Abstract
The brown alga, Cladosiphon okamuranus (Okinawa mozuku), is economically one of the most important edible seaweeds, and is cultivated for market primarily in Okinawa, Japan. C. okamuranus constitutes a significant source of fucoidan, which has various physiological and biological activities. To facilitate studies of seaweed biology, we decoded the draft genome of C. okamuranus S-strain. The genome size of C. okamuranus was estimated as ∼140 Mbp, smaller than genomes of two other brown algae, Ectocarpus siliculosus and Saccharina japonica. Sequencing with ∼100× coverage yielded an assembly of 541 scaffolds with N50 = 416 kbp. Together with transcriptomic data, we estimated that the C. okamuranus genome contains 13,640 protein-coding genes, approximately 94% of which have been confirmed with corresponding mRNAs. Comparisons with the E. siliculosus genome identified a set of C. okamuranus genes that encode enzymes involved in biosynthetic pathways for sulfated fucans and alginate biosynthesis. In addition, we identified C. okamuranus genes for enzymes involved in phlorotannin biosynthesis. The present decoding of the Cladosiphon okamuranus genome provides a platform for future studies of mozuku biology.
Keywords: brown alga, Cladosiphon okamuranus, genome decoding, genes for enzymes of polysaccharide synthesis
1. Introduction
Taxonomically, brown algae (Phaeophyceae) belong to the Stramenopiles and include multicellular species.1 They are photosynthetic organisms with chloroplasts surrounded by four membranes,2 suggesting that they originated from a symbiotic relationship between two eukaryotes.3 Most brown algae contain the pigment, fucoxanthin, which is responsible for the distinctive greenish-brown color that gives brown algae their common name. Phaeophytes include many types of seaweed in the Northern Hemisphere, and they are important members of marine ecosystems, both because they create habitats for other organisms and because they provide food.4
Cladosiphon okamuranus (Chordariales, Phaeophyceae),5 Okinawa mozuku in Japanese, is one of the important edible seaweeds. In Okinawa, C. okamuranus has been cultivated for more than 35 years by several fishermen’s associations, including those in Onna and Chinen Villages. This cultivation history has established several strains of mozuku that have similar morphology and texture. It was reported in the 36th annual report of the Japanese Cabinet Office that approximately 20 kilotons of mozuku (C. okamuranus and Nemacystus decipiens (‘Itomozuku’)) are produced annually, yielding approximately 4 billion Japanese yen in 2006. In addition, C. okamuranus and N. decipiens are sources of fucoidan,6 a sulfated polysaccharide found in the cell-wall matrix of brown algae that has anti-coagulant, anti-thrombin-like, and tumor-suppressant activities.7 Brown algae also produce alginates.8,9
Due to their biological significance, genomes of two species of brown algae have been decoded: Ectocarpus siliculosus (Order Ectocarpales10) and Saccharina japonica (Order Laminariales11). The genome size of former is approximately 214 Mbp with 16,256 predicted protein-coding genes, while that of the latter is 545 Mbp with 18,733 predicted protein-coding genes. Several genetic features of the two brown algae have been characterized to understand their biology.10,11 A close phylogenetic relationship between Ectocarpales and Chordariales has been reported.5,12 Given its importance for fisheries, food, and possible pharmaceuticals, we decoded the draft genome of Cladosiphon okamuranus S-strain, (Order Chordariales).
2. Materials and methods
2.1. Strain and DNA extraction
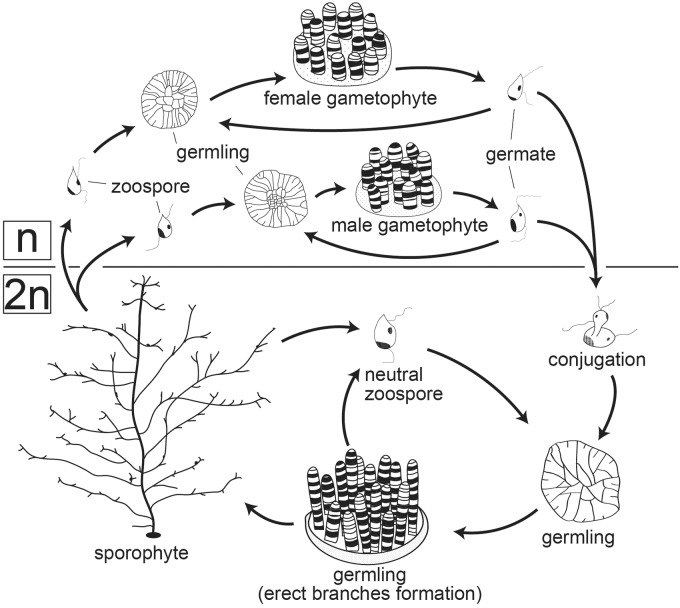
The S-strain of Cladosiphon okamuranus (‘Shikenjo-kabu’) has been maintained as a stock culture at the Okinawa Prefectural Fisheries Research and Extension Center, Okinawa, Japan. It is cultivated at 22.5° C with a 12-h light-dark cycle in sea water containing 0.5% KW21 (Daiichi Seimo Co., Ltd). The life cycle of C. okamuranus includes both haploid (n) and diploid (2n) generations (Fig. 1). 2n germlings mature into sporophytes that are harvested for market. For DNA extraction, 2n germlings of C. okamuranus were frozen in liquid nitrogen and crushed to powder with a frozen-cell-crusher, Cryo-Press (Microtec Co., Ltd). Genomic DNA was extracted from the powder using an extraction kit, DNA-Suisui-VS (Rizo Co., Ltd).
Figure 1.
A diagram showing the life cycle of the brown alga, Cladosiphon okamuranus. The alga has n and 2n generations. Cladosiphon okamuranus is cultivated and sporophytes are harvested for market. Genomic DNA was extracted from 2n germlings, while RNA was extracted from 2n germlings and 2n sporophytes.
2.2. Genome sequencing and assembly
The Illumina platform (Miseq and Hiseq 2500) was used for sequencing.13 Libraries were prepared according to slight modifications of protocols provided by the manufacturer. Fragmented genomic DNA was further purified using Blue Pippin (Sage Science). A paired-end library consisting of clones ∼720 bp was prepared for the Miseq using a TruSeq DNA PCR-Free LT Sample Prep Kit (Illumina), and 3-kb and 8-kb mate-pair libraries were prepared for the Hiseq 2500 using a Nextera Mate Pair Sample Prep Kit (Illumina), respectively (Supplementary Table S1). Longer reads were obtained by using more reagent kits for the Hiseq. K-mer counting and estimation of genome size were performed using JELLYFISH 2.2.0 software.14,15
Adapter sequences were trimmed from all reads using Trimmomatic-0.30.16 Paired-end reads of high quality (quality value ≥ 20) were assembled de novo using Newbler 2.9 (GS Assembler) to create contigs. Then subsequent scaffolding of the Newbler output was performed using SSPACE 3.0,17 based on Illumina mate-pair information. Gaps inside scaffolds were closed using GapCloser 1.12.18 Diploid sequences of gap-closed scaffolds were merged with Haplomerger-2-20151124.19 CEGMA 2.5 software20 was used to evaluate genome assembly. The mitochondrial genome was generated with the IDBA_UD 1.1.1 assembler.21
2.3. Transcriptome analyses
RNA was isolated from sporophytes (2–5 cm) and 2n germlings (Fig. 1). Total RNA was extracted following the instructions of the manufacturer using DNase and an RNeasy Plant mini kit (QIAGEN). Transcriptome libraries were prepared using a TruSeq Stranded mRNA Library Prep kit (Illumina). RNAs were sequenced as per the manufacturer’s instructions for the Illumina Hiseq 2500. Only sequences of high quality (quality value ≥ 20) were assembled, using Velvet 1.2.1022 and Oases 0.2.08.23
2.4. Gene model prediction
A set of gene model predictions (C. okamuranus Gene Model ver. 1) was generated using AUGUSTUS 3.2.1.24 AUGUSTUS was trained on the 9120 transcriptome contigs recommended by PASA 2.0.225 for this purpose. Gene models were produced by running AUGUSTUS on a repeat-masked genome, produced with RepeatMasker 4.0.6 (http://ftp.genome.washington.edu/RM/RepeatMasker.html) and refined with PASA.
2.5. Transposable elements and repetitive sequences
Repetitive sequences were detected as described previously.26 Tandem repeats were detected using Tandem Repeat Finder (version 4.04),27 and then classified using RepeatModeler-1.1.8 (http://www.repeatmasker.org/RepeatModeler.html). A de novo repeats library was generated with RepeatScout (version 1.0.5).28 Transposons and SINE in the scaffold were identified with CENSOR (version 4.2)29 using Blast searches against the Repbase TE library (version 16.05).30
2.6. Gene annotation and identification
Three approaches, individually or in combination, were used to annotate protein-coding genes in the C. okamuranus genome. The primary approach for identification of putative C. okamuranus orthologous genes was reciprocal BLAST analysis. This was carried out using mutual best hits of genes of E. siliculosus10 against the C. okamuranus gene models (BLASTP) or the assembly (TBLASTN). A second approach used for genes encoding proteins with one or more specific protein domains, was to screen the models against the Pfam database (Pfam-A.hmm, release 24.0; http://pfam.sanger.ac.uk),31 which contains approximately 11,000 conserved domains using HMMER (hmmer3).32
In the case of complex multigene families, a third annotation method was employed; sets of related sequences were subjected to phylogenetic analyses in order to more precisely determine orthologous relationships between proteins of C. okamuranus and E. siliculosus. For this purpose, amino acid sequences were aligned using MAFFT33 with default options. Gaps and ambiguous areas were excluded using SeaView version 4.5.334 manually. Based on alignment datasets, phylogenetic trees were constructed by the maximum likelihood method. A maximum likelihood phylogenetic tree was constructed with MEGA 5.235 using the best model with 1,000 bootstrap duplications.
2.7. Genome browser
A genome browser has been established for the assembled genome sequences using the JavaScript-based Genome Browser (JBrowse) 1.11.6.36 The assembled sequence and gene models are accessible at http://marinegenomics.oist.jp/gallery/.
2.8. Gene expression analysis
RNA-seq reads were aligned to the genome using TopHat-2.0.9,37 while FPKM values were calculated from the aligned results using Cufflinks v2.0.0.38 Gene expression levels were visualized with R 3.2.2 (https://www.r-project.org/) using the pheatmap 1.0.8 package.39,40
3. Results and discussion
3.1. Genome sequencing and assembly
The Illumina Miseq (average library size, 720 bp) generated a total of 9.75 gigabase pairs (Gbp) of paired-end sequence data (read length, 309 ×2 bases) and the Hiseq2500 generated 5.79 Gbp of mate-pair sequence data (read length, 283 ×2 bases); 5.02 Gbp for the 3-kb library (physical coverage, 177×) and 0.78 Gbp for the 8-kb library (physical coverage, 73×), respectively. A total of 15.6 Gbp of sequence data was obtained (Supplementary Table S1). As mentioned below, the genome size of Cladosiphon okamuranus was estimated as approximately 140 Mb (Supplementary Fig. S1A). Total reads of 15.6 Gbp correspond to approximately 100x sequencing coverage of the genome.
Illumina whole-genome, shotgun paired-end reads were assembled de novo with Newbler. The assembler generated 31,858 contigs with an N50 size of 21.7 kb (Table 1). The longest contig was 943.8 kb and approximately 40% of the sequences were covered by contigs >2 kb. Subsequent scaffolding of the 31,858 contigs of Newbler output was performed with SSPACE using Illumina mate-pair sequence information (Supplementary Table S1). Gaps inside scaffolds were closed with GapCloser. As a result, the final assembly contained 732 scaffolds with an N50 size of 536 kb. The total scaffold length reached 170 Mb.
Table 1.
Comparison of draft genome assemblies of three species of brown algae, Cladosiphon okamuranus (Order Chordariales), Ectocarpus siliculosus (Order Ectocarpales), and Saccharina japonica (Order Laminariales)
| Cladosiphon okamuranusa | Ectocarpus siliculosusb | Saccharina japonicac | |
|---|---|---|---|
| Genome size (Mb) | 140 | 214 | 545 |
| Total assembled length (Mb) | 129.9 | 195.8 | 537 |
| Number of scaffolds | 541 | 1,561 | 13,327 |
| N50 scaffold size (kb) | 416 | 504 | 252 |
| Number of contigs | 31,858 | 14,043 | 29,670 |
| N50 Contig size (bp) | 21,705 | 32,862 | 58,867 |
| Number of genes | 13,640 | 16,256 | 18,733 |
| GC Contents (%) | 54 | 54 | 50 |
| Repeated sequences (%) | 4.1 | 22.7 | 39 |
| Average gene length (bp) | 7,696 | 6,859 | 9,587 |
| Average coding sequence length (bp) | 2,004 | 1,563 | — |
| Average number of introns per gene | 8.26 | 6.98 | — |
| Average intron length (bp) | 522 | 703.8 | 1,057 |
| Cegma Partial (%) | 91.9 | 91.5 | 79.0 |
| Cegma Completeness (%) | 84.3 | 77.4 | 45.6 |
Bacterial contaminant scaffolds were identified in the genome of an Ectocarpus siliculosus strain.10 To detect scaffolds of likely bacterial origin, 732 scaffolds were classified into one large group (129.9 Mb containing 541 scaffolds) and seven smaller groups (3.6–8.0 Mb) using MaxBin software,41 which bins assembled microbial scaffolds, based on tetranucleotide frequencies, scaffold coverage levels and marker sequences. Using transcriptome mapping of C. okamuranus (See later) and similarities to Ectocarpus genes, it was confirmed that the large group contained Cladosiphon okamuranus scaffolds. All scaffolds of smaller groups had no mapped transcriptomes that showed exon–intron structures. For 541 scaffolds of C. okamuranus, the number of contigs was 2,774 and the N50 of contigs was 88 kb. The longest scaffold was 2.8 Mbp, and the scaffold N50 was 416 kb (Table 1). Approximately 87% of scaffold sequences were covered with contigs of >20 kb. Chloroplast and mitochondrial genome sequences were included in scaffold IDs, Cok_S_s60 (599,436 bp) and Cok_S_s1074 (36,249 bp), respectively (data not shown).
CEGMA analysis was carried out to evaluate the accuracy of the assembled genome (129.9 Mb). CEGMA reported 91.9% sequences (CEGMA partial) with an average degree of completeness for all sequences of 84.3%. On the other hand, CEGMA completeness values for genome sequences of E. siliculosus and S. japonica are 77.4% and 45.6% (Table 1), respectively, suggesting the assembled genome of C. okamuranus is the highest quality brown algal genome to date.
3.2. Genome size
The genome size of C. okamuranus was estimated by K-mer analysis (K-mer = 25). The peak appeared around 47 (Supplementary Fig. S1A). The estimated genome size was ∼140 Mb. The C. okamuranus genome is the smallest phaeophyte genome sequenced to date (Table 1). The genome size of E. siliculosus was estimated to be 214 Mbp while that of S. japonica was 545 Mb (Table 1).10,11
3.3. GC content
GC content of the C. okamuranus genome was estimated to be ∼54% (Supplementary Fig. S1B and Table 1). GC contents for E. siliculosus and S. japonica were 54% and 50%, respectively; thus, the three brown algae are similar in this regard (Table 1).
3.4. RNAseq, clustering, and mapping
Transcriptomic analysis is essential to determine which genes are expressed by a given organism. RNAs extracted from germlings and sporophytes were sequenced on an Illumina Hiseq 2500 (read length, 329 ×2 bases from both samples) (Supplementary Table S1). About 44.8 giga nucleotide reads were obtained.
An assembly of transcriptome sequences yielded 59,590 contigs (92.8 mega nucleotides (nts)) with an N50 size of 2,231 nts. Of those, 58,095 (97.4%) had a blat alignment (with default settings) to the assembled genome sequences (129.9 Mb). 37,299 of the 59,590 contigs had predicted ORFs from start to stop codons of at least 450 nts. Of these putatively full-length RNA-seq contigs, 36,648 (98.6%) had a blat alignment to assembled scaffolds (129.9 Mb). These data were used to produce gene models for annotation.
3.5. Gene modeling
Assembled RNAseqs of C. okamuranus, algae EST datasets available on NCBI and putative protein coding loci found with PASA, were incorporated as AUGUSTUS ‘hints.’ The number of gene models was 13,640 (Table 1). This is somewhat smaller than the 16,256 predicted genes of E. siliculosus and the 18,733 of S. japonica, respectively, although this value is comparable. The 12,813 genes of C. okamuranus were supported by mRNA sequences.
The average (arithmetic mean) length of C. okamuranus genes that included both exons and introns, was 7,696 bp, and that of exons translated to form proteins (coding sequence) was 2,004 bp. The average lengths of C. okamuranus 5′ and 3′UTRs were 911 bp and 459 bp, respectively. Cock et al.10 noted that E. siliculosus genes are intron-rich, with an average number of introns per gene of 6.98 and an average intron length of 704 bp (Table 1). This was the case in the C. okamuranus genome; the average number of introns per gene was 8.26 and the average intron length was 522 bp (Table 1).
3.6. Transposable elements and other genomic components
We examined the proportion of transposable elements and repetitive elements in the assembled mozuku genome. DNA transposons and retrotransposons apparently account for 0.539% and 2.491% of the C. okamuranus genome, respectively (Supplementary Table S2). DNA transposons included hAT (0.104% of the assembled sequences), EnSpm (0.090%), Helitron (0.084%), PIF-Harbinger (0.056%) and others, while retrotransposons included LTR retrotransposons such as Gypsy (1.096%), Copia (0.752%) and Bel_Pao (0.115%), and non-LTR retrotransposons such as CR1 (0.040%) and I (0.018%). The percentages of LINE (long interspersed nuclear elements) such as L1 (0.123%), Tx1, and Jockey, and SINE (short interspersed nuclear elements) were smaller in the C. okamuranus genome, compared to other brown algal genomes.
Including unclassified repeats, 4.1% of the C. okamuranus genome was composed of repetitive sequences (Table 1). This ratio is lower than in the two other brown algae, 22.7% for E. siliculosus and 39% for S. japonica, respectively. It may be that the high frequency of repeated sequences in brown algal genomes is not universal among the Phaeophyceae.
3.7. Identification of Cladosiphon okamuranus genes
In order to examine the utility of the assembled genome as a platform for future studies in mozuku biology, we searched for genes in the assembled C. okamuranus genome. By comparing genes found in the E. siliculosus genome,10 we found the corresponding mozuku genes associated with transcription factors, signaling molecules, and enzymes involved in polysaccharide biosynthesis and phlorotannin biosynthesis.
3.7.1 Transcription factors
Transcription factors play pivotal roles in many biological processes. We examined transcription factors (TFs) by searching conserved protein domains using hmmer3 and the Pfam database (e-value cutoff < e − 5). The number of domains that have been identified in transcription factors was determined and compared with those in the E. siliculosus genome. These domains include HSF, Myb, bZIP, Zinc Finger, bHLH, CCAAT-binding, homeobox, AP2-EREBP, Nin-like, TAF, E2F-DP, CBF/NF-Y/archaeal and Sigma-70 r2/r3/r4 (Table 2).
Table 2.
Number of transcription factors (TFs) in the brown algae, Cladosiphon okamuranus and Ectocarpus siliculosus
| Species | ||||
|---|---|---|---|---|
| Family | Cladosiphon okamuranus | Ectocarpus siliculosus | ||
| Myb | 58 | (27.1%)a | 59 | (22.7%) |
| CBF/NF-Y/archaeal | 35 | (16.3%) | 42 | (16.2%) |
| Zn_finger, C2H2-type | 23 | (10.7%) | 30 | (11.5%) |
| Zn_finger, CCCH-type | 22 | (10.3%) | 24 | (9.2%) |
| bZIP | 17 | (7.9%) | 23 | (8.8%) |
| HSF | 13 | (6.1%) | 22 | (8.5%) |
| TAF | 10 | (4.7%) | 18 | (6.9%) |
| Sigma-70 r2/r3/r4 | 9 | (4.2%) | 10 | (3.8%) |
| Zn_finger, TAZ-type | 7 | (3.3%) | 7 | (2.7%) |
| Nin-like | 5 | (2.3%) | 1 | (0.4%) |
| bHLH | 3 | (1.4%) | 2 | (0.8%) |
| Homeobox | 3 | (1.4%) | 7 | (2.7%) |
| AP2-EREBP | 3 | (1.4%) | 4 | (1.5%) |
| E2F-DP | 3 | (1.4%) | 4 | (1.5%) |
| CXC/tesmin | 3 | (1.4%) | 4 | (1.5%) |
| Zn_finger, GATA-type | 0 | (0.0%) | 1 | (0.4%) |
| Fungal TRF | 0 | (0.0%) | 2 | (0.8%) |
| Total | 214 | 260 | ||
aPercentage of the family within the brown algal genome.
This analysis showed that the C. okamuranus genome contains 214 transcription factor genes (Table 2), fewer than the 260 found in the E. siliculosus genome. However, the ratio of transcription factor genes to total genes was ∼1.6% in both genomes. The most abundant TF in both genomes was the Myb family, with 58 and 59 genes in the C. okamuranus and E. siliculosus genomes, respectively. The next most abundant TFs were CBF/NF-Y/archaeal (35), Zinc Finger C2H2-type (23) and Zinc Finger CCCH-type (22). The C. okamuranus genome likely contains three genes for bHLH domains, three for homeobox domains and 17 genes for bZIP domains. The expression level of bZIP genes seems to be similar in germlings and sporophytes, whereas some of CBF/NF-Y/archaeal genes may have different expression levels (Fig. 2A). Expression and function of phaeophyte TF genes should be expressly examined in future studies.
Figure 2.
Heat maps compared with gene expression levels of germlings and sporophytes of Cladsiphon okamuranus based on FPKM values. Gene IDs for the genome browser are indicated by sXXX_XXXXX. (A) Expression levels of transcription factors shown in Table 2. (B) Expression levels of receptor kinase genes shown in Supplementary Fig. S2. (C) Expression levels of C. okamuranus genes potentially contributing to biosynthetic pathways of sulfated fucans, alginates, and phlorotannin shown in Figs. 3- -5 and Supplementary Table S3.
3.7.2 Signaling molecules
Cell–cell signaling molecules play pivotal roles in organismal development and cellular physiological activity. Cock et al.10 specifically analyzed membrane-spanning receptor kinase genes, since receptor kinases have been shown to function in developmental processes in both animals and green plants. They showed that Ectocarpus receptor kinases form a monophyletic clade, as in the case of animals and plants, suggesting that brown algae evolved independently of other taxa. The present domain analysis, using the Pfam database, showed that the C. okamuranus and E. siliculosus genomes contain 286 and 338 genes for protein kinase domains, which are both equivalent to about 2.1% of total genes.
Cock et al.10 also identified five genes for stramenopile-lineage-specific receptor kinases in E. siliculosus.10 The C. okamuranus genome likely contains at least two genes for brown algal-specific receptor kinases (Cok_S_s144_12294 and Cok_S_s043_7434), which have orthologous relationships with E. siliculosus genes (Supplementary Fig. S2). Three other genes are additional candidates (Cok_S_s126_11689, Cok_S_s097_10483 and Cok_S_s044_7515). All five genes are found on different scaffolds. RNA-seq reads demonstrate that Cok_S_s144_12294 is expressed in both germlings and sporophytes (Fig. 2B).
3.7.3. Genes associated with biosynthesis of polysaccharides
Genes encoding enzymes for polysaccharide metabolism in brown algae have been predicted in the E. siliculosus genome.9 Using informatics methods, we investigated gene families for biosynthesis of sulphated fucans and alginates, compounds characteristic of brown algae. Genes encoding enzymes involved in biosynthetic pathways are apparently conserved between C. okamuranus and E. siliculosus, although downstream enzymes of the polysaccharide biosynthesis pathways are likely to be expanded in each lineage.
3.7.3.1 Fucoidans
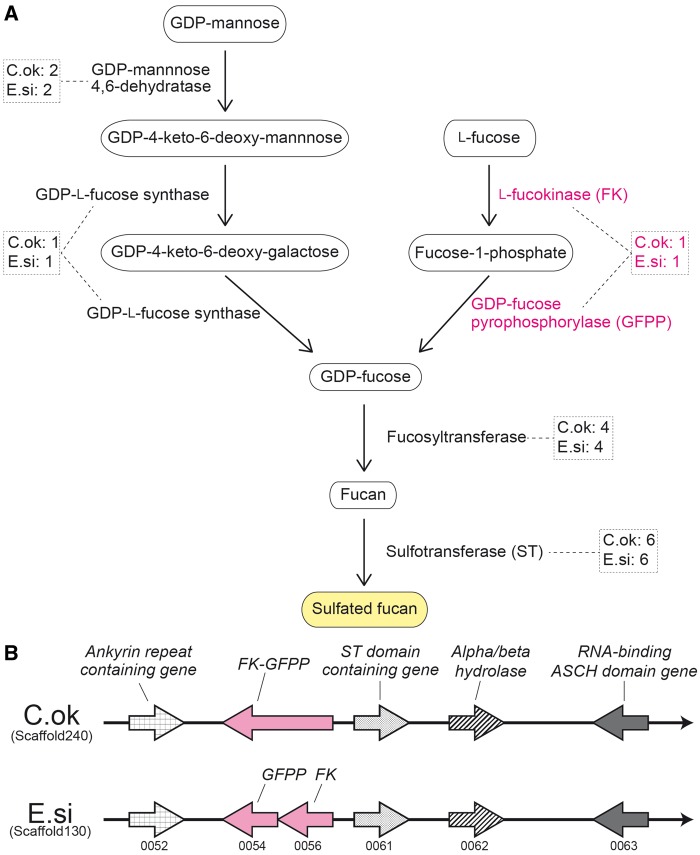
Fucoidans are a family of sulfated homo- and heteropolysaccharides of brown algae that include L-fucose residues. The family comprises a broad spectrum of polysaccharides, from compounds with high uronic acid content and very little fucose and sulfate, to almost pure α-l-fucan, in which the dominant monosaccharide is fucose. GDP-mannose and l-fucose are original sources of GDP-fucose, which is then transformed from fucan to sulfated fucan (Fig. 3A). Seven enzymes are likely involved in this pathway (Fig. 3A).
Figure 3.
Identification of all genes in the predicted biosynthetic pathway of sulfated fucans in the draft genome of Cladosiphon okamuranus. (A) A schematic representation of the biosynthetic pathway of sulphated fucans in brown algae, based on the description of Michel et al.9 Genes encoding each of the enzymes in the genomes of Cladosiphon okamuranus (C.ok) and Ectocarpus siliculosus (E.si), and the number of identified genes is shown in broken squares. (B) The location of candidate genes for l-fucosinase (FK) and GDP-fucose pyrophosphorylase (GFPP) in the C. okamuranus genome, suggesting the presence of an enzymatic gene cluster having shared synteny with E. siliculosus (http://bioinformatics.psb.ugent.be/orcae/overview/Ectsi). Two enzymatic genes that contain sulphotransferase and hydrolase domains are clustered with a gene (magenta) for a bifunctional enzyme with FK and GHMP-kinase domains. Genes coloured grey encode conserved proteins with RNA-binding domains.
Examination of genes for enzymes in both the C. okamuranus and E. siliculosus genomes identified two genes encoding GDP-mannose 4,6-dehydratase and one for GDP-l-fucose synthase (Fig. 3A and Supplementary Table S3). The presence of a candidate gene (FK-GFPP) for a bifunctional enzyme possessing both l-fucokinase and GDP-fucose pyrophosphorylase activities was identified in the C. okamuranus genome (Fig. 3A). These enzyme genes were expressed at germling and sporophyte stages (Fig. 2C). Comparison of the C. okamuranus and E. siliculosus genomes revealed syntenic localization of enzyme genes, suggesting the presence of an enzymatic gene cluster in both species (Fig. 3B). Two other enzymatic genes that contain a sulfotransferase domain and a hydrolase domain are clustered with FK-GFPP. The 3′ region of FK-GFPP contains an ankyrin-repeat domain (Fig. 3B).
In addition, the C. okamuranus and E. siliculosus genomes contain four genes for fucosyltransferase, and six genes for sulfotransferase, respectively (Fig. 3A). Identification of genes that encode enzymes involved in sulfated fucan biosynthesis suggests that C. okamuranus provides a good source of fucoidans, which should be further characterized in future studies.
3.7.3.2. Alginates
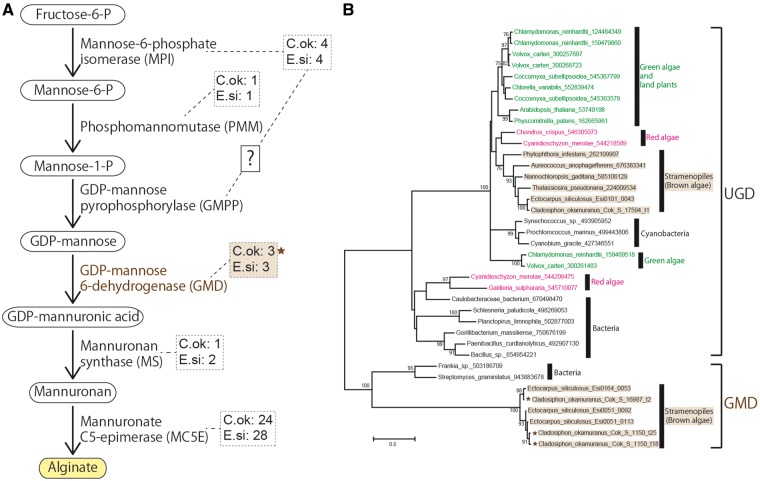
Alginates are cell-wall constituents of brown algae. They are chain-forming heteropolysaccharides consisting of blocks of mannuronic acid and glucuronic acid. Composition of the blocks varies by species and by the part of the thallus from which the extraction is made. Enzymes involved in the alginate biosynthetic pathway, including GDP-mannose 6-dehydrogenase (GMD) and mannuronate C5-epimerase (MC5E), have been identified in genomes of E. siliculosus9 and C. okamuranus (Fig. 4A and Supplementary Table S3). The number of enzymatic genes is comparable in both species. Some of MC5E-coding genes likely have different expression levels in germlings and sporophytes (Fig. 2C). These expression data will be useful for functional analyses of these enzymes.
Figure 4.
Predicted genes related to the alginate biosynthetic pathway in brown algae and conservation of a key enzyme, GDP-mannose 6-dehydrogenase (GMD). (A) A schematic representation of the alginate biosynthetic pathway in brown algae.9 As in Fig. 3, identified gene numbers in broken squares are followed by species identifiers, C.ok and E.si. (B) A molecular phylogenetic analysis of GMD revealing the conservation of a key enzyme for the alginate biosynthesis pathway between Cladosiphon okamuranus and Ectocarpus siliculosus. The maximum-likelihood (ML) tree was constructed with a WAG + G model. Numbers at nodes indicate more than 70% bootstrap support. Brown stars indicate Cladosiphon GMD proteins that were presumably transferred horizontally from bacteria. UDP glucose 6-dehydrogenase (UGD) is potentially involved in fucoidan biosynthesis, rather than alginate biosynthesis.
Previous studies showed that E. siliculosus9 and S. japonica11 possess three genes for GMDs of possible bacterial origin. Examination of the C. okamuranus genome and further molecular phylogenetic analyses indicated that the C. okamuranus genome also contains three GMD genes for this key enzyme (Fig. 4A and B and Supplementary Table S3). Twenty-four MC5E genes of C. okamuranus have homology to E. siliculosus genes (Supplementary Table S3). Our survey suggests that MC5E genes are independently expanded in each lineage.
3.7.3.3. Phlorotannin biosynthesis
Brown algal phenols attract considerable attention due to the wide variety of biological activities and potential health benefits of phlorotannins.42,43 Phlorotannins are known only from brown algae and are structural analogs of condensed tannins, such as anthocyanidins and other flavonoid derivatives, a diverse class of metabolites with a vast array of functions in terrestrial plants.44 They probably serve multiple ecological functions in brown algae, such as antifouling substances45 and chemical defenses against herbivory,46 in addition to providing UV protection for intertidal seaweeds.47 Their chemical structures are based on aryl-aryl and/or diaryl ether linkages of phloroglucinol (1,3,5-trihydroxybenzene) units and are rather complex. Polymerization leads to a wide range of molecular masses (10 and 100 kDa).48,49
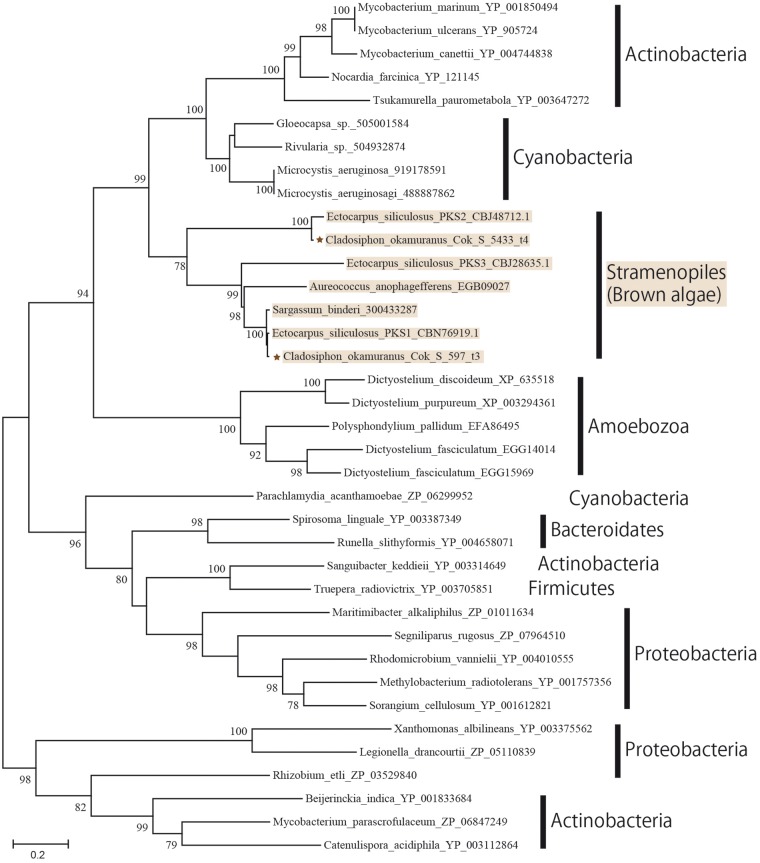
Polyketide synthase (PKS) is a key gene for phlorotannin biosynthesis.50 It has recently been reported that E. siliculosus PKS1 was classified as a Type-III PKS.50 We surveyed PKS genes in the C. okamuranus genome and found two candidate genes (Fig. 5 and Supplementary Table S3). Molecular phylogenetic analysis indicated that C. okamuranus Cok_S_597_t3 is the ortholog of E. siliculosus PKS1, a key gene in phlorotannin biosynthesis. E. siliculosus has three type III PKS genes. On the other hand, C. okamuranus has only two (PKS1 and PKS2) and has probably lost PKS3. PKS1 and PKS2 are expressed in germlings and sporophytes of C. okamuranus (Fig. 2C). Type III PKS genes of brown algae probably originated with bacteria.
Figure 5.
A molecular phylogenetic tree of two proteins similar to type III PKS in the Cladosiphon okamuranus genome. The ML tree was constructed with a WAG + G + I model. Proteins are indicated by brown stars. Other protein sequences refer to Meslet-Cladiere et al.50.
4. Data availability
A genome browser has been established at: http://marinegenomics.oist.jp/gallery/. Gene annotations from domain searches and Blast2GO51 are available at: (http://marinegenomics.oist.jp/gallery/).
Accession numbers
All Illumina reads (Supplementary Table S1) are available from DRA accession no. DRA004654. Genome sequences of the S-strain have been deposited with accession nos. BDDF01000001-01004525 (contigs) and DF977685-DF978416 (scaffolds).
Supplementary Material
Acknowledgements
We wish to thank all members of Marine Genomics Unit and the DNA Sequencing Section of OIST for their support of this work. We also thank Dr. Steven Aird for editing the manuscript.
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported by OIST internal funding for the Marine Genomics Unit.
References
- 1.Adl S.M., Simpson A.G., Farmer M.A., et al. 2005, The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol., 52, 399–451. [DOI] [PubMed] [Google Scholar]
- 2.Keeling P.J. 2009, Chromalveolates and the evolution of plastids by secondary endosymbiosis. J Eukaryot. Microbiol., 56, 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Cavalier-Smith T. 1999, Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol., 46, 347–66. [DOI] [PubMed] [Google Scholar]
- 4.Van Den Hoek C., Mann D.G., Jahns H.M. 1995, Algae: An Introduction to Phycology. Cambridge University Press, Cambridge.
- 5.Yoshida T., Suzuki M., Yoshinaga K. 2015, Checklist of marine algae of Japan (revised in 2015). Jpn. J. Phycol (Sôrui)., 63, 129–89. [Google Scholar]
- 6.Tako M., Nakada T., Hongou F. 1999, Chemical characterization of fucoidan from commercially cultured nemacystus decipiens (Itomozuku). Biosci. Biotechnol. Biochem., 63, 1813–5. [DOI] [PubMed] [Google Scholar]
- 7.Baba M., Snoeck R., Pauwels R., de Clercq E. 1988, Sulfated polysaccharides are potent and selective inhibitors of various enveloped viruses, including herpes simplex virus, cytomegalovirus, vesicular stomatitis virus, and human immunodeficiency virus. Antimicrob. Agents Chemother., 32, 1742–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin T.Y., Hassid W.Z. 1966, Pathway of algnic acid synthesis in the marine brown alga, Fucus gardneri Silva. J. Biol. Chem., 241, 5284–97. [PubMed] [Google Scholar]
- 9.Michel G., Tonon T., Scornet D., Cock J.M., Kloareg B. 2010, The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol., 188, 82–97. [DOI] [PubMed] [Google Scholar]
- 10.Cock J.M., Sterck L., Rouze P., et al. 2010, The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature, 465, 617–21. [DOI] [PubMed] [Google Scholar]
- 11.Ye N., Zhang X., Miao M., et al. 2015, Saccharina genomes provide novel insight into kelp biology. Nat. Commun., 6, 6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siemer B.L., Stam W.T., Olsen J.L., Pedersen P.M. 1998, Phylogenetic relationships of the brown algal orders Ectocarpales, Chordariales, Dictyosiphonales, and Tilopteridales (Phaeophyceae) based on RUBISCO large subunit and spacer sequences. J. Phycol., 34, 1038–48. [Google Scholar]
- 13.Bentley D.R. 2006, Whole-genome re-sequencing. Curr. Opin. Genet. Dev., 16, 545–52. [DOI] [PubMed] [Google Scholar]
- 14.Marcais G., Kingsford C. 2011, A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics, 27, 764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirakawa H., Shirasawa K., Kosugi S., et al. 2014, Dissection of the octoploid strawberry genome by deep sequencing of the genomes of Fragaria species. DNA Res., 21, 169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolger A.M., Lohse M., Usadel B. 2014, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boetzer M., Henkel C.V., Jansen H.J., Butler D., Pirovano W. 2011, Scaffolding pre-assembled contigs using SSPACE. Bioinformatics, 27, 578–9. [DOI] [PubMed] [Google Scholar]
- 18.Li R., Fan W., Tian G., et al. 2010, The sequence and de novo assembly of the giant panda genome. Nature, 463, 311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang S., Chen Z., Huang G., et al. 2012, HaploMerger: reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res., 22, 1581–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parra G., Bradnam K., Korf I. 2007, CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics, 23, 1061–7. [DOI] [PubMed] [Google Scholar]
- 21.Peng Y., Leung H.C., Yiu S.M., Chin F.Y. 2012, IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics, 28, 1420–8. [DOI] [PubMed] [Google Scholar]
- 22.Zerbino D.R., Birney E. 2008, Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res., 18, 821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz M.H., Zerbino D.R., Vingron M., Birney E. 2012, Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics, 28, 1086–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanke M., Diekhans M., Baertsch R., Haussler D. 2008, Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics, 24, 637–44. [DOI] [PubMed] [Google Scholar]
- 25.Haas B.J., Delcher A.L., Mount S.M., et al. 2003, Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res., 31, 5654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi T., Kawashima T., Koyanagi R., et al. 2012, Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res., 19, 117–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benson G. 1999, Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res., 27, 573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price A.L., Jones N.C., Pevzner P.A. 2005, De novo identification of repeat families in large genomes. Bioinformatics, 21 Suppl 1, i351–8. [DOI] [PubMed] [Google Scholar]
- 29.Jurka J., Klonowski P., Dagman V., Pelton P. 1996, CENSOR – a program for identification and elimination of repetitive elements from DNA sequences. Comput. Chem., 20, 119–21. [DOI] [PubMed] [Google Scholar]
- 30.Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. 2005, Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res., 110, 462–7. [DOI] [PubMed] [Google Scholar]
- 31.Finn R.D., Mistry J., Schuster-Bockler B., et al. 2006, Pfam: clans, web tools and services. Nucleic Acids Res., 34, D247–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eddy S.R. 1998, Profile hidden Markov models. Bioinformatics, 14, 755–63. [DOI] [PubMed] [Google Scholar]
- 33.Katoh K., Misawa K., Kuma K., Miyata T. 2002, MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res., 30, 3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gouy M., Guindon S., Gascuel O. 2010, SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol., 27, 221–4. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011, MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol., 28, 2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H. 2009, JBrowse: a next-generation genome browser. Genome Res., 19, 1630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C., Pachter L., Salzberg S.L. 2009, TopHat: discovering splice junctions with RNA-Seq. Bioinformatics, 25, 1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C., Williams B.A., Pertea G., et al. 2010, Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol., 28, 511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraskov A., Stogbauer H., Andrzejak R.G., Grassberger P. 2005, Hierarchical clustering using mutual information. Europhys. Lett., 70, 278–84. [Google Scholar]
- 40.Szekely G.J., Rizzo M.L. 2005, Hierarchical clustering via joint between-within distances: Extending Ward's minimum variance method. J. Classif., 22, 151–83. [Google Scholar]
- 41.Wu Y.W., Tang Y.H., Tringe S.G., Simmons B.A., Singer S.W. 2014, MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome, 2, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Connan S., Stengel D.B. 2011, Impacts of ambient salinity and copper on brown algae: 2. Interactive effects on phenolic pool and assessment of metal binding capacity of phlorotannin. Aquat. Toxicol., 104, 1–13. [DOI] [PubMed] [Google Scholar]
- 43.Thomas N.V., Kim S.K. 2011, Potential pharmacological applications of polyphenolic derivatives from marine brown algae. Environ. Toxicol. Pharmacol., 32, 325–35. [DOI] [PubMed] [Google Scholar]
- 44.Dakora F.D. 1995, Plant flavonoid: biological molecules for useful exploitation. Aust. J. Plant Physiol., 22, 87–99. [Google Scholar]
- 45.Sieburth J.M., Conover J.T. 1965, Slicks associated with Trichodesmium Blooms in the Sargasso Sea. Nature, 205, 830–1. [Google Scholar]
- 46.Pavia H., Toth B.G. 2000, Inducible chemical resistance to herbivory in the brown seaweed Ascophyllum nodosum. Ecology, 81, 3215–25. [Google Scholar]
- 47.Amsler C.D., Fairhead V.A. 2006, Defensive and sensory chemical ecology of brown algae. Adv. Botanical Res., 43, 1–91. [Google Scholar]
- 48.Boettcher A.A., Targett N.M. 1993, Role of polyphenolic molecular size in reduction of assimilation efficiency in Xiphister mucosus. Ecology, 74, 891–903. [Google Scholar]
- 49.Le Lann K., Connan S., Stiger-Pouvreau V. 2012, Phenology, TPC and size-fractioning phenolics variability in temperate Sargassaceae (Phaeophyceae, Fucales) from Western Brittany: native versus introduced species. Mar. Environ. Res., 80, 1–11. [DOI] [PubMed] [Google Scholar]
- 50.Meslet-Cladiere L., Delage L., Leroux C.J., et al. 2013, Structure/function analysis of a type iii polyketide synthase in the brown alga Ectocarpus siliculosus reveals a biochemical pathway in phlorotannin monomer biosynthesis. Plant Cell, 25, 3089–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gotz S., Arnold R., Sebastian-Leon P., et al. 2011, B2G-FAR, a species-centered GO annotation repository. Bioinformatics, 27, 919–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.