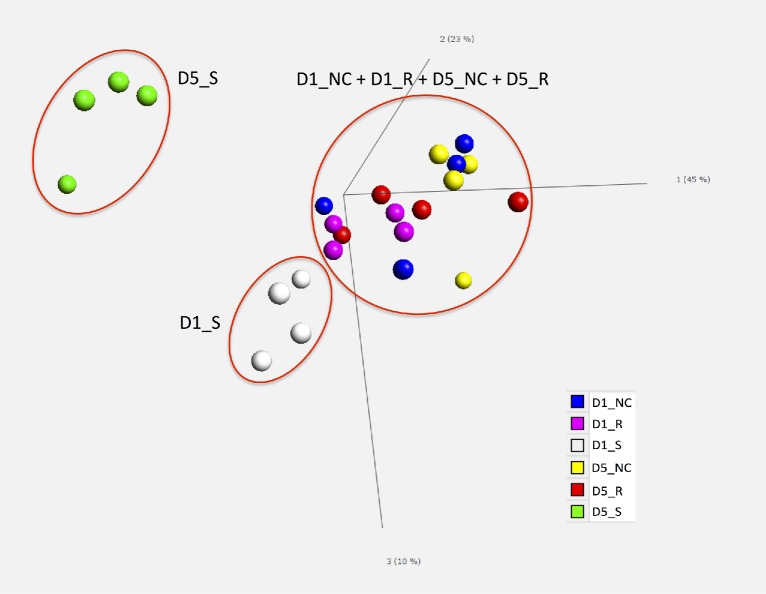
After alignment, the average number of reads for all samples was 12.53 million using HTseq counting. The average transcriptome coverage, i.e., the number of detected transcripts over the total annotated transcripts, was 85.89%. To further explore the relationship among the total 24 samples, PCA was used to cluster similar samples in multivariate space. The PCA results showed that the 5 dpi susceptible birds were distinct from the other 5 treatment groups (Figure 3). Additionally, 1 dpi susceptible birds differed slightly from the 4 groups: 1 dpi resistant, 5 dpi resistant, 1 dpi non-challenged, and 5 dpi non-challenged birds. Variability among replicates in each treatment group was low and the clear separation of the different groups indicated that susceptible birds possess a unique characteristic expression pattern that was greatly different from resistant and from non-challenged birds.
Figure 3.
Principal component plot. Across the entire data set of differentially expressed genes, principal component analysis (PCA) was used to test the spatial distribution of the 6 treatment groups. Different colors are used to illustrate each treatment group. D1_NC, 1 d post infection (dpi) non-challenged birds; D1_R, 1 dpi resistant birds; D1_S, 1 dpi susceptible birds; D5_NC, 5 dpi non-challenged birds; D5_R, 5 dpi resistant birds; D5_S, 5 dpi susceptible birds.
Analysis of Differentially Expressed (DE) Genes
From a total of 16,693 detected transcripts, 2,484 transcripts were novel. After keeping genes with read counts above one count per million for at least 3 samples in at least one treatment group and removing the other low-expression reads, 11,585 transcripts were statistically analyzed. Comparisons of gene expression with respect to treatment, time, and pathology effects were carried out to identify candidate genes that respond to APEC infection. Nine total contrasts were constructed for interesting 2-way comparisons. The numbers of up-regulated DE transcripts were greater than those of down-regulated ones for most of the 9 contrasts (Table 2).
Table 2.
Numbers of significantly differentially expressed genes (FDR < 5% & FC > 1.5).
| Contrast | # of DE genes | # of ↑ DE genes | # of ↓ DE genes |
|---|---|---|---|
| 1 dpi susceptible vs. 1 dpi non-infected birds | 158 | 89 | 69 |
| 1 dpi susceptible vs. 1 dpi resistant birds | 23 | 19 | 4 |
| 1 dpi resistant vs. 1 dpi non-infected birds | 4 | 2 | 2 |
| 5 dpi susceptible vs. 5 dpi non-infected birds | 3,061 | 2,162 | 899 |
| 5 dpi susceptible vs. 5 dpi resistant birds | 3,816 | 2,640 | 1,176 |
| 5 dpi resistant vs. 5 dpi non-infected birds | 3 | 0 | 3 |
| 5 dpi vs. 1 dpi susceptible birds | 2,563 | 2,089 | 474 |
| 5 dpi vs. 1 dpi resistant birds | 3 | 3 | 0 |
| 5 dpi vs. 1 dpi non-infected birds | 2 | 2 | 0 |
Note: FC, fold change; dpi, days post infection; #, number; DE, differentially expressed.
Tests for 4 comparisons (pair-wise contrasts) identified large numbers of DE genes: 1 dpi susceptible vs. 1 dpi non-infected birds, 5 dpi susceptible vs. 5 dpi non-infected birds, 5 dpi susceptible vs. 5 dpi resistant birds, and 5 dpi vs. 1 dpi susceptible birds (Table 2). However, tests for the other comparisons detected only a few DE genes (N < 25). There were 158 DE genes detected comparing 1 dpi susceptible vs. 1 dpi non-infected birds. Thousands of DE genes were identified when comparing 5 dpi susceptible vs. 5 dpi non-infected birds; 5 dpi susceptible vs. 5 dpi resistant birds; and 5 dpi vs. 1 dpi susceptible birds. These results indicate that there were large differences between 5 dpi susceptible birds and 5 dpi resistant birds, and between 5 dpi susceptible birds and 5 dpi non-challenged birds. However, resistant birds differed little from the non-infected birds. The transcriptomic response of susceptible birds greatly increased over time post infection, whereas a time-related response increase did not occur in resistant and non-infected birds.
Significant GO Terms Analysis
To provide sufficient genes for common biological process analysis, the 4 comparisons with the largest numbers of DE genes were used for further analysis. The false discovery rate was controlled at 5% for all the significant GO terms and pathways in the following results and discussion.
In the contrast of 1 dpi susceptible vs. non-challenged birds, the DE genes were mainly involved in these top 3 significant GO terms: defense response to bacterium, defense response, and response to bacterium. The comparisons of 5 dpi susceptible vs. 5 dpi non-infected birds and of 5 dpi susceptible vs. 5 dpi resistant birds had the top significant GO terms of immune response, toll-like receptor signaling pathway, T/B cell activation, and T-cell lineage commitment. With passage of time post infection (5 dpi vs. 1 dpi), the susceptible birds’ response mainly focused on natural killer cell differentiation, myeloid progenitor cell differentiation, lymphoid progenitor cell differentiation, and lymphocyte differentiation GO terms.
Significant Pathways Analysis
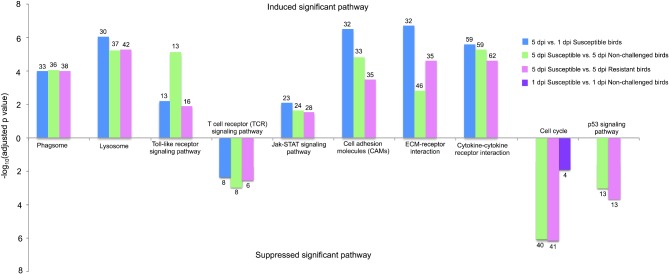
These 4 comparisons also had significantly changed pathways in response to APEC infection. Generally, phagosome, lysosome, toll-like receptor (TLR) signaling pathway, Jak-STAT signaling pathway, cell adhesion molecules (CAMs), ECM-receptor interaction, and cytokine-cytokine receptor interaction were dramatically induced in these 3 comparisons: 5 dpi susceptible vs. 5 dpi non-challenged birds, 5 dpi susceptible vs. 5 dpi resistant birds, and 5 dpi vs. 1 dpi susceptible birds. Moreover, T-cell receptor (TCR) signaling pathway was strongly suppressed in the above 3 contrasts. Also, cell cycle and p53 signaling pathways were significantly suppressed in the contrast of 5 dpi susceptible vs. 5 dpi non-challenged birds and of 5 dpi susceptible vs. 5 dpi resistant birds. Cell cycle also was detected in 1 dpi susceptible vs. 1 dpi non-challenged birds. Figure 4 showed the DE genes that were involved in the significant pathways in the 4 contrasts. Detailed information of DE genes of the significant pathways for the 4 contrasts was displayed in Table S1-S4. These results indicate that compared to resistant birds, susceptible birds extensively initiate their pathways of immune response, signal transduction, and signal molecules and interaction to resist APEC infection. However, the T-cell differentiation and proliferation and cell growth are significantly impaired in susceptible birds.
Figure 4.
Significantly changed pathways in different contrasts. The upper bar chart means the significantly induced pathways while the lower bar chart indicates the significantly suppressed pathways. The Y axis is adjusted P-value, which is processed by the –log 10. The number on the bar chart represents the numbers of the significantly differentially expressed genes that are involved in the induced or suppressed pathways. dpi, d post infection.
Validation of RNAseq Data
To validate the RNAseq data, qPCR was performed on the following 11 genes selected from immune related genes that were significantly DE in RNAseq: IL7, IL7R, LCK, ZAP70, CD3Z, IL18, IL8, IFNGR, NOD1, LIG4, TLR6. The qPCR results for 10 of 11 selected genes conformed to the same direction of fold change and significance as those in RNAseq data (Table 3). A close correlation (93.42%) in the expression level was between qPCR results and RNAseq data. Only one gene, CD3Z, was not significantly DE in the qPCR experiment; however, the CD3Z expression pattern in the qPCR experiment conformed to the same direction as for RNAseq (Table 3).
Table 3.
Quantitative PCR validation.
| Gene | Contrast | qPCR | RNA-seq |
|---|---|---|---|
| IL7 | 5 dpi susceptible vs. 5 dpi resistant birds | +3.05* | +2.36* |
| 5 dpi vs. 1 dpi susceptible birds | +4.17** | +2.36* | |
| 5 dpi susceptible vs. 5 dpi non-infected birds | +4.16* | +3.14** | |
| IL7R | 5 dpi susceptible vs. 5 dpi resistant birds | +4.30** | +2.55** |
| 5 dpi vs. 1 dpi susceptible birds | +5.11** | +2.66** | |
| LCK | 1 dpi susceptible vs. 1 dpi resistant birds | −2.11* | −1.56** |
| 5 dpi susceptible vs. 5 dpi non-infected birds | −3.33* | −1.66** | |
| ZAP70 | 5 dpi susceptible vs. 5 dpi resistant birds | −3.42* | −1.87** |
| 5 dpi vs. 1 dpi susceptible birds | −3.51* | −2.45** | |
| 5 dpi susceptible vs. 5 dpi non-infected birds | −1.89 | −2.13** | |
| CD3Z | 5 dpi susceptible vs. 5 dpi resistant birds | −1.94 | −1.83* |
| 5 dpi vs. 1 dpi susceptible birds | −1.57 | −2.45** | |
| 5 dpi susceptible vs. 5 dpi non-infected birds | +4.40** | +2.89* | |
| IL18 | 5 dpi susceptible vs. 5 dpi resistant birds | +4.39** | +5.17** |
| 5 dpi vs. 1 dpi susceptible birds | +5.43** | +2.93* | |
| 5 dpi susceptible vs. 5 dpi non-infected birds | +3.00* | +4.11** | |
| IL8 | 5 dpi susceptible vs. 5 dpi resistant birds | +3.38** | +3.94** |
| 5 dpi vs. 1 dpi susceptible birds | +2.95** | +2.91** | |
| 5 dpi susceptible vs. 5 dpi non-infected birds | +1.96* | +1.67** | |
| IFNGR | 5 dpi susceptible vs. 5 dpi resistant birds | +1.86* | +1.74** |
| 5 dpi vs. 1 dpi susceptible birds | +1.98* | +1.59** | |
| NOD1 | 5 dpi susceptible vs. 5 dpi resistant birds | +2.07* | +1.68** |
| 5 dpi susceptible vs. 5 dpi non-infected birds | −4.29* | −1.91* | |
| LIG4 | 5 dpi susceptible vs. 5 dpi resistant birds | −3.53* | −1.99** |
| 5 dpi vs. 1 dpi susceptible birds | −3.82* | −2.10** | |
| 5 dpi susceptible vs. 5 dpi non-infected birds | +3.45* | +4.56** | |
| TLR6 | 5 dpi susceptible vs. 5 dpi resistant birds | +2.96* | +3.12** |
| 5 dpi vs. 1 dpi susceptible birds | +4.11** | +3.14** |
Note: Fold change between contrasts presented in third and fourth column. + values indicate higher expression in the first group, − values indicate higher expression in the second group.
** means P-value < 0.01 in qPCR and RNA-seq, * represents P-value < 0.05 in qPCR and RNA-seq.
DISCUSSION
The novel experimental design of the current study enabled characterization of the resistance and susceptibility mechanisms of different phenotype birds in response to APEC infection through chicken thymus transcriptome analysis. The PCA of the thymus transcriptome of different phenotype birds, together with the identified DE genes in different contrasts (Figure 3 and Table 2), demonstrated that it was appropriate to classify the challenged birds as resistant or susceptible birds based upon their total lesion scores.
Nakamura et al. (1985) demonstrated that marked atrophy of the thymus and bursa were observed in natural colibacillosis of broiler chickens, and the relative weights of the thymus and bursa were dramatically decreased at 1 dpi (Nakamura et al., 1986). Histologically, the T and B lymphocytes were greatly depleted in the thymus and bursa, respectively, after 1 dpi in colibacillosis of white Leghorn (Nakamura et al., 1986). These results indicate T and B cells have important functions in bacteria infection. Thus, the primary lymphoid tissues (bone marrow, bursa, and thymus) have critical importance to understand how the host's primary immune organs respond to systemic APEC infection. Transcriptome analyses of bone marrow and bursa have been published on investigations of the earliest phases of immune response to systemic APEC infection (Sun et al., 2015a; 2015b), as well as the combined analysis of bone marrow, bursa, and thymus to investigate primary lymphoid tissues’ interaction or cooperation (Sun et al., 2016). To date, however, the gene expression patterns in the thymus of resistant and susceptible birds under systemic APEC infection have not been reported. The thymus is an essential primary lymphoid organ, providing an appropriate environment for T cell precursor development, differentiation, and maturation (Rose, 1979) and unique pathway changes were identified in thymus transcriptome analysis, compared to the results of combined analysis of primary lymphoid tissues.
In the current study, the TLR signaling pathway, lysosome pathway, CAMs, and TCR signaling pathway were the major response mechanisms in the thymus after APEC infection. In the comparison of combined analysis of primary lymphoid tissues (Sun et al., 2016), TLR and CAM were the unique pathway changes in the thymus. The TLR is the major innate immune response modulator for chicken resistance to APEC infection. TLRs can recognize pathogen-associated molecular patterns (PAMPs) to trigger inflammatory cascades (Martinon and Tschopp, 2005; Akira et al., 2006).
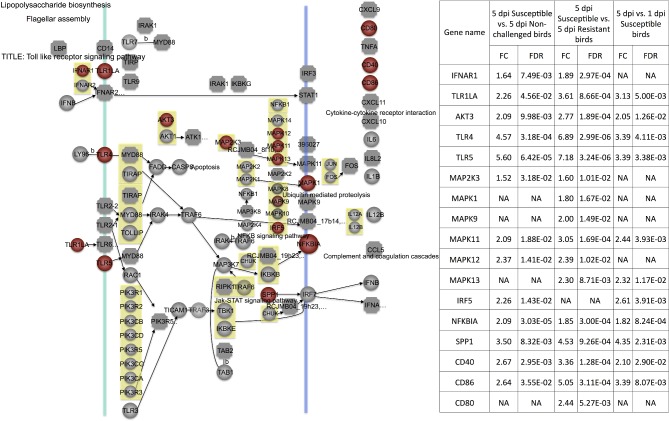
The TLR4 protein bound to Gram-negative bacteria can interact with TIR-domain-containing adaptor proteins (MyD88, MAL, and IRAK4) to transmit signals, inducing MAPK (mitogen-activated protein kinases) signaling pathway activation and inflammatory cytokines (Akira et al., 2001; Werling and Jungi, 2003; Akira, 2006; Sutterwala et al., 2006). Moreover, TLR6 can use the same signaling pathway as TLR4 (Figure 5). TLR5 can bind to flagellin to activate cytokine IL8 expression and inflammatory response (Hayashi et al., 2001). In the current study, TLR6 (TLR1LA), TLR4, TLR5, and IL8 were all over-expressed in 5 dpi susceptible vs. 5 dpi non-challenged and 5 dpi susceptible vs. 5 dpi resistant birds (Figure 5). These results may indicate that susceptible birds attempt to trigger high levels of activation of the innate immune response to resist the systemic APEC infection, compared to resistant and non-challenged birds.
Figure 5.
Toll-like receptor (TLR) signaling pathway. Red = significantly up-regulated differentially expressed genes. Green = significantly down-regulated differentially expressed genes. dpi, d post infection; FC, fold change; FDR, false discovery rate; NA, none available.
MAPK and ERK have important functions in signal transduction under cellular stresses (Davis, 1993; Kyriakis and Avruch, 1996). Currently, controversial evidence showed signal transduction pathway JNK and MAPK had a complex role in transmitting a distinct cellular effect in different cell lineages (Huh et al., 2004). For example, MAPK signaling was activated when pathogenic bacteria invaded (Watanabe et al., 2001). Activation of ERK1/2, JNK, and p38 MAPK was induced in the infection of epithelial cell lines with Listeria monocytogenes, Salmonella enterica, or enteropathogenic Escherichia coli (EPEC) (Chen et al., 1996; Hobbie et al., 1997; Czerucka et al., 2001). In our study, the p38 (MAPK11, MAPK12, and MAPK13), ERK (MAPK1), and JNK (MAPK9) genes were over-expressed in 5 dpi susceptible vs. 5 dpi non-challenged birds and 5 dpi susceptible vs. 5 dpi resistant birds. It seems that susceptible birds activated signaling transduction pathways to protect cell survival under systemic APEC, compared to resistant and non-challenged birds.
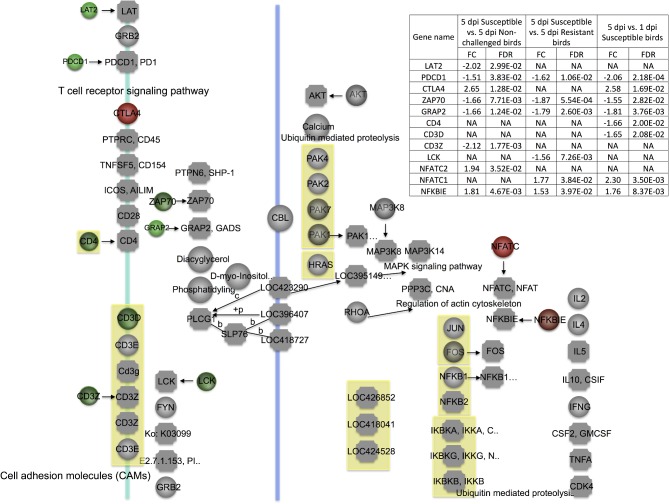
Moreover, the TLR signaling pathway also produced the costimulatory molecules (CD40, CD80, and CD86) to stimulate T cells (Melief et al., 2002; Severa et al., 2007). In the current study, CD40, CD80, and CD86 were more highly expressed in 5 dpi susceptible birds than in 5 dpi non-challenged and 5 dpi resistant birds (Figure 6). The same phenomenon was also observed in the contrasts of 5 dpi vs. 1 dpi susceptible birds (Figure 6). Additionally, TLR6 (TLR1LA), TLR4, TLR5, MAPK1, and CD40 also were significantly changed in bone marrow in 5 dpi susceptible vs. 5 dpi non-challenged birds (Sun et al., 2015a). TLR4 and CD40 were also DE in bone marrow in 5 dpi susceptible vs. 5 dpi resistant birds (Sun et al., 2015a). These genes might be potential biomarkers for chicken host response to APEC infection.
Figure 6.
T-cell receptor (TCR) signaling pathway. Red = significantly up-regulated differentially expressed genes. Green = significantly down-regulated differentially expressed genes. dpi, d post-infection; FC, fold change; FDR, false discovery rate; NA, none available.
CD40 also was involved in the significantly changed pathways CAMs. Here, the CAMs pathway was strongly induced in the thymus in 5 dpi susceptible vs. 5 dpi resistant birds and 5 dpi susceptible vs. 5 dpi non-challenged birds. The VCAM1, ITGB1, and ITGA6 genes were all more highly expressed in 5 dpi susceptible birds than in 5 dpi non-challenged and 5 dpi resistant birds in the current study, strongly suggesting important roles of these genes. The highly induced CAMs pathway, together with previous reports of thymus atrophy and T lymphocyte depletion under colibacillosis (Nakamura et al., 1985; Nakamura et al., 1986), indicates that CAMs might be the major local tissue repair mechanism after APEC infection.
As the thymus provides the essential environment for T-cell development and maturation, many distinct stages of T-cell development were marked with changes in gene expression under APEC infection. The TCR signaling is a critical requisite signal to initiate T-cell selection, proliferation, activation, and response magnitude in mice one d after Listeria infection (Zehn et al., 2009). The interaction between antigen peptide and MHC complexes can activate the TCR signal to trigger a complex downstream series of signaling cascades that can result in a variety of outcomes (Anderson et al., 1996; Kannan et al., 2012). The proximal signaling events include activation of Src tyrosine kinase Lck, phosphorylation of ITAMs in the TCR/CD3 complex, recruitment and activation of ZAP70, phosphorylation of LAT, recruitment of a variety of signaling molecules, and the activation of NFAT and NF-kB (Irving and Weiss, 1991; Chan et al., 1992; Letourneur and Klausner, 1992; Bubeck et al., 1996; Zhang et al., 1998; Smith-Garvin et al., 2009). In the present study, the key genes (CD3Z, LAT, ZAP70, GRAP2, and VAV) in the TCR signal had reduced expression levels in the 3 contrasts of 5 dpi susceptible vs. 5 dpi non-infected birds, 5 dpi susceptible vs. 5 dpi resistant birds, and 5 dpi vs. 1 dpi susceptible birds (Figure 6). Deficiency of PDCD1, a co-inhibitory receptor expressed on T cells, can promote autoimmunity (Latchman et al., 2004; Keir et al., 2006; Hirahara et al., 2012). This gene was also down-regulated in 5 dpi susceptible birds compared to 5 dpi resistant or 5 dpi non-challenged birds. Collectively, the TCR signal was deeply impaired in susceptible birds, which indicates T-cell proliferation, activation, differentiation, and maturation are significantly impaired by APEC infection in susceptible birds. Moreover, CD3Z was also significantly DE in bone marrow in 5 dpi susceptible compared to 5 dpi resistant birds (Sun et al., 2015a), indicating this gene is a positive marker of resistance in birds.
Expression of NFATC can result in T-cell anergy and NFKBIE can inhibit NF-kB transactivation (Whiteside et al., 1997; Heissmeyer et al., 2004). These 2 genes both exhibited higher expression in 5 dpi susceptible birds, indicating damage of the TCR signal. T cells are activated not only by antigen presentation signals but also by co-stimulatory molecules for negative and positive regulatory signal transduction pathways (De Koker et al., 2011). CTLA4 can interact with CD80 or CD86 to terminate T-cell activation and result in cell-cycle arrest (Alegre et al., 2001). In the current study, expression of CTLA4 and CD86 were increased in 5 dpi susceptible vs. 5 dpi non-challenged birds and 5 dpi vs. 1 dpi susceptible birds. These results suggest that APEC infection suppresses T-cell activation in susceptible birds.
Moreover, IL7 exerts a significant impact on naive T-cell survival, proliferation, and homeostasis in mammals (Hsu and Mountz, 2010; Vicente et al., 2010; Hong et al., 2012). Hsu and Mountz (2010) reported that the interaction between IL7 and IL7R could lead to proliferation and progression of T cells. IL7 and IL7R also play pivotal roles in the development of γδ T cells (Watanabe et al., 1991; Plum et al., 1993). IL7R can also be highly expressed in CD4+ and CD8+ cells and correlated with T-cell activation status in chickens (Van Haarlem et al., 2009). IL7 signaling is a negative-feedback loop (IL-7R → CD8 → TCR ⊣ IL-7R) that drives cell-intrinsic IL7R and TCR oscillatory signaling (Huang and August, 2015). In the present study, IL7 and IL7R had increased expression levels in 5 dpi susceptible vs. 5 dpi resistant birds and 5 dpi vs. 1 dpi susceptible birds. These results suggest that the over-expressed IL7 and IL7R may down-regulate TCR signaling.
Conclusion
The current study provides novel evidence that, in susceptible birds, T-cell development, activation, and cell cycle progression are impaired by APEC infection through reduced expression of regulatory genes in TCR signaling, while the innate immune response is activated through cross-talk among multiple signaling pathways. Infection with APEC induces very few transcriptomic differences between challenged-resistant and non-challenged birds. Taken together, the transcriptome analysis of thymus tissue during APEC infection demonstrates that both T-cell development and immune response mechanisms concurrently contribute to avian resistance to APEC infection. Moreover, many genes, especially TLR4, CD40, CD3Z, were identified as potential markers for host resistance to APEC infection. The CAM pathway might be a major local tissue repair mechanism after APEC infection. These findings contribute to the knowledge of the transcriptomic response in the thymus of genes that are involved in the earliest phases of the immune response to APEC, including those that drive the subsequent cellular immune reaction. The current study is foundational to the identification of genetic variation that differentiates birds that are susceptible or resistant to the pathological effects of APEC.
Abbreviation
DE, differentially expressed; APEC, avian pathogenic Escherichia coli; ExPEC, extraintestinal pathogenic Escherichia coli; dpi, day post infection; PCA, principal component analysis; GO, gene ontology; TLR, toll-like receptor; CAM, cell adhesion molecule; TCR, T-cell receptor.
Competing Interests
The authors declare that there were no competing interests regarding the publication of this paper.
Authors’ Contributions
HS isolated RNA from tissues and generated cDNA libraries, analyzed data of the RNAseq experiment, conducted qPCR validation, and wrote the manuscript. PL, LKN, and SJL conceived the concept, participated in the animal experiments, and revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the assistance of members of the Nolan and Lamont labs in collecting tissues for this study, as well as grant support from the United States Department of Agriculture National Institute of Food and Agriculture, Hatch Project number 5203 and Grant no. 2008-35604-18805 from the USDA National Institute of Food and Agriculture Microbial Genome Program.
SUPPLEMENTARY DATA
Table S1. Differentially expressed genes that are involved in the significantly changed pathways in the contrast of 1 d post infection (dpi) susceptible vs. 1 dpi non-challenged birds.
Table S2. Differentially expressed genes that are involved in the significantly changed pathways in the contrast of 5 d post infection (dpi) vs. 1 dpi susceptible birds.
Table S3. Differentially expressed genes that are involved in the significantly changed pathways in the contrast of 5 dpi susceptible vs. 5 dpi non-challenged birds.
Table S4. Differentially expressed genes that are involved in the significantly changed pathways in the contrast of 5 dpi susceptible vs. 5 dpi resistant birds.
Supplementary data are available at PSCIEN online.
REFERENCES
- Akira S., Takeda K., Kaisho T. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- Akira S. TLR Signaling. Curr. Top. Microbiol. Immunol. 2006;311:1–16. doi: 10.1007/3-540-32636-7_1. [DOI] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alegre M. L., Frauwirth K. A., Thompson C. B. T-cell regulation by CD28 and CTLA4. Nat. Rev. Immunol. 2001;1:220–228. doi: 10.1038/35105024. [DOI] [PubMed] [Google Scholar]
- Alizadeh A. A., Eisen M. B., Davis R. E., Ma C., Lossos I. S., Rosenwald A., Boldrick J. C., Sabet H., Tran T., Yu X., Powell J. I., Yang L., Marti G. E., Moore T., Hudson Jr. J., Lu L., Lewis D. B., Tibshirani R., Sherlock G., Chan W. C., Greiner T. C., Weisenburger D. D., Armitage J. O., Warnke R., Levy R., Wilson W., Grever M. R., Byrd J. C., Botstein D., Brown P. O., Staudt L. M. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Anderson G., Moore N. C., Owen J. J., Jenkinson E. J. Cellular interactions in thymocyte development. Annu. Rev. Immunol. 1996;14:73–99. doi: 10.1146/annurev.immunol.14.1.73. [DOI] [PubMed] [Google Scholar]
- Bahar R., Hartmann C. H., Rodriguez K. A., Denny A. D., Busuttil R. A., Dollé M. E., Calder R. B., Chisholm G. B., Pollock B. H., Klein C. A., Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- Bergeron C. R., Prussing C., Boerlin P., Daignault D., Dutil L., Reid-Smith R. J., Zhanel G. G., Manges A. R. Chicken as reservoir for extraintestinal pathogenic Escherichia coli in humans, Canada. Emerg. Infect. Dis. 2012;18:415–421. doi: 10.3201/eid1803.111099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck W. J., Fu C., Jackman J. K., Flotow H., Wilkinson S. E., Williams D. H., Johnson R., Kong G., Chan A. C., Findell P. R. Phosphorylation of SLP-76 by the ZAP-70 protein-tyrosine kinase is required for T-cell Receptor function. J. Biol. Chem. 1996;271:19641–19644. doi: 10.1074/jbc.271.33.19641. [DOI] [PubMed] [Google Scholar]
- Call D. R., Brockman F. J., Chandler D. P. Detect-ing and genotyping Escherichia coli O157:H7 using multiplexed PCR and nucleic acid microarrays. Int. J. Food Microbiol. 2001;67:71–80. doi: 10.1016/s0168-1605(01)00437-8. [DOI] [PubMed] [Google Scholar]
- Chan A. C., Iwashima M., Turck C. W., Weiss A. ZAP-70: a 70 kd protein-tyrosine kinase that associates with the TCR zeta chain. Cell. 1992;71:649–662. doi: 10.1016/0092-8674(92)90598-7. [DOI] [PubMed] [Google Scholar]
- Chen L. M., Hobbie S., Galan J. E. Requirement of Cdc42 for Salmonella-induced cytoskeletal and nuclear responses. Science. 1996;274:2115–2118. doi: 10.1126/science.274.5295.2115. [DOI] [PubMed] [Google Scholar]
- Czerucka D., Dahan S., Mograbi B., Rossi B., Rampal P. Implication of mitogen-activated protein kinases in T84 cell responses to enteropathogenic Escherichia coli infection. Infect Immun. 2001;69:1298–1305. doi: 10.1128/IAI.69.3.1298-1305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. J. The mitogen-activated protein kinase signal transduction pathway. J. Biol. Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- De Koker S., Lambrecht B. N., Willart M. A., van Kooyk Y., Grooten J., Vervaet C., Remon J. P., De Geest B. G. Designing polymeric particles for antigen delivery. Chem. Soc. Rev. 2011;40:320–329. doi: 10.1039/b914943k. [DOI] [PubMed] [Google Scholar]
- Dho-Moulin M., Fairbrother J. M. Avian pathogenic Escherichia coli (APEC) Vet. Res. 1999;30:299–316. [PubMed] [Google Scholar]
- Ewers C., Li G., Wilking H., Kiessling S., Alt K., Antáo E. M., Laturnus C., Diehl I., Glodde S., Homeier T., Böhnke U., Steinrück H., Philipp H. C., Wieler L. H. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: How closely related are they? Int. J. Med. Microbiol. 2007;297:163–176. doi: 10.1016/j.ijmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- Heissmeyer V., Macian F., Im S. H., Varma R., Feske S., Venuprasad K., Gu H., Liu Y. C., Dustin M. L., Rao A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 2004;5:255–265. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- Hirahara K., Ghoreschi K., Yang X. P., Takahashi H., Laurence A., Vahedi G., Sciumè G., Hall A. O., Dupont C. D., Francisco L. M., Chen Q., Tanaka M., Kanno Y., Sun H. W., Sharpe A. H., Hunter C. A., O'Shea J. J. Interleukin-27 priming of T cells controls IL-17 production in trans via induction of the ligand PD-L1. Immunity. 2012;36:1017–1030. doi: 10.1016/j.immuni.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie S., Chen L. M., Davis R. J., Galan J. E. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J. Immunol. 1997;159:5550–5559. [PubMed] [Google Scholar]
- Hong C., Luckey M. A., Park J. H. Intrathymic IL-7: The where, when, and why of IL-7 signaling during T cell development. Semin. Immunol. 2012;24:151–158. doi: 10.1016/j.smim.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H. C., Mountz J. D. Metabolic syndrome, hormones, and maintenance of T cells during aging. Curr. Opin. Immunol. 2010;22:541–548. doi: 10.1016/j.coi.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., August A. The signaling symphony: T cell receptor tunes cytokine-mediated T cell differentiation. J. Leukoc. Biol. 2015;97:447–485. doi: 10.1189/jlb.1RI0614-293R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh J. E., Kang K. S., Chae C., Kim H. M., Ahn K. S., Kim S. H. Roles of p38 and JNK mitogen-activated protein kinase pathways during cantharidin-induced apoptosis in U937 cells. Biochem. Pharmaco. 2004;67:1811–1818. doi: 10.1016/j.bcp.2003.12.025. [DOI] [PubMed] [Google Scholar]
- Irving B. A., Weiss A. The cytoplasmic domain of the T cell receptor zeta chain is sufficient to couple to receptor-associated signal transduction pathways. Cell. 1991;64:891–901. doi: 10.1016/0092-8674(91)90314-o. [DOI] [PubMed] [Google Scholar]
- JanBen T., Schwarz C., Preikschat P., Voss M., Philipp H. C., Wieler L. H. Virulence-associated genes in avian pathogenic Escherichia coli (APEC) isolated from internal organs of poultry having died from colibacillosis. Int. J. Med. Microbiol. 2001;291:371–378. doi: 10.1078/1438-4221-00143. [DOI] [PubMed] [Google Scholar]
- Johnson T. J., Kariyawasam S., Wannemuehler Y., Mangiamele P., Johnson S. J., Doetkott C., Skyberg J. A., Lynne A. M., Johnson J. R., Nolan L. K. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 2007;189:3228–3236. doi: 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir S. M. L. Avain colibacillosis and salmonellosis: A closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. Inter. J. Env. Res. Pub. Health. 2010;7:89–114. doi: 10.3390/ijerph7010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan A., Huang W., Huang F., August A. Signal transduction via the T cell antigen receptor in naive and effector/memory T Cells. Int. J. Biochem. Cell Biol. 2012;44:2129–134. doi: 10.1016/j.biocel.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir M. E., Liang S. C., Guleria I., Latchman Y. E., Qipo A., Albacker L. A., Koulmanda M., Freeman G. J., Sayegh M. H., Sharpe A. H. Tissue expression of PD-L1 mediates peripheral T cell tolerance. J. Exp. Med. 2006;203:883–895. doi: 10.1084/jem.20051776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmett K., Humphrey T., Rushton S., Close A., Wigley P., Williams N. J. A longitudinal study simultaneously exploring the carriage of APEC virulence associated genes and the molecular epidemiology of faecal and systemic E. coli in commercial broiler chickens. PLoS ONE. 2013;8:e67749. doi: 10.1371/journal.pone.0067749. doi:10.1371/ journal.pone.0067749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmett K., Williams N. J., Chaloner G., Humphrey S., Wigley P., Humphrey T. The contribution of systemic Escherichia coli infection to the early mortalities of commercial broiler chickens. Avian Pathol. 2014;43:37–42. doi: 10.1080/03079457.2013.866213. [DOI] [PubMed] [Google Scholar]
- Kranis A., Gheyas A. A., Boschiero C., Turner F., Yu L., Smith S., Talbot R., Pirani A., Brew F., Kaiser P., Hocking P. M., Fife M., Salmon N., Fulton J., Strom T. M., Haberer G., Weigend S., Preisinger R., Gholami M., Qanbari S., Simianer H., Watson K. A., Woolliams J. A., Burt D. W. Development of a high density 600 K SNP genotyping array for chicken. BMC Genomics. 2013;14:59. doi: 10.1186/1471-2164-14-59. doi: 10.1186/1471-2164-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis J. M., Avruch J. Sounding the alarm: Protein kinase cascades activated by stress and inflammation. J. Biol. Chem. 1996;271:24313–24316. doi: 10.1074/jbc.271.40.24313. [DOI] [PubMed] [Google Scholar]
- Lanz R., Kuhnert P., Boerlin P. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 2003;91:73–84. doi: 10.1016/s0378-1135(02)00263-8. [DOI] [PubMed] [Google Scholar]
- Latchman Y. E., Liang S. C., Wu Y., Chernova T., Sobel R. A., Klemm M., Kuchroo V. K., Freeman G. J., Sharpe A. H. PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc. Natl. Acad. Sci. USA. 2004;101:10691–1096. doi: 10.1073/pnas.0307252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavric M., Maughan M. N., Bliss T. W., Dohms J. E., Bencina D., Keeler C. L., Narat M. Gene expression modulation in chicken macrophages exposed to mycoplasma synoviae or Escherichia coli. Vet. Microbiol. 2008;126:111–121. doi: 10.1016/j.vetmic.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Letourneur F., Klausner R. D. Activation of T cells by a tyrosine kinase activation domain in the cytoplasmic tail of CD3 epsilon. Science. 1992;255:79–82. doi: 10.1126/science.1532456. [DOI] [PubMed] [Google Scholar]
- Li X., Chiang H. I., Zhu J., Dowd S. E., Zhou H. Characterization of a newly development chicken 44 K Agilent microarray. BMC Genomics. 2008;9:60. doi: 10.1186/1471-2164-9-60. doi: 10.1186/1471-2164-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges A. R., Johnson J. R. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin. Infect. Dis. 2012;55:712–719. doi: 10.1093/cid/cis502. [DOI] [PubMed] [Google Scholar]
- Martinon F., Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005;26:447–454. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Melief C. J., Van Der Burg S. H., Toes R. E., Ossendorp F., Offringa R. Effectve therapeutic anticancer vaccines based on precision guiding of cytolytic T lymphocytes. Immunol. Rev. 2002;188:177–182. doi: 10.1034/j.1600-065x.2002.18816.x. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Maeda M., Imada Y., Imada T., Sato K. Pathology of spontaneous colibacillosis in a broiler flock. Vet. Pathol. 1985;22:592–597. doi: 10.1177/030098588502200614. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Imada Y., Maeda M. Lymphocytic depletion of bursa of Fabricius and thymus in chickens inoculated with Escherichia coli. Vet. Pathol. 1986;23:712–717. doi: 10.1177/030098588602300610. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Milos P. M. RNA Sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum J., De-Smedt M., Leclercq G. Exogenous IL-7 promotes the growth of CD3-CD4-CD8-CD44+CD25+/CD25- precursor cells and blocks the differentiation pathway of TCR-alpha beta cells in fetal thymus organ culture. J. Immunol. 1993;150:2706–2716. [PubMed] [Google Scholar]
- Redmond S. B., Tell T. M., Coble D., Mueller C., Palic D., Andreasen C. B., Lamont S. J. Differential splenic cytokine responses to dietary immune modulation by diverse chicken lines. Poult. Sci. 2010;89:1635–1641. doi: 10.3382/ps.2010-00846. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Siek K. E., Giddings C. W., Doetkott C., Johnson T. J., Fakhr M. K., Nolan L. K. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology. 2005;151:2097–2110. doi: 10.1099/mic.0.27499-0. [DOI] [PubMed] [Google Scholar]
- Rose M. E. The immune system in birds. J. R. Soc. Med. 1979;72:701–705. doi: 10.1177/014107687907200914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. T., Scherf U., Eisen M. B., Perou C. M., Rees C., Spellman P., Iyer V., Jeffrey S. S., Van de Rijn M., Waltham M., Pergamenschikov A., Lee J. C., Lashkari D., Shalon D., Myers T. G., Weinstein J. N., Botstein D., Brown P. O. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biology programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Russo T. A., Johnson J. R. Medical and economic impact of extraintestinal infections due to Escherichia coli: Focus on an increasingly important endemic problem. Microbes Infect. 2003;5:449–456. doi: 10.1016/s1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- Sandford E. E., Orr M., Balfanz E., Bowerman N., Li X., Zhou H., Johnson T. J., Kariyawasam S., Liu P., Nolan L. K., Lamont S. J. Spleen transcriptome response to infection with avian pathogenic Escherichia coli in broiler chickens. BMC Genomics. 2011;12:469. doi: 10.1186/1471-2164-12-469. doi: 10.1186/1471-2164-12-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandford E. E., Orr M., Shelby M., Li X., Zhou H., Johnson T. J., Kariyawasam S., Liu P., Nolan L. K., Lamont S. J. Leukocyte transcriptome from chickens infected with avian pathogenic Escherichia coli identifies pathways associated with resistance. Results Immunol. 2012;2:44–53. doi: 10.1016/j.rinim.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severa M., Remoli M. E., Giacomini E., Annibali V., Gafa V., Lande R., Tomai M., Salvetti M., Coccia E. M. Sensitization to TLR7 agonist in IFNβ-preactivated dendritic cells. J. Immunol. 2007;178:6208–6216. doi: 10.4049/jimmunol.178.10.6208. [DOI] [PubMed] [Google Scholar]
- Smith-Garvin J. E., Koretzky G. A., Jordan M. S. T Cell Activation. Annu. Rev. Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stordeur P., Bree A., Mainil J., Moulin-Schouleur M. Pathogenicity of pap-negative avian Escherichia coli isolated from septicaemic lesions. Microb. Infect. 2004;6:637–645. doi: 10.1016/j.micinf.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Sun H., Liu P., Nolan L. K., Lamont S. J. Avian pathogenic Escherichia coli (APEC) infection alters bone marrow transcriptome in chickens. BMC Genomics. 2015a;16:690. doi: 10.1186/s12864-015-1850-4. doi: 10.1186/s12864-015-1850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Liu P., Nolan L. K., Lamont S. J. Novel pathways revealed in Bursa of Fabricius transcriptome in response to extraintestinal pathogenic Escherichia coli (ExPEC) infection. PLoS ONE. 2015b;10:e0142570. doi: 10.1371/journal.pone.0142570. doi:10.1371/journal. pone.0142570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Bi R., Liu P., Nolan L. K., Lamont S. J. Combined analysis of primary lymphoid tissues’ transcriptomic response to extraintestinal Escherichia coli (ExPEC) infection. Dev. Comp. Immunol. 2016;57:99–106. doi: 10.1016/j.dci.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Sutterwala F. S., Ogura Y., Szczepanik M., Lara-Tejero M., Lichtenberger G. S., Grant E. P., Bertin J., Coyle A. J., Galán J. E., Askenase P. W., Flavell R. A. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Van Haarlem D. A., Van Kooten P. J. S., Rothwell L., Kaiser P., Vervelde L. Characterisation and expression analysis of the chicken interleukin-7 receptor alpha chain. Dev. Comp. Immunol. 2009;33:1018–1026. doi: 10.1016/j.dci.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Vicente R., Swainson L., Marty-Gres S., De Barros S. C., Kinet S., Zimmermann V. S., Taylor N. Molecular and cellular basis of T cell lineage commitment. Semin. Immunol. 2010;22:270–275. doi: 10.1016/j.smim.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent C., Boerlin P., Daignault D., Dozois C. M., Dutil L., Galanakis C., Reid-Smith R. J., Tellier P. P., Tellis P. A., Ziebell K., Manges A. R. Food reservoir for Escherichia coli causing urinary tract infections. Emerg. Infect. Dis. 2010;16:88–95. doi: 10.3201/eid1601.091118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., de Caestecker M. P., Yamada Y. Transcriptional cross-talk between Smad, ERK1/2, and p38 mitogen-activated protein kinase pathways regulates transforming growth factor-beta-induced aggrecan gene expression in chondrogenic ATDC5 cells. J. Biol. Chem. 2001;276:14466–14473. doi: 10.1074/jbc.M005724200. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Sudo T., Minato N., Ohnishi A., Katsura Y. Interleukin 7 preferentially supports the growth of gamma delta T cell receptor-bearing T cells from fetal thymocytes in vitro. Int. Immunol. 1991;3:1067–1075. doi: 10.1093/intimm/3.11.1067. [DOI] [PubMed] [Google Scholar]
- Werling D., Jungi T. W. Toll-like receptors linking innate and adaptive immune response. Vet. Immunol. Immunopathol. 2003;91:1–12. doi: 10.1016/s0165-2427(02)00228-3. [DOI] [PubMed] [Google Scholar]
- Whiteside S. T., Epinat J. C., Rice N. R., Israel A. I kappa B epsilon, a novel member of the I kappa B family, controls RelA and cRel NF-kappa B activity. EMBO. J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Chen S., White D. G., Zhao S., McDermott P., Walker R., Meng J. Characterization of multiple-antimicrobial-resistant Escherichia coli isolates from diseased chickens and swine in China. J. Clin. Microbiol. 2004;42:3483–3489. doi: 10.1128/JCM.42.8.3483-3489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehn D., Lee S. Y., Bevan M. J. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Sloan-Lancaster J., Kitchen J., Trible R. P., Samelson L. E. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.