Abstract
Awamori is a traditional distilled beverage made from steamed Thai-Indica rice in Okinawa, Japan. For brewing the liquor, two microbes, local kuro (black) koji mold Aspergillus luchuensis and awamori yeast Saccharomyces cerevisiae are involved. In contrast, that yeasts are used for ethanol fermentation throughout the world, a characteristic of Japanese fermentation industries is the use of Aspergillus molds as a source of enzymes for the maceration and saccharification of raw materials. Here we report the draft genome of a kuro (black) koji mold, A. luchuensis NBRC 4314 (RIB 2604). The total length of nonredundant sequences was nearly 34.7 Mb, comprising approximately 2,300 contigs with 16 telomere-like sequences. In total, 11,691 genes were predicted to encode proteins. Most of the housekeeping genes, such as transcription factors and N-and O-glycosylation system, were conserved with respect to Aspergillus niger and Aspergillus oryzae. An alternative oxidase and acid-stable α-amylase regarding citric acid production and fermentation at a low pH as well as a unique glutamic peptidase were also found in the genome. Furthermore, key biosynthetic gene clusters of ochratoxin A and fumonisin B were absent when compared with A. niger genome, showing the safety of A. luchuensis for food and beverage production. This genome information will facilitate not only comparative genomics with industrial kuro-koji molds, but also molecular breeding of the molds in improvements of awamori fermentation.
Keywords: Aspergillus luchuensis, kuro (black) koji mold, genome sequence
1.Introduction
Aspergillus luchuensis is a kuro (black) koji mold that is used widely for brewing Japanese traditional spirits, awamori, in the Okinawa islands, which are located in the most southern part of Japan.1–3 Recently, A. luchuensis has also been employed to produce shochu, a distilled beverage manufactured in the Kyushu islands. A. luchuensis produces vast amounts of enzymes, which facilitate the maceration and saccharification of raw materials, such as rice, wheat, and sweet potato. Similar to A. niger, A. luchuensis produces an abundance of citric acid, which maintains the fermentation mash at a low pH to prevent contamination by wild microorganisms. Interestingly, the glycosidases produced by A. luchuensis have higher catalytic activity in a more acidic pH range than those secreted by A. oryzae, which is used for brewing Japanese sake, rice wine.4,5 These characteristics enable highly effective and reliable fermentation in warm latitudes, such as Okinawa and the Kyushu islands. There are approximately 50 awamori brewers in Okinawa who originally used their own strains, but recently, they have employed koji-seed (conidiospores) supplied by companies. The market size of awamori is 13,000 Myen (130 Mdollar), and the total market size of Japanese traditional spirits, including Shochu, is 470,000 Myen (4,700 Mdollar). Furthermore, the high potential of its secretory enzymes and the safety of A. luchuensis make this microorganism extremely important for modern biotechnology.
Historically, A. luchuensis was isolated primarily from awamori koji in the Okinawa islands by Inui in 1901,6 who stated that A. luchuensis is the major fermentation agent in awamori production and that it possesses uniseriate conidial heads. In the same year, Usami also isolated two major kuro koji molds from awamori koji and reported that black Aspergillus No. 1 must be A. luchuensis Inui.7 In 1913, Nakazawa obtained α and β strains from awamori koji, and the α strain is an important mold for awamori fermentation.8 He designated the α strain as A. awamori and rejected A. luchuensis as a kuro koji mold because the α strain possessed biseriate conidial heads, although its morphological characteristics were similar to those of A. luchuensis. These kuro koji molds exhibit clear differences from standard A. niger strains, where the conidial surface is smooth and unable to assimilate nitrate in its early culture stage.9,10 In 1980, Al-Musallam revised the taxonomy of black Aspergillus, and A. awamori was synonymized as A. niger var. awamori based on NRRL4948 from the Instituto Ozwaldo Cruz, Brazil, rather than Nakazawa’s α strain.11 A recent phylogenetic analysis of several known genes, including the RTS of ribosomal DNA, demonstrated that the A. awamori strains in NRIB and NITE (NBRC) formed two major clusters, one of which co-clustered with A. niger, whereas the other clade co-clustered with A. kawachii and industrial kuro koji molds.1 Thus, the classification of kuro koji molds is in dispute. In 2012, Hong et al. re-described A. luchuensis as an industrially important black Aspergillus in East Asia with biseriate conidial heads, and A. kawachii was synonymized as A. luchuensis.12,13 Extrolite analysis of A. luchuensis strains showed that they do not produce mycotoxins, and thus, they can be considered safe for use in food and beverage fermentation. Hong et al. also proposed that A. awamori is a cryptic phylogenetic species in the section Nigri, and they rejected the species name, A. awamori.
In this study, we sequenced A. luchuensis NBRC 4314 (RIB 2604), which was received from Usami as A. awamori No. 1, and it was stored at the IFO (NBRC).14 The genome size was approximately 34.7 Mb, which is almost the same as that of A. niger CBS 513.88, and the average shared identity at the nucleic acid level was 88.9%. We identified some genes that are important for awamori fermentation, and we analyzed the protein N-and O-glycosylation systems in detail. We also determined the mating locus and confirmed the nonproduction of mycotoxins.
2. Materials and methods
2.1. Strain and DNA preparation
A. luchuensis NBRC 4314 (RIB 2604) was used as the DNA donor. Genomic DNA preparation and removal of mitochondrial DNA were performed as described previously.15,16
2.2. Genome sequencing
The genome of A. luchuensis was sequenced using the whole-genome shotgun (WGS) approach by accumulating raw sequence reads with a depth of coverage of approximately 5.7×. The linkage between contigs was analyzed by fingerprinting and PCR, as well as by optical mapping (OpGen).
2.3 Gene prediction and annotation
GeneDecoder17 and GlimmerHMM18 were used for gene prediction. Automated annotation was performed using NCBI BLASTP with the A. niger CBS 513.88 genomic database.19 Interproscan20 was used for functional analysis of the predicted proteins. The number of genes in each cluster of orthologous group (COG)21 category was analyzed by BLASTP using the amino acid sequences in the COG set with bit scores ≥60. Comparative analysis of A. luchuensis and A. niger was conducted with MUMmer tools.22 AntiSmash23 was used to search for secondary metabolite biosynthesis gene clusters.
Results and discussion
3.1. Genome sequence and gene prediction
The A. luchuensis genome was sequenced using the WGS approach. The 34.7 Mb genome was predicted to contain 11,691 genes that encoded proteins with a length greater than 100 amino acid residues (Table 1). The genome was confirmed as comprising eight chromosomes (chromosomes 1–8 in decreasing order of size).
Table 1.
Properties of the genomes of Aspergillus luchuensis NBRC 4314, A. niger CBS 513.88, and A. kawachii NBRC 4308
| A. luchuensis | A. niger | A. kawachii | |
|---|---|---|---|
| Genome size (Mb) | 34.7 | 33.9 | 36.6 |
| G + C content (%) | 49.7 | 50.4 | 49.9 |
| Gene models | 11,691 | 13,160 | 11,255 |
| Protein length (amino acids) | 483.6 | 466.4 | 500.1 |
| Exons per gene | 2.8 | 3.7 | 3.1 |
| Exon length (bp) | 660.2 | 533.0 | 660.1 |
| Intron length (bp) | 113.8 | 94.6 | 80.7 |
Genes that encoded polypeptides longer than 100 amino acids were used in the analysis.
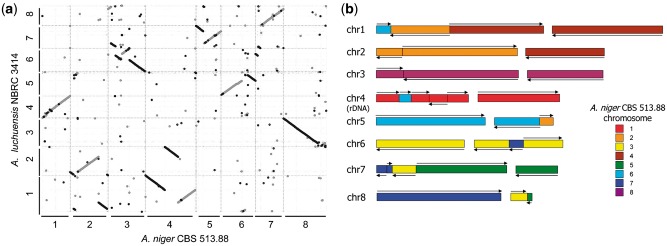
The average shared identity at the nucleic acid level between A. luchuensis NBRC 4314 and A. niger CBS 513.88 was estimated as 88.9% using the dnadiff wrapper script in the MUMmer 3.0 package. MUMmer also showed that the correspondence between the chromosomes of A. luchuensis and A niger was extremely low compared with that between those of A. niger CBS 513.88 and A. niger ATCC 1015,24 which were almost the same (Fig. 1). For example, chromosome 1 of A. luchuensis appeared to be composed of portions of chromosome 6, 2 and 4 of A. niger. These results are consistent with the suggestion that A. luchuensis is a different species from A. niger.
Figure 1.
(A) Dot-plot alignments of chromosomes in Aspergillus luchuensis NBRC 4314 and A. niger CBS 513.88 using MUMmer 3.0. (B) Schematic representation of A. luchuensis NBRC 4314 chromosomes. The colored areas refer to the chromosomes assigned to A. niger CBS 513.88.
The COG classification detected no major differences between A. niger and A. oryzae (Table 2). For instance, most of the transcription factors were found in the genomes of A. luchuensis, as well as A. niger, and A. oryzae (Supplementary Table S1).25,26
Table 2.
Gene numbers for each COG in Aspergillus luchuensis NBRC 4314, A. niger CBS 513.88, and A. kawachii NBRC 4308
| A. luchuensis | A. niger | A. kawachii | ||
|---|---|---|---|---|
| Information storage and processing | ||||
| J | Translation, ribosomal structure, and biogenesis | 308 | 302 | 300 |
| A | RNA processing and modification | 210 | 203 | 207 |
| K | Transcription | 240 | 238 | 244 |
| L | Replication, recombination, and repair | 193 | 193 | 190 |
| B | Chromatin structure and dynamics | 93 | 88 | 91 |
| Cellular processes and signaling | ||||
| D | Cell cycle control, cell division, chromosome partitioning | 159 | 156 | 152 |
| Y | Nuclear structure | 26 | 27 | 26 |
| V | Defense mechanisms | 50 | 57 | 49 |
| T | Signal transduction mechanisms | 370 | 370 | 372 |
| M | Cell wall/membrane/envelope biogenesis | 93 | 97 | 92 |
| N | Cell motility | 1 | 1 | 1 |
| Z | Cytoskeleton | 107 | 111 | 110 |
| W | Extracellular structures | 4 | 4 | 5 |
| U | Intracellular trafficking, secretion, and vesicular transport | 282 | 273 | 284 |
| O | Posttranslational modification, protein turnover, chaperones | 453 | 456 | 444 |
| Metabolism | ||||
| C | Energy production and conversion | 393 | 396 | 390 |
| G | Carbohydrate transport and metabolism | 285 | 284 | 291 |
| E | Amino acid transport and metabolism | 386 | 380 | 373 |
| F | Nucleotide transport and metabolism | 87 | 87 | 89 |
| H | Coenzyme transport and metabolism | 114 | 116 | 113 |
| I | Lipid transport and metabolism | 384 | 356 | 373 |
| P | Inorganic ion transport and metabolism | 144 | 143 | 142 |
| Q | Secondary metabolite biosynthesis, transport, and catabolism | 461 | 446 | 433 |
| Poorly characterized | ||||
| R/Sa | General function prediction only/function unknown | 1,417 | 1,448 | 1,401 |
| Xb | Gene with unknown function | 6,161 | 7,663 | 5,806 |
| COG hit | 5,530 | 5,497 | 5,446 | |
| Total | 11,691 | 13,160 | 11,252 | |
Genes that encoded polypeptides longer than 100 amino acids were used in the analysis. Genes that shared homology with ≥2 COGs were counted redundantly in each COG.
aGenes that shared homology with COGs other than R/S were excluded.
bGenes that shared no homology with any COGs including the genes sharing homology with R/S.
COG, cluster of orthologous group.
3.2. Glycosidases and citric acid production
During the fermentation of awamori and shochu, the most important role of the kuro koji mold A. luchuensis is to produce sufficient glycolytic enzymes to convert starch in materials, such as glucose, as well as generating citric acid, which maintains the fermentation mash at a low pH. Previously, it was reported that A. luchuensis secretes a characteristic acid-stable α-amylase (AA1_SCon_015_0343), which can function at an acidic pH due to citric acid production. This acid-stable α-amylase has a starch-binding domain in the C-terminus. By contrast, A. oryzae possesses three copies of the taka-amylase gene, where the amyA gene comprises an amy gene cluster with the amyR (transcription factor of amylase genes) and agdA (α-glucosidase) genes. A. luchuensis was also predicted to possess an amy gene cluster comprising AA1_SCon_020_0630 (amyR), AA1_SCon_020_0632 (α-glucosidase), and AA1_SCon_020_0632.5 (α-amylase). Interestingly, the homology between AA1_SCon_020_0632.5 and the amyA gene product (taka-amylase) was relatively low at 73.5%, but a gene almost identical to taka-amylase was found on contig AA1_SCon_049.Con002, which was not assigned to the chromosome. Similarly, A. kawachii has a taka-amylase orthologue, AKAW_11452, in addition to an amy gene cluster and acid-stable α-amylase. Three other putative α-amylase genes (AA1_SCon_008_0403, AA1_SCon_018_0661, and AA1_SCon_011_0021) were also found, which shared 39%–46% identity with taka-amylase according to a BLASTP search. They possessed three conserved amino acid residues in the catalytic active site of α-amylase. A. oryzae possesses two glucoamylase genes, where glaA is expressed in submerged culture and glaB is expressed in solid-state culture.27 We found that A. luchuensis contained only one glucoamylase gene (AA1_SCon_031_0271), which had a starch-binding domain in the C-terminus, whereas A. oryzae lacks this domain. More detailed studies are required to clarify the functions of these glycolytic enzymes in awamori and shochu fermentation.
During the production of awamori and shochu koji (solid-state culture of Aspergillus mold on grain), the temperature is typically increased to 40 °C in the early cultivation stages to increase the activity of amylase and then lowered to 30 °C in the later stages to promote the production of citric acid. However, the method employed for citric acid production is poorly understood in A. luchuensis, and the detailed mechanism is also unclear in A. niger. Kirimura reported that a cyanide- and antimycin A-insensitive and salicylhydroxamic acid-sensitive respiratory pathway catalyzed by an alternative oxidase (AOX1) functions in mitochondria, where the cytochrome pathway and inhibition of AOX1 dramatically decreased citric acid production in A. niger, although mycelial growth was not affected.28 The orthologue of AOX1, AA1_SCon_015_0483, may also play an important role in citric acid production by A. luchuensis. The members of the glycolytic system and tricarboxylic acid cycle in A. luchuensis are listed in Supplementary Table S2. More detailed functional studies of these enzymes are required.
3.3 Protease genes
Genome sequencing of A. luchuensis NBRC 4314 showed that this strain contained almost the same protease genes as A. niger CBS 513.88. We estimated the putative amino acid sequences from the A. luchuensis genome sequence based on the fact that the amino acid sequences around the catalytic residues in the active sites of proteases are conserved.29 Black Aspergillus, A. luchuensis, and A. niger possess many acidic proteases. The typical acidic proteases, aspartic endopeptidases,30,31 and serine-type carboxypeptidases32,33 have an optimum pH of below pH 4.0, and we found 10 and 13 of each, respectively, in the A. luchuensis genome. In addition, A. niger and A. oryzae possess 10 and 11 aspartic protease genes, respectively, and 12 serine carboxypeptidase genes. Each of the 12 serine-type carboxypeptidase genes in A. luchuensis had orthologs in the other two Aspergillus strains, and one additional gene in A. luchuensis was a homologue of gene corresponding to ocpB.34 These two genes might have been generated by gene duplication. The sedolysin family enzymes are also acidic proteolytic enzymes, and the optimum pH range for these enzymes in A. fumigatus was reported as pH 5.5–6.0.35 A. luchuensis and A. niger have seven sedolysin family enzyme genes, whereas A. fumigatus Af 239 and A. oryzae RIB 40 have only four and five genes, respectively. The neutral and alkaline protease genes were not as abundant as the acidic proteases. Three serine endopeptidase (kexin,36 an oryzin37 ortholog, and a putative vacuole enzyme38) genes were found in the A. luchuensis genome, similar to other Aspergillus strains. Among the metalloendopeptidases, there was one thermolysin-type enzyme gene,39 but deuterolysins40 were not found, similar to the A. niger genome. The genes of theses enzyme have been found in A. sojae, A. oryzae,41 and A. fumigatus,42 and even Penicillium.43 As described above, A. luchuensis and A. niger share similar protease genes and black Aspergillus strains might differ genetically from other Aspergillus strains. However, in terms of glutamic peptidase genes,44 A. luchuensis appeared to possess both A. niger-type and A. oryzae-type genes. A. niger and A. oryzae possess three glutamic peptidase genes, and A. luchuensis has five genes. The phylogenetic tree of the genes for these enzymes in the three Aspergillus strains is shown in Figure 2. AA1_SCon_006_0020- and AA1_SCon_019_0031-type genes are found in all three Aspergillus strains. A. niger lacks a counterpart of AA1_SCon_039_0035, whereas it is present in A. oryzae. By contrast, genes corresponding to AA1_SCon_011_0044 are found in A. niger but not in A. oryzae. AA1_SCon_006_0811 is an orthologue of AA1_Scon_019_0031. These results might indicate that glutamic peptidase genes have been amplified by gene duplication in Aspergillus strains, but A. niger and A. oryzae possess a reduced number of unnecessary genes, whereas they have been conserved in A. luchuensis. Thus, Aspergillus strains might be divided into A. oryzae and black Aspergillus groups and black Aspergillus might have lost the deuterolysin genes. A. luchuensis might have maintained glutamic peptidase genes that resemble the genes in A. oryzae. Glutamic peptidases are also acidic proteases, so maintaining genes for both types of enzyme might have improved the ability in acidic condition.
Figure 2.
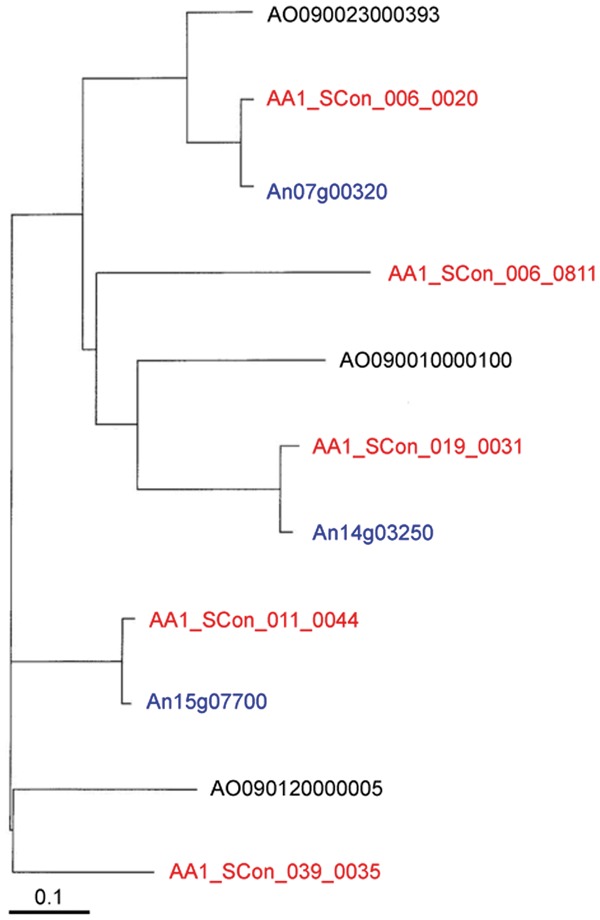
Phylogenetic tree of glutamic peptidase of Aspergillus luchuensis NBRC 4314 (red), A. niger CBS 513.88 (blue), and A. oryzae RIB 40 (black).
3.4. Protein N-and O-glycosylation
A. luchuensis and other Aspergillus species produce large amounts of extracellular proteins, such as glucoamylase and α-amylase. During the secretion process, the proteins are glycosylated in the endoplasmic reticulum (ER) and the Golgi apparatus. Protein glycosylation is believed to play critical roles in various cell activities, such as quality control in secretory proteins, cell wall integrity, environmental adaptation, and pathogenicity in some pathogenic fungi. These roles are related to the function, protein stability, and appropriate localization of secretory proteins, and antigenicity is modulated by protein glycosylation. Analysis of the A. luchuensis genome found that the genes involved in the protein N- and O-glycosylation are conserved relative to those in yeast, Saccharomyces cerevisiae, although the N- and O-glycan structures attached to the proteins differ between these two members of Ascomycota.45,46
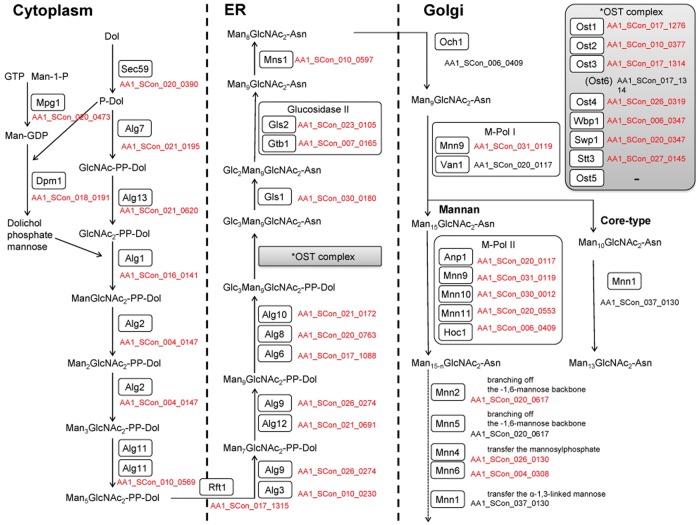
The N-glycan structure in S. cerevisiae is characterized as a high mannose type. Protein N-glycosylation is initiated in the ER by an oligosaccharyltransferase (OST) complex using Glc3Man9GlcNAc2-PP-Dolichol as a sugar donor. The OST complex comprises at least eight subunits in yeast. The orthologue of the catalytic subunit, Stt2, in the OST complex has been characterized in A. fumigatus.47 All of the genes that encode the seven subunits other than yeast-specific OST5 are conserved in A. luchuensis (Fig. 3). After the transfer of Glc3Man9GlcNAc2 to Asn residues in a polypeptide, three glucose residues and one mannose residue are removed by the actions of the Gls2–Gtb1 complex and Mns1, respectively. This process is crucial for the quality control of secretory proteins in the ER and for the ER-associated degradation of misfolded proteins via recognition by lectins. The orthologues of yeast GlS2, GTB1, and MNS1 have been functionally characterized in A. oryzae.48,49 Thus, the genes (AA1_SCon_023_0105, AA1_SCon_007_0165, and AA1_SCon_010_0597) responsible for the trimming of N-glycans in the ER are well conserved in A. luchuensis. Glycoproteins that enter the Golgi apparatus are subjected to further elongation with N-glycans. Yeast glycoproteins localized in the cell wall and the periplasmic space contain a long branched polymer with approximately 200 mannoses and phosphate in the N-glycans. However, the N-glycans in Aspergillus species are Man5-24GlcNac2-N, which are shorter in size than those found in yeast.50,51 The differences in the structure of N-glycans between these fungi are due to the presence of the long α-1,6-mannan and a unique phosphorylation pattern in yeast. The long α-1,6-mannan and phosphate residues in N-glycans are synthesized via the actions of Och1 and Mnn6, respectively (Fig. 3). The A. fumigatus OCH1 orthologue compensates for the defective OCH1 in yeast.52 A. luchuensis also possesses an OCH1 orthologue (AA1_SCon_006_0409), so this fungus may have the ability to elongate N-glycans in the same manner as yeast. Given that A. luchuensis possesses all of the genes involved in the elongation of N-glycans in the same manner as yeast and that the genes are expressed according to microarray analysis (Supplementary Table S3), then it is likely that the degradation of N-glycans by α-mannosidases will yield short N-glycans in Aspergillus species. S. cerevisiae possesses at least two genes, DCW1 and DFG5, encoding putative GPI-anchored α-1,6-mannosidases.53 We found that the A. luchuensis genome contained 11 ORFs with good hits against Dcw1 according to BLASTP searches using S. cerevisiae sequence datasets (Supplementary Table S4). Microarray data showed that all of these genes were expressed in the solid-state culture, koji. Among the 11 putative α-mannosidases, 10 α-mannosidases possessed the predicted signal sequence, thereby implying that these are secretory proteins. Four α-mannosidases, that is, AA1_SCon_019_0007, AA1_SCon_027_0100, AA1_SCon_016_0076, and AA1_SCon_020_0838, possessed a potential GPI-modification site as found in yeast, similar to Dcw1 and Dfg5, whereas no GPI-modification site was predicted in the other seven α-mannosidases. Thus, the localizations and functions of these α-mannosidases may differ from those in yeast, Dcw1 and Dfg5. Experimentally based functional characterizations of these mannosidases will allow us to understand the reason why Aspergillus species contain short N-glycans compared with those in yeast.
Figure 3.
Putative N-glycosylation pathway in Aspergillus luchuensis. The proteins involved in N-glycosylation in A. luchuensis were identified by BLASTP search using the amino acid sequences of the components of the N-glycosylation pathway in Saccharomyces cerevisiae.45,46 The proteins detected as reciprocal best BLAST hits are shown in red. The orthologues of Stt3, Gls2, Gtb1, Mns1, and Och1 were also confirmed by BLASTP analysis using the functionally characterized genes from A. oryzae and A. fumigatus. See Supplementary Table S3 for details of the BLASTP results. Asn, asparagine; Dol, dolichol; ER, endoplasmic reticulum; GDP, guanosine diphosphate; GlcNAc, N-acetylglucosamine; GTP, guanosine triphosphate; Man, mannose; OST, oligosaccharyltransferase; P, phosphate.
O-glycans attached to proteins from Aspergillus spp. are characterized by the presence of branching forms of oligosaccharides. The O-glycans comprise Man1-O, Manα1-2Man1-O, Manα1-6Man1-O, Man1-6(Glc1-3)Man1-O, Man1-6(Galp1-3)Man1-O, Man1-6Man1-6Glc1-3Man1-O, and Manα1-2(Manα1-6)Man1-O.54 The O-glycan found in the glycoproteins of S. cerevisiae possesses a linear chain with up to five mannose residues, the structure of which is Manα1-3Manα1-3Manα1-2Manα1-2Man1-O. Protein O-glycosylation is initiated in the ER by the action of the protein O-mannosyltransferase (Pmt) using dolichol phosphate mannose as a sugar donor.55 Seven pmt genes are present in S. cerevisiae, which are classified into three protein subfamilies, i.e., Pmt1, Pmt2, and Pmt4 subfamilies. Each member of the Pmt4 subfamily forms a dimer complex with proteins from the same subfamily, whereas each member of the Pmt1 subfamily forms a dimer complex with Pmt2 subfamily members. The simultaneous disruption of more than three pmt genes yields a lethal phenotype in yeast. However, filamentous fungi only possess three pmt genes from each of the Pmt subfamilies. The three pmt genes characterized in A. nidulans and A. fumigatus play crucial roles in hyphal development, morphology, and asexual conidiation in these fungi.56–60 Our genome analysis demonstrated that A. luchuensis also possesses three pmt genes, among which pmtA has been characterized (Supplementary Table S5).61 The disruption of pmtA in A. awamori (luchuensis) does not affect the extracellular secretion level of glucoamylase, but it has negative effects on the growth rate, cell morphology, and conidia formation. After initial mannosylation by Pmt in the ER, further glycosylation occurs due to the action of sugar transferases in the Golgi apparatus during the secretory process. Multiple α-1,2- and α-1,3-mannosyltransferases have been characterized in yeast, but a sugar transferase responsible for this elongation process has not been characterized in Aspergillus spp. Our genome analysis of A. luchuensis detected three orthologues of yeast α-1,2 mannosyltransferase62 (Fig. 4).
Figure 4.
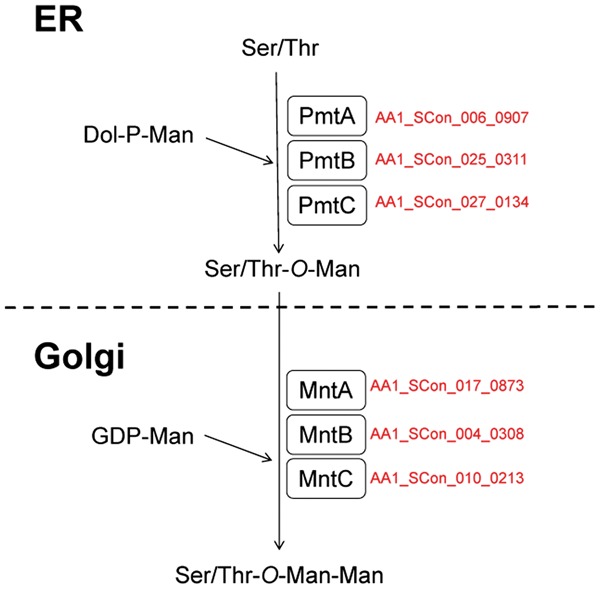
Putative O-glycosylation pathways in Aspergillus luchuensis. To identify the orthologues involved in O-glycosylation in A. luchuensis, BLASTP search was performed using the amino acid sequences of the components of the O-glycosylation pathway in A. nidulans as queries (Oka et al. 2004, Goto 2007, Goto et al. 2009). The protein names of the reciprocal best BLAST hits for the ORFs in A. nidulans are shown in red. See Supplementary Table S5 for details of the BLAST results. Dol, dolichol; ER, endoplasmic reticulum; GDP, guanosine diphosphate; Man, mannose; P, phosphate.
It has been reported that Galf residues are included specifically in the N- and O-glycans in Aspergillus spp. as well as closely related filamentous fungi.51,63 A single 1,2-linked Galf residue is present at the nonreducing terminus in N-glycans and terminally β-1,5-linked Galf residues are present in O-mannose-type glycans. The Galf residues present in pathogenic A. fumigatus are immunogenic in mammals, and they are assumed to be involved in pathogenicity in humans, so the inhibition and detection of this sugar’s biosynthesis might be exploited in the development of chemotherapy and diagnostics related to Aspergillus infections.64 The gfsA gene that encodes galactofuranosyltransferases involved in the addition of Galf to O-mannose-type glycans has been identified in A. nidulans and A. fumigatus.65 We found that A. luchuensis genome possesses three homologues of the A. nidulans gfs (Supplementary Table S5).
3.5. Mating-type loci
A. luchuensis is considered to be asexual, and it might be heterothallic in the same manner as A. oryzae. In heterothallic ascomycetes, the mating type is determined by the alternative presence of either of two genes called idiomorphs, which occupy the same locus in the chromosome, but that shares no sequence similarity with each other. The MAT1-1 gene encodes a protein with an alpha box domain, and the MAT1-2 gene encodes a protein with a high mobility group (HMG) domain. We found the MAT1-2 HMG gene (AA1_SCon_025_0346) in the A. luchuensis NBRC 4314 genome. To investigate the possibility of breeding A. luchuensis strains by mating, we searched for the opposite mating type gene in 28 strains classified as A. luchuensis by PCR using the primer set: MAT1-1F-An (5′-GCGGCCACTGAACAGTTTCATTGCT-3′) and MAT1-1R-An (5′-TGATGGAGTATGCCTTGGCTACGATG-3′). Interestingly, we found no MAT1-1 strains, and all 28 of the A. luchuensis strains that we analyzed had the MAT1-2 mating type gene. Thus, our previous efforts to mate A. luchuensis were unsuccessful because we tried to cross strains with the same mating type. It was also suggested that if we could isolate strains with MAT1-1, this would allow the improvement of A. luchuensis strains by sexual crossing.
3.6. Safety and nonproductivity of mycotoxins
The secondary metabolites produced by filamentous fungi, especially mycotoxins, are most important from a safety viewpoint. A. niger has been reported to produce two types of mycotoxin, i.e., ochratoxin A (OTA) and fumonisin B.66,67 Previously, we reported that the synthesis of OTA by A. niger is mediated partly by polyketide synthase, which is encoded by the An15g07920 gene, and we showed that A. luchuensis lacks this gene based on PCR and Southern analyses.1 Interestingly, in the genome of A. niger CBS 513.88, An15g07920 forms a gene cluster with the cytochrome P450 and nonribosomal peptide synthase genes, whereas A. niger ATCC 1015 lacks this gene cluster. Genome sequencing demonstrated A. luchuensis has also lost a 21-kb region of the OTA cluster in a similar manner to both A. niger ATCC 1015 and A. kawachii IFO 4308.3,24 Thus, these results show clearly that A. luchuensis lacks the ability to produce OTA.
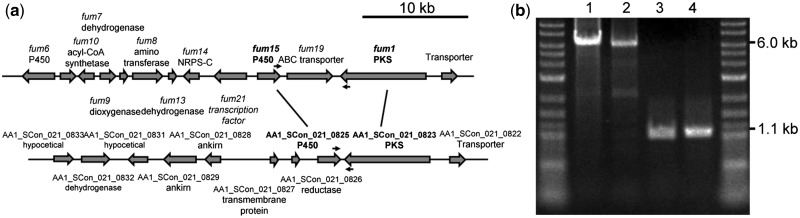
The fumonisins are a group of polyketide-derived mycotoxins, which were first isolated in 1988 from Fusarium verticillioides.68 In contrast to the OTA gene cluster, the genomes of both A. niger CBS 513.88 and ATCC 1015 include putative homologues of the F. verticillioides fumonisin gene cluster, and the actual production of fumonisin B2 has been confirmed. The fumonisin gene cluster of A. niger contains at least 14 fum genes. We searched the A. luchuensis genome for orthologues of A. niger fumonisin biosynthesis cluster genes using bidirectional best hit analysis. Only the fum1 and fum15 gene products shared high identity with AA1_SCon_021_0823 (68% identity, polyketide synthase) and AA1_SCon_021_0825 (72% identity, cytochrome P450) . The other FUM proteins shared low identities of 20%–43%, and these orthologues were distributed throughout the A. luchuensis genome. Analyses of the secondary metabolite biosynthesis gene clusters using antiSMASH only predicted that AA1_SCon_021_0823 and AA1_SCon_021_0825 comprised a gene cluster (Fig. 5A). These results indicate that A. luchuensis lacks most of the fum genes and productivity of fumonisin, even if these two gene products are functional. Interestingly, the orthologues of fum1 and fum15 lie next to each other in opposite directions in the A. luchuensis genome, whereas the fum9 gene is located between fum1 and fum15 genes in A. niger. According to this feature, we designed a primer set, i.e., Fum1_cons_primer (5′-GGCGGCATTGAGATCAGCACATTGGA-3′) and Fum15_cons_primer (5′-GAAGGTAACCCGCACAGTAACTGCCAGGCC-3′), to amplify a 1.1-kb gene fragment from the A. luchuensis genome as a template and a 6.0-kb fragment from A. niger (Fig. 5B). Using this primer set, all of the A. luchuensis strains had the same 1.1-kb signal, thereby indicating that A. luchuensis has the same genome structure, and it lacks the ability to produce fumonisin (data not shown). Recently, Shimizu et al. reported that disruption of the fum8 gene, which encodes α-oxoamine synthase, resulted in the loss of fumonisin B2 from A. niger, but A. luchuensis appears to lack this gene.69 Susca et al. suggested that fumonisin production was once more widespread among black aspergilli and that nonrandom partial deletion of this cluster has occurred multiple times based on partial fumonisin gene cluster homologues identified in several black Aspergillus species genomes.70
Figure 5.
(A) The fumonisin gene cluster in A. niger CBS 513.88 and the genes around fum1 and fum15 homologues in Aspergillus luchuensis NBRC 3414. The predicted proteins are also indicated. Fum1_cons_primer and Fum15_cons_primer are indicated by black arrows. (B) Results of PCR amplification using the primer set Fum1_cons_primer and Fum15_cons_primer: lane 1, A. niger CBS 513.88; lane 2, A. niger ATCC 1015; lane 3, A. luchuensis NBRC 4314; and lane 4, A. kawachii genomic DNA as a template.
The ex-type strain of A. luchuensis is NBRC 4281 (RIB 2642), but we used NBRC 4314 in the present study, which is the oldest strain, and it was isolated in 1901. We also confirmed that Nakazawa’s α strain was stored as JCM 2261 (IAM 2112)71 and that it belongs to A. luchuensis. JCM 22320 (IAM 2351) was reported by Sakaguchi in 1949, and it is now known that the kuro koji mold that survived World War II is also A. luchuensis.72 In 1975, Sugama isolated two types of kuro koji mold from awamori koji (awamori type and acid-producing saitoi type), and we confirmed that both of these strains belong to A. luchuensis.73 Tukahara reported that ISH1 (awamori kin, a high glycosidase-producing strain) and ISH2 (saitoi kin, a high acid-producing strain), which are the strains used most widely for awamori fermentation at present, are also A. luchuensis according to next-generation sequencing74 Thus, A. luchuensis had been used historically and widely in awamori production, which clearly indicates that A. luchuensis is most appropriate for making awamori. The genome sequence obtained in this study will provide a platform to help understand the important characteristics of A. luchuensis, such as citric acid production. A transformation system and an efficient gene-targeting system for A. luchuensis have already been established.75 We hope that more advanced molecular biological research will be conducted using the kuro koji mold, A. luchuensis.
The sequences and annotations reported in this article have been deposited in DDBJ/EMBL/GenBank under the accession nos BCWF01000001-BCWF01000044. Information about the sequences and gene annotations are also available in the genome database of microorganisms sequenced at NITE (DOGAN; http://www.bio.nite.go.jp/dogan/top).
Supplementary Material
Acknowledgements
The authors thank Gen Okada (Japan Collection of Microorganisms, RIKEN BioResource Center) and Sayaka Ban (Biological Resource center, National Institute of Technology and Evaluation) for details of their research and information about Aspergillus strains, as well as Ryoko Ikeda and Risa Hayashi (National Research Institute of Brewing) for technical assistance.
Accession numbers
BCWF01000001-BCWF01000044
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
References
- 1.Yamada O., Takara R., Hamada R., Hayashi R., Tsukahara M., Mikami S. 2011, Molecular biological researches of Kuro-koji molds, their classification and safety, J. Biosci. Bioeng., 112, 233–7. [DOI] [PubMed] [Google Scholar]
- 2.Futagami T., Mori K., Wada S., et al. 2014, Transcriptomic Analysis of Temperature Responses of Aspergillus kawachii during Barley Koji Production, Appl. Environ. Microbiol., 81, 1353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Futagami T., Mori K., Yamashita A., et al. 2011, Genome Sequence of the White Koji Mold Aspergillus kawachii IFO 4308, Used for Brewing the Japanese Distilled Spirit Shochu, Eukaryot. Cell, 10, 1586–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machida M., Yamada O., Gomi K. 2008, Genomics of Aspergillus oryzae. learning from the history of Koji mold and exploration of its future, DNA Res., 15, 173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneko A., Sudo S., Takayasu-Sakamoto Y., Tamura G., Ishikawa T., Oba T. 1996, Molecular cloning and determination of the nucleotide sequence of a gene encoding an acid-stable α-amylase from Aspergillus kawachii, J. Ferment. Bioeng., 81, 292–8. [Google Scholar]
- 6.Inui T., 1901, Ryukyu awamori hakko kin chyosa houkoku. J. Chem. Soc. Japan, 4, 1421–30 (in Japanese). [Google Scholar]
- 7.Usami K., 1901, Awamori-shu jyozo kenkyu houkoku. J. Chem. Soc. Japan, 4, 1437–61 (in Japanese). [Google Scholar]
- 8.Nakazawa R., 1913, On awamori-koji fungus (1st report). Government Research Institute, Formosa, Report, 2, 93–7 (in Japanese). [Google Scholar]
- 9.Sakaguchi K., Iizuka H., Yamazaki S., 1950, A study on black Aspergilli., J. Agric. Chem. Soc. Japan., 24, 138–42 (in Japanese). [Google Scholar]
- 10.Murakami H., 1979, Classification system of the black Aspergilli. Taxonomic studies on Japanese industrial strains of the Aspergillus (Part 32). J. Brew. Soc. Japan., 74, 849–53 (in Japanese). [Google Scholar]
- 11.Al-Musallam A., 1980, Revision of the black Aspergillus species. Ph.D. Thesis, Utrecht University.
- 12.Hong S. B., Lee M. A., Kim D.H., et al. 2013, Aspergillus luchuensis, an industrially important black Aspergillus in East Asia. PLoS ONE, e63769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong S. B., Yamada O., Samson R. A., 2014, Taxonomic re-evaluation of black koji molds, Appl. Microbiol. Biotechnol., 98, 555–61. [DOI] [PubMed] [Google Scholar]
- 14.Naganishi H., 1927, Awamori-kin no tohka-so ni tsuite., Soc. Biosci. Bioeng. Japan, 5, 155–84. [Google Scholar]
- 15.Iimura Y., Gomi K., Uzu H., Hara S. 1987, Transformation of Aspergillus oryzae through plasmid-mediated complementation of the methionine-auxotrohpic mutation, Agric. Biol. Chem., 51, 323–8. [Google Scholar]
- 16.Watson J., Thompson W. F., 1986, Purification and restriction endonuclease analysis of plant nuclear DNA, Methods Enzymol., 118, 57–75. [Google Scholar]
- 17.http://www.cbrc.jp/cbrc/team/researcher/asai/index.ja.html
- 18.Majoros W.H., Pertea M., Salzberg S.L., 2004, TigrScan and GlimmerHMM: two open-source ab initio eukaryotic gene-finders, Bioinformatics, 20, 2878–9. [DOI] [PubMed] [Google Scholar]
- 19.Pel H. J., Winde J. H., Archer D. B., et al. 2007, Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88, Nat. Biotechnol., 25, 221–31. [DOI] [PubMed] [Google Scholar]
- 20.Zdobnov E.M., Apweiler R., 2001, InterProScan-an integration platform for the signature-recognition methods in InterPro, Bioinformatics, 17, 847–8. [DOI] [PubMed] [Google Scholar]
- 21.Tatusov R. L., Fedorova N. D., Jackson J. D., et al. 2003, The COG database: an updated version includes eukaryotes. BMC Bioinformatics, 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtz S., Phillippy A., Delcher A. L., et al. 2004, Versatile and open software for comparing large genomes, Genome Biol., 5, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medema M. H., Blin K., Cimermancic P., et al. 2011, antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters, Nucl. Acids Res., 39, W339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen M. R., Salazar M. P., Schaap P. J. et al. 2011, Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88, Genome Res., 21, 885–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galagan J. E., Calvo S,E., Cuomo C., et al. 2005, Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae, Nature, 438, 1105–15. [DOI] [PubMed] [Google Scholar]
- 26.Machida M., Asai K., Sano M., et al. 2005, Genome sequencing and analysis of Aspergillus oryzae, Nature, 438, 1157–61. [DOI] [PubMed] [Google Scholar]
- 27.Ishida H., Hata Y., Ichikawa E., Kawato A., Suginami K., Imayasu S. 1998, Regulation of the glucoamylase-encoding gene (glaB), expressed in solid-state culture (koji) of Aspergillus oryzae, J. Ferment. Bioeng., 86, 301–7. [DOI] [PubMed] [Google Scholar]
- 28.Kirimura K., Yoda M., Shimizu H., et al. 2000, Contribution of cyanide-insensitive respiratory pathway, catalyzed by the alternative oxidase, to citric acid production in Aspergillus niger, Biosci. Biotechnol. Biochem., 64, 2034–9. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashsi T., Abe K., Asai, et al. 2009, Genomics of Aspergillus oryzae, Biosci. Biotechnol. Biochem., 71, 646–70. [DOI] [PubMed] [Google Scholar]
- 30.Ichishima E. 1970, Purification and mode of assay for acid proteinase of Aspergillus saitoi, Methods Enzymol. 19, 397–406. [Google Scholar]
- 31.Takeuchi M., Ogura K., Hamamoto T., Kobayashi Y. 1995, Molecular cloning and sequence analysis of a gene encoding an aspartic proteinase from Aspergillus oryzae, Adv. Exp. Med. Biol., 326, 577–80. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi M., Ushijima T., Ichishima E. 1982, A new acid carboxypeptidase, O-1, from Aspergillus oryzae, Curr. Microbiol., 7, 19–23. [Google Scholar]
- 33.Chiba Y., Midorikawa T., Ichishima E. 1995, Cloning and expression of the carboxypeptidase gene from Aspergillus saitoi and determination of the catalytic residues by site-directed mutagenesis, Biochem. J., 308, 405–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morita H., Okamoto A., Yamagata, et al. 2009, Heterologous expression and characterization of CpI, OcpA, and novel serine-type carboxypeptidase OcpB from Aspergillus oryzae, Appl. Microbiol. Biotechnol., 85, 335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reichard U., Léchenne B., Asif A. R., a. et al. 2006, Sedolisins, a new class of secreted proteases from Aspergillus fumigatus with endoprotease or tripeptidyl-peptidase activity at acidic pHs, Appl. Environ. Microbiol., 72, 1739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizutani O., Nojima A., Yamamoto M., et al. 2004, Disordered cell integrity signaling caused by disruption of the kexB gene in Aspergillus oryzae, Eukaryot. Cell, 3, 1036–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa Y. 1970, Alkaline proteinases from Aspergillus, Methods Enzymol., 19, 581–91. [Google Scholar]
- 38.Moehle C. M., Tizard R., Lemmon S. K., Smart J., Jones E. W. 1987, Protease B of the lysosomelike vacuole of the yeast Saccharomyces cerevisiae is homologous to the subtilisin family of serine proteases, Mol. Cell Biol., 7, 4390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakadai T., Nasuno S., Iguchi N. 1973, Purification and properties of neutral proteinase I from Aspergillus oryzae, Agric. Biol. Chem., 37, 2695–701. [Google Scholar]
- 40.Sekine H. 1972, Neutral protease I and II of Aspergillus sojae, Agric. Biol. Chem., 36, 198–206. [Google Scholar]
- 41.Tatsumi H., Murakami S., Tuji, et al. 1991, Cloning and expression in yeast of a cDNA clone encoding Aspergillus oryzae neutral protease II, a unique metalloprotease, Mol. Gen. Genet., 228, 97–103. [DOI] [PubMed] [Google Scholar]
- 42.Ramesh M. V., Sirakova T. D., Kolattukudy P. E. 1995, Cloning and characterization of the cDNAs and genes (mep20) encoding homologous metalloproteinases from Aspergillus flavus and A. fumigatus, Gene, 165, 121–5. [DOI] [PubMed] [Google Scholar]
- 43.Gripon J. C., Hermier J. 1974, Le système protéolytique de Penicillium roqueforti. III. Purification, propriétés et spécificité d’une protéase inhibée par l’E.D.T.A, Biochimie, 56, 1324–32. [PubMed] [Google Scholar]
- 44.Fujinaga M., Cherney M. M., Oyama H., Oda K., James M. N. 2004, The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum, Proc. Natl Acad. Sci. USA, 101, 3364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jigami Y. 2008, Yeast glycobiology and its application, Biosci. Biotechnol. Biochem., 3, 637–48. [DOI] [PubMed] [Google Scholar]
- 46.Deshpande N., Wilkins M. R., Packer N., Nevalainen H. 2008, Protein glycosylation pathways in filamentous fungi. Glycobiology, 8, 626–37. [DOI] [PubMed] [Google Scholar]
- 47.Li K., Ouyang H, Lü Y., Liang J., Wilson I. B., Jin C. 2011, Repression of N-glycosylation triggers the unfolded protein response (UPR) and overexpression of cell wall protein and chitin in Aspergillus fumigatus, Microbiology, 7, 1968–79. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe T., Totani K., Matsuo I., Maruyama J., Kitamoto K., Ito Y. 2009, Genetic analysis of glucosidase II beta-subunit in trimming of high-mannose-type glycans Glycobiology, 8, 834–40. [DOI] [PubMed] [Google Scholar]
- 49.Akao T., Yahara A., Sakamoto K., Yamada O., Akita O., Yoshida T. 2012, Lack of endoplasmic reticulum 1,2-α-mannosidase activity that trims N-glycan Man9GlcNAc2 to Man8GlcNAc2 isomer B in a manE gene disruptant of Aspergillus oryzae, J. Biosci. Bioeng., 4, 438–41. [DOI] [PubMed] [Google Scholar]
- 50.Maras M., van Die I., Contreras R., van den Hondel C. A. 1990, Filamentous fungi as production organisms for glycoproteins of bio-medical interest, Glycoconj. J., 16, 99–107. [DOI] [PubMed] [Google Scholar]
- 51.Wallis G. L., Easton R. L., Jolly K., Hemming F. W., Peberdy J. F. 2001, Galactofuranoic-oligomannose N-linked glycans of alpha-galactosidase A from Aspergillus niger, Eur. J. Biochem., 268, 4134–143. [DOI] [PubMed] [Google Scholar]
- 52.Lambou K., Perkhofer S., Fontaine T., Latge J. P. 2010, Comparative functional analysis of the OCH1 mannosyltransferase families in Aspergillus fumigatus and Saccharomyces cerevisiae, Yeast, 8, 625–36. [DOI] [PubMed] [Google Scholar]
- 53.Kitagaki H., Wu H., Shimoi H., Ito K. 2002, Two homologous genes, DCW1 (YKL046c) and DFG5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae, Mol. Microbiol., 46, 1011–22. [DOI] [PubMed] [Google Scholar]
- 54.Goto M. 2007, Protein O-glycosylation in fungi: diverse structures and multiple functions, Biosci. Biotechnol. Biochem., 6, 1415–27. [DOI] [PubMed] [Google Scholar]
- 55.Willer T., Valero M. C., Tanner W., Cruces J., Strahl S. 2003, O-mannosyl glycans: from yeast to novel associations with human disease Curr. Opin. Struct. Biol., 13, 621–30. [DOI] [PubMed] [Google Scholar]
- 56.Oka T., Hamaguchi T., Sameshima Y., Goto M., Furukawa K. 2004, Molecular characterization of protein O-mannosyltransferase and its involvement in cell-wall synthesis in Aspergillus nidulans, Microbiology, 6, 1973–82. [DOI] [PubMed] [Google Scholar]
- 57.Zhou H., Hu H., Zhang L., et al. 2007, O-Mannosyltransferase 1 in Aspergillus fumigatus (AfPmt1p) is crucial for cell wall integrity and conidium morphology, especially at an elevated temperature, Eukaryot, Cell, 6, 2260–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goto M., Harada Y., Oka T., Matsumoto S., Takegawa K., Furukawa K. 2009, Protein O-mannosyltransferases B and C support hyphal development and differentiation in Aspergillus nidulans, Eukaryot. Cell, 10, 1465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kriangkripipat T., Momany M. 2009, Aspergillus nidulans protein O-mannosyltransferases play roles in cell wall integrity and developmental patterning, Eukaryot. Cell, 8, 1475–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mouyna I., Kniemeyer O., Jank T., et al. 2010, Members of protein O-mannosyltransferase family in Aspergillus fumigatus differentially affect growth, morphogenesis and viability, Mol. Microbiol., 76, 1205–1221. [DOI] [PubMed] [Google Scholar]
- 61.Oka T., Sameshima Y., Koga T., Kim H., Goto M., Furukawa K. 2005, Protein O-mannosyltransferase A of Aspergillus awamori is involved in O-mannosylation of glucoamylase I, Microbiology, 151, 3657–3667. [DOI] [PubMed] [Google Scholar]
- 62.Häusler A., Ballou L., Ballou C. E., Robbins P. W. 1992, Yeast glycoprotein biosynthesis: MNT1 encodes an alpha-1,2-mannosyltransferase involved in O-glycosylation, Proc. Natl Acad. Sci. US A, 89, 6846–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leitao E. A., Bittencourt V. C., Haido R. M., et al. 2003, Beta-galactofuranose-containing O-linked oligosaccharides present in the cell wall peptidogalactomannan of Aspergillus fumigatus contain immunodominant epitopes, Glycobiology, 13, 681–692. [DOI] [PubMed] [Google Scholar]
- 64.Tefsen B., Ram A. F., van Die I., Routier F. H. 2012, Galactofuranose in eukaryotes: aspects of biosynthesis and functional impact, Glycobiology, 22, 456–469. [DOI] [PubMed] [Google Scholar]
- 65.Komachi Y., Hatakeyama S., Motomatsu H., et al. 2013, gfsA gene encodes a novel galactofuranosyltransferase involved in the biosynthesis of galactofuranose antigen of O-glycan in Aspergillus nidulans and Aspergillus fumigatus. Mol. Microbiol. 90, 1054–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nielsen K. F., Mogensen J. M., Johansen M., Larsen T. O., Frisvad J. C. 2009, Review of secondary metabolites and mycotoxins from the Aspergillus niger group, Anal Bioanal Chem, 395, 1225–1242. [DOI] [PubMed] [Google Scholar]
- 67.Frisvad J. C., Smedsgaard J., Samson R. A., Larsen T. O., Thrane U. 2007, Fumonisin B2 production by Aspergillus niger, J. Agric. Food Chem., 55, 9727–9732. [DOI] [PubMed] [Google Scholar]
- 68.Gelderblom W. C. A., Jaskiewicz K., Marasas W. F. O., et al. 1988, Fumonisins-novel mycotoxins with cancer-promoting activity produced by Fusarium moniliforme, Appl. Environ. Microbiol., 54, 1806–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shimizu K., Nakagawa H., Hashimoto R., et al. 2015, The a-oxoamine synthase gene fum8 is involved in fumonisin B2 biosynthesis in Aspergillus niger, Mycoscience, 56, 301–308. [Google Scholar]
- 70.Susca A., Proctor R. H., Butchko, et al. 2014, Variation in the fumonisin biosynthetic gene cluster in fumonisin-producing and nonproducing black aspergilli, Fungal Genet. Biol., 73, 39–52. [DOI] [PubMed] [Google Scholar]
- 71.http://www.jcm.riken.jp/cgi-bin/jcm/jcm_number?JCM=2261.
- 72.Sakaguchi K., Iizuka H., Yamazaki S. 1946, A study on kuro koji molds (I), J. Appl. Mycol., 3, 53–63 (Japanese). [Google Scholar]
- 73.Sugama S., Nishiya S., Ohba T., et al. 1975, Research of awamori koji, J. Chem. Soc. Japan, 70, 595–598 (in Japanese). [Google Scholar]
- 74.Tukahara M., Nezuo M., 2012, Comparative genomics of Kuro-koji molds used for production of Awamori, traditional distilled spirit in Okinawa, Biosci. Indust., 70, 273–277 (in Japanese). [Google Scholar]
- 75.Takahashi T., Mizutani O., Shiraishi Y., Yamada O. 2011, Development of an efficient gene-targeting system in Aspergillus luchuensis by deletion of the non-homologous end joining system, J. Biosci. Bioeng., 112, 529–534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.