Abstract
Transmissible spongiform encephalopathies arise as a consequence of infection of the central nervous system by prions, where neurons and glial cells are regarded as primary targets. Neuronal loss and gliosis, associated with the accumulation of misfolded prion protein (PrP), are hallmarks of prion diseases; yet the mechanisms underlying such disorders remain unclear. Here we introduced a cell system based on primary cerebellar cultures established from transgenic mice expressing ovine PrP and then exposed to sheep scrapie agent. Upon exposure to low doses of infectious agent, such cultures, unlike cultures originating from PrP null mice, were found to accumulate de novo abnormal PrP and infectivity, as assessed by mouse bioassay. Importantly, using astrocyte and neuron/astrocyte cocultures, both cell types were found capable of sustaining efficient prion propagation independently, leading to the production of proteinase K-resistant PrP of the same electrophoretic profile as in diseased brain. Moreover, contrasting with data obtained in chronically infected cell lines, late-occurring apoptosis was consistently demonstrated in the infected neuronal cultures. Our results provide evidence that primary cultured neural cells, including postmitotic neurons, are permissive to prion replication, thus establishing an approach to study the mechanisms involved in prion-triggered neurodegeneration at a cellular level.
Transmissible spongiform encephalopathies (TSE), which include Creutzfeldt–Jakob disease in humans, bovine spongiform encephalopathy in cattle, and scrapie in sheep, are fatal neurodegenerative disorders caused by prions, a class of unconventional agents that targets the CNS in mammals. A hallmark of prion diseases is the accumulation of abnormal prion protein (PrPSc), a misfolded form of the cellular PrP (PrPc). Transmissibility is believed to stem from the ability of the prion isoform to promote the conformational transition from PrPc to PrPSc. Biologically distinct prion strains can propagate in a same host, presumably through the perpetuation of different specific PrPSc conformers (1–3).
Although it seems clear that neuronal dysfunction must lie at the root of the clinical disorders observed in these diseases, it is still obscure what triggers neurodegeneration and what role nonneuronal cells may play in this process. There is ample evidence to support a primary role of the neurons in prion propagation and neuropathogenesis into the CNS. Intra- or perineuronal PrPSc deposition, spongiform vacuolation involving cell soma and processes, and neuronal loss are typical histopathological changes observed in TSE-affected brain tissues (4, 5). Transgenic mice with PrP expression specifically targeted to neurons have been obtained that turned out to be fully susceptible to prion disease (6). More recently, it was shown that an acute neuron-targeted depletion of PrP in the brain of mice with ongoing infection is able to prevent neuronal loss and progression to disease and even to reverse early spongiform change, despite marked accumulation of PrPSc in nonneuronal cells (7). However, several data also argue for a crucial involvement of glial cells, such as astrocytes and microglial cells, in TSE pathogenesis (8–10). Unlike neuronal loss, astrogliosis appears to occur in an extremely consistent fashion in affected brains and can even precede spongiosis. Transgenic mice expressing PrP specifically in astrocytes developed a typical disease upon prion intracerebral inoculation and accumulated PrPSc at high levels in their brain (11), indicating that infection of neurons was not mandatory to mediate susceptibility to TSE.
The study of prion propagation in the various permissive cell culture systems available to date did not greatly contribute to furthering our understanding of the events leading to neurodegeneration. Immortalized neuronal and neuroglial cell lines persistently infected by prions generally show no overt signs of cytotoxicity, although producing readily detectable amounts of PrPSc and infectivity (12–15). Various phenotypical alterations have been reported (16, 17), yet it remains uncertain whether similar dysfunctions may affect TSE-injured postmitotic neurons. This calls for renewed efforts toward the development of primary cell culture systems in which the individual contribution of various CNS cells to prion propagation and its effects could be evaluated. Indeed, infection of cultured primary neurons from embryonic or neonatal mice and rat has proved a valuable model to study the pathogenesis of neurotropic viruses, in particular to distinguish between injuries caused by the host response and those caused by the infectious agent itself (18–20).
Here we demonstrate that brain-derived primary cells maintained in cultures can enter an infected state after exposure to sheep and also to mouse-adapted scrapie agent. We show that both neurons and astrocytes can sustain active prion propagation, leading to progressive neuronal loss, thus arguing that TSE models based on primary cells may represent a relevant approach toward a better understanding of the mechanisms underlying prion-induced neurodegeneration.
Materials and Methods
Primary Cell Culture. Primary cultures were established from tg338 mice overexpressing ovine PrPc on a mouse PrP0/0 background (21). Tg338 mice are homozygous for the transgene, a 125-kb DNA fragment including the V136R154Q171 ovine PrP allele, and overexpression is ≈10-fold.
Cerebellar granule neurons (CGN) were extracted from 6-day-old mice by mechanical and enzymatic dissociation. They were plated at a density of 1,900 cells/mm2 on plastic culture wells coated with 20 μg/ml polyd-lysine (PDL) and incubated at 37°C with 6% CO2. Cells were cultured in DMEM-glutamax I (GIBCO) containing 10% FCS (BioWhittaker), 20 mM KCl, penicillin, and streptomycin (GIBCO), and completed with N2 and B27 (GIBCO). The medium was supplemented weekly with 10 μM of the antimitotics uridine and fluorodeoxyuridine (Sigma) and glucose to maintain a concentration of 1 mg/ml. If necessary, the cerebellar preparation was preplated in dishes coated with 1 μg/ml PDL. Cells were left for 5–10 min to allow astrocyte sedimentation, and the supernatant containing ≈99% neurons was plated as described above. As negative controls, CGN cultures were established from PrP0/0 mice [Zurich I (22)].
Cerebellar astrocytes (CAS) were prepared as described above and left for 8 days. To discard the neuronal population, the cells were shifted to DMEM, 10% FCS, and penicillin–streptomycin, without KCl. The medium was changed weekly, and astrocytes were grown to confluence before use.
Prion Infection of Cultured Cells. The material used for infection was prepared from the brain of terminally ill tg338 mice inoculated with the 127S scrapie strain (21) or from MovS2 cells, an immortalized Schwann cell line originally derived from tg338 mice and infected with the 127S strain [ScMov cells (14)]. Ten percent brain homogenates were prepared in PBS and stored at –20°C until use. Mov cell material was prepared as follows: Cell culture medium from 7 days confluent Mov or ScMov cells was collected, centrifuged for 30 min at 4,200 × g to remove cellular debris, sonicated for 10 min, and stored at 4°C until use. The remaining confluent ScMov adherent cells were scraped in PBS, frozen/thawed three times, and stored at –20°C until use.
Two days after plating, primary neuronal or astrocytes cell cultures were exposed to either (i) 0.1% or 0.01% brain homogenates previously sonicated for 10 min in a cup-horn apparatus [multiplicity of infection (moi) ≈2 and 0.2 ID50/cell; or (ii) ScMov crude cell extracts diluted and sonicated in DMEM (moi ≈1–2 ID50/cell). For infection conducted with Mov cell culture medium, growing astrocytes cultures or freshly dissociated neurons were exposed to 1/2 dilution of ScMov supernatant in the relevant cell culture medium. Infection of CGN cultures established from tga20 (23) was also performed by using 0.1% brain homogenate from terminally ill mice infected with 139A mouse strain (originating from the R. Carp Laboratory; Staten Island, NY).
Immunofluorescence. Cells were plated on polyd-lysine-coated glass coverslips. Fixed cells (10 min at room temperature in PBS containing 4% paraformaldehyde and 4% sucrose) were permeabilized (5 min with PBS and 0.1% Triton X-100) and, if necessary, treated 5 min with 3 M guanidine thiocyanate (GdnSCN) to expose PrPSc epitopes as described (24). PrP was detected with mAb ICSM18 and with mAb ICSM35 in GdnSCN-treated cultures (25). Neuronal and astrocytic populations were specifically labeled by using mAb anti-NeuN (Chemicon) and anti-glial fibrillary acidic protein polyclonal antibody (DAKO), respectively. Microglia was labeled with anti-CD11b polyclonal antibody (Sigma). Cells were then incubated with appropriate FITC- or Alexa-conjugated secondary antibodies and nuclear marker 4′,6-diamidino-2-phenylindole (Sigma) and finally mounted in Fluoromount (Sigma).
PrP Immunoblot. Protein concentration in cell lysates was measured by the BCA protein assay (Pierce), and 40 μg of protein was treated or not with proteinase K (PK; 7.5 μg/mg of protein, Euromedex, Mundolsheim, France) for 30 min at 37°C. All samples were then supplemented with 1 mM Pefabloc, methanol precipitated for 1 h at –20°C, and centrifuged at 16,000 × g for 10 min. Pellets were resuspended in sample buffer, boiled, subjected to SDS/PAGE electrophoresis on 15% tricine gels, and electrotransferred onto nitrocellulose membranes. PrP was visualized with either ICSM18 or SAF 84.3 mAb, both directed to PrP C-terminal half. Western blots were revealed with an enhanced chemiluminescence detection system (ECL, Amersham Pharmacia). Densitometric analysis of PrP was performed by using scion image analysis software. For enzymatic deglycosylation of PrP, 15-μl aliquots of samples were digested with 5,000 units of recombinant PNGase F (New England Biolabs) in 1% Nonidet P-40 and appropriate buffer. Samples were incubated for 2 h at 37°C, mixed with an equal volume of sample buffer, and analyzed by immunoblot.
Mouse Bioassay. Cell-associated infectivity was measured by bioassay on tg338 mice (21). CGN cells were either scraped in PBS, centrifuged and resuspended in 5% sterile glucose, or lysed and further diluted in 5% glucose, as indicated in Table 1. CAS cells were scraped as above. Cell material was freeze–thawed three times and sonicated before injection. Twenty microliters was inoculated intracerebrally to 6-week-old mice. Animals showing scrapie signs were killed at terminal stage, and PK-resistant abnormal PrP (PrPres) in their brain was analyzed by immunoblot (21).
Table 1. Bioassay in ovine PrP tg338 mice of cell material from neuron- and astrocyte-enriched cultures infected by sheep prion.
| Inoculum
|
||||
|---|---|---|---|---|
| Cultures* | Days p.e. | Number of cells inoculated i.c.† | Days to death (n/n0)‡ | Log titer (total ID50)§ |
| ScCGN338 | 14 | 9.103 | 74 ± 1.2 (5/5) | 3.7 |
| 14 | 5.104 | 70 ± 0.4 (5/5) | 4.2 | |
| 28 | 9.103 | 70 ± 1.5 (5/5) | 4.2 | |
| 28 | 5.104 | 67 ± 1.1 (5/5) | 4.7 | |
| ScCGN0/0 | 14 | 9.103 | 89 ± 1.9 (5/5) | 1.5 |
| 14 | 5.104 | 88 ± 2.3 (5/5) | 1.6 | |
| 28 | 9.103 | 88 ± 1.9 (5/5) | 1.6 | |
| ScCAS338 | 14 | 1.105 | 72 ± 1.0 (6/6) | 3.9 |
| 28 | 1.105 | 71 ± 1.2 (6/6) | 4.2 | |
| ScCAS0/0 | 14 | 1.105 | 94 ± 2.2 (6/6) | 1.3 |
| 28 | 1.105 | 98 ± 2.3 (6/6) | 1.1 | |
CGN and CAS cultures were exposed to infectious inoculum consisting of ScMov culture medium.
Cell homogenates (5.104 and 1.105) or lysates (9.103) were injected intracerebrally (see Materials and Methods).
Mean days ± SEM; n, number of terminally ill animals; n0, number of animals inoculated.
As estimated from a dose—response curve constructed with infected Mov cell homogenate (45).
Quantification of Apoptosis by Immunocytochemistry. Scrapie- or mock-infected CGN cells were fixed at weekly intervals postexposure (p.e.) and stained with 4′,6-diamidino-2-phenylindole nuclear marker. Cultures were labeled with anti-GFAP and with anti-PrP ICSM35 antibodies to check for abnormal PrP accumulation. Neuronal apoptosis rate was determined by counting GFAP-negative cells with fragmented nuclei and expressed as a percentage of total number of neurons. Apoptotic death was confirmed by colocalization of fragmented nuclei and terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling-positive cells (Promega, according to the manufacturer's instructions). Double-blind quantification was performed.
Results
Primary Cultures of Mouse Cerebellar Cells Expressing Sheep PrP. In an attempt to set up a cell system in which the susceptibility to prion infection of defined primary neural cells could be assessed, we chose to culture granular neurons derived from the cerebellum of tg338 mice. This mouse line overexpresses ovine PrP from natural regulatory sequences under a mouse PrP0/0 background (21). Primary culture of CGN provides a means to obtain highly enriched and homogenous populations of postmitotic neurons, which can be maintained for >1 month (26, 27). Typically, cultures were mainly composed of neurons (>95%) up to 2 weeks after seeding, with 4–5% astrocytes and <1% microglial cells. After 2 weeks, the CGN started to slowly degenerate, with a constant neuronal loss of 15–20% per week. After 4 weeks, 40–50% of the initially seeded neurons were alive, the absolute number of astrocytes remaining the same, due to constant antimitotic pressure. No difference in cell survival was detected between cultures from tg338 (CGN338) and PrP0/0 (CGN0/0), in which the neurons showed similar differentiation with a complex dendritic arborization, one long branched axon and synaptic buttons (data not shown). Cultures of CAS (>99% astrocytes) were also established from tg338 and PrP0/0 (CAS338 and CAS0/0, respectively) and were maintained up to 1 month.
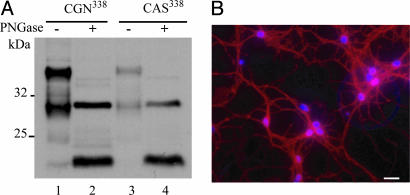
Because PrP can be rate-limiting for prion infection in vivo (28), we checked its level and pattern of expression in the cultures thus obtained. Immunoblotting performed on CGN338 cell material revealed a typical PrPc pattern, with a relatively high proportion of truncated forms assumed to correspond to the C1 fragment (29), reminiscent of cerebellum PrPc (25). A similar pattern together with a 3- to 5-fold lower expression level was observed in CAS338 cultures (Fig. 1A). Immunofluorescence analysis on CGN338 revealed a neuronal PrP staining pattern that was ubiquitous and uniform along the cell soma, axon, and synapses (Fig. 1B), consistent with in vivo observations (30).
Fig. 1.
Ovine PrP expression in primary neural cells established from tg338 mouse cerebellum. (A) PrPc expression in neuron-enriched primary cultures (CGN338) and in astrocyte cultures (CAS338) as assessed by Western blotting. Forty micrograms of protein was loaded in each lane and probed with anti-PrP mAb ICSM18. Aliquots were deglycosylated by PNGase F treatment (lanes 2 and 4). (B) Fixed and permeabilized CGN338 cultures stained with anti-PrP mAb ICSM18 (red) and the nuclear marker 4′,6-diamidino-2-phenylindole (blue).
Scrapie-Exposed CGN Cultures Accumulate PrPSc and Infectivity. We next examined the permissiveness of CGN338 cultures to infection by sheep prion. As a source of infectious agent, we used a strain of natural sheep scrapie that can propagate in permissive cell lines expressing ovine PrP (14, 31) and in tg338 mice, leading to death within 60 days (21). Unlike established cell systems involving actively dividing cells, neuron cultures do not allow washing out of the inoculum or subpassage cells. Therefore, CGN338 cultures were exposed to relatively low doses of inoculum, and CGN0/0 cultures were inoculated in parallel, thus making it possible to monitor the levels of input PrPSc and infectivity throughout the experiment.
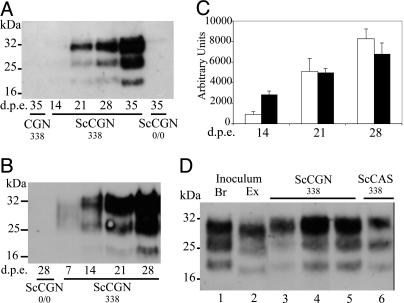
Various inocula, prepared from either steady-state-infected Mov cultures [ScMov; (14)] or terminally diseased tg338 mouse brain, were tested. Whatever the inoculum, a gradual accumulation of PrPres was observed reproducibly in exposed CGN338 cultures (Fig. 2C). Typically, PrPres was readily detected from 21 and 14 days after exposure to ScMov culture medium or cell extract, respectively (Fig. 2 A and B). The earlier PrPres accumulation with cell extract inoculum is consistent with a moi (≈1 ID50/cell) ≈100-fold higher than with culture medium. Importantly, PrPres failed to be detected in exposed CGN0/0 cultures at both earlier (7 days; not shown) and later time points (Fig. 2 A and B). Also worth noting, the PrPres band size in scrapie-exposed CGN338 (ScCGN338) cultures clearly differed from that in ScMov inoculum (Fig. 2D). Altogether, these findings led us to conclude that authentic conversion of PrPc into PrPres was taking place in ScCGN338 cultures. Use of 0.01% infected brain homogenate instead of ScMov-derived inoculum gave similar results in terms of kinetics and level of PrPres production (data not shown). Finally, we also exposed CGN cultures established from tga20-overexpressing mouse PrP to 139A mouse-adapted prion and observed accumulation of PrPres at similar levels (data not shown).
Fig. 2.
Accumulation of PrPres in CGN338 cultures exposed to sheep scrapie agent as assessed by Western blotting. (A) Kinetics of PrPres accumulation between 14 and 35 days p.e. to ScMov culture medium (moi ≈ 0.01 ID50/cell). Tg338 and PrP0/0 CGN cultures were exposed to either ScMov culture medium (designated ScCGN338 and ScCGN0/0, respectively) or mock-infected Mov culture medium (CGN338). (B) PrPres accumulation kinetics between 7 and 28 days p.e. to ScMov cell extract (moi ≈ 1–2ID50/cell). (A and B) Note the absence of detectable PrPres signal in ScCGN0/0 cultures. (C) PrPres accumulation in ScCGN338 cultures after infection with ScMov culture medium (white bars) or ScMov cell extract (black bars). Immunoblotted PrPres was quantified in three independent experiments (mean ± SEM). (D) Comparison of electrophoretic patterns of PrPres produced in ScCGN338 and of inoculum-derived PrPres. Tg338-infected brain homogenate (lane 1, Br) or cell-propagated scrapie (lane 2; ScMov cell extract, Ex) were used as inoculum. Note the faster-moving bands in ScMov PrPres compared to Br, ScCGN, and ScCAS. Whatever the inoculum used (lanes 3 and 6, ScMov culture medium; lane 4, ScMov cell extract; lane 5, brain), PrPres accumulated in ScCGN338 and astrocyte-enriched ScCAS338 cultures had the same electrophoretic pattern, similar to that in the brain of scrapie-infected tg338 mice (Br). (A, B, and D) Forty micrograms of protein was PK-treated and PrPres was revealed by using mAb SAF 84.3.
To examine whether prion infectivity increased concomitantly with PrPres, homogenates were prepared from ScCGN338 and ScCGN0/0 cultures at 14 and 28 days p.e. to ScMov supernatants and then intracerebrally inoculated to indicator tg338 mice (Table 1). All mice died with typical scrapie symptoms and accumulated PrPres in their brain (10/10 tested). A stable background infectivity assumed to represent remnant input infectivity was found in PrPres-negative ScCGN0/0 cultures at the same time points, consistent with a higher sensitivity of mouse bioassay over biochemical detection. However, ScCGN338 culture extracts at 14 days p.e. contained markedly higher (≈160- to 400-fold) levels of infectivity, which further increased at 28 days p.e. in agreement with the dynamics of PrPres accumulation. From these results, it was concluded that CGN cultures sustained an active propagation of the infectious agent.
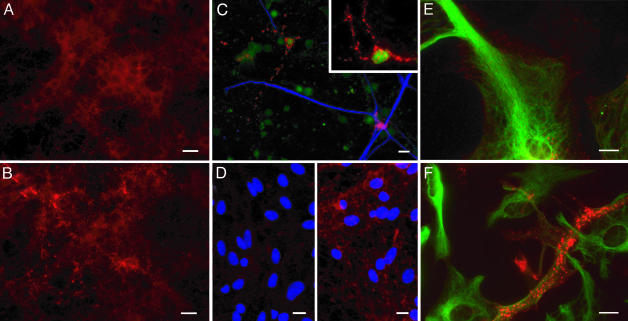
Prion Propagation in Exposed Cultures Involves both Neurons and Astrocytes. To document the nature and proportion of cells accumulating PrPSc, ScCGN cultures fixed at various times p.e. were treated with GdnSCN to expose PrPSc-associated epitopes (14, 24) and examined after PrP immunofluorescent labeling. The aspect of PrP staining was found to strikingly differ in infected and mock-infected cultures; whereas the soma and cell processes stained rather uniformly in CGN control cultures, bright and punctuated PrP labeling was predominant in ScCGN cultures (Fig. 3 A and B). Such abnormal PrP staining was detectable as early as 7 days p.e. and involved a gradually increasing proportion of cells with time. Importantly, it was observed in both NeuN- and GFAP-positive cells (Fig. 3C).
Fig. 3.
Abnormal PrP immunodetection in CGN and CAS infected cultures. PrP was detected by using mAb ICSM35 after permeabilization and GdnSCN denaturation (red). (A and B) Mock-infected (A) or infected (B) CGN338 cultures were PrP-labeled at 14 days after exposure to 0.01% tg338 brain homogenate. (C) To better visualize individual infected cells, 1% CGN338 were diluted in 99% CGN0/0 and exposed to ScMov culture medium (14 days p.e.); green, NeuN (neuronal marker); blue, GFAP (astrocytic marker). (D) Mock-infected (Left) or infected (Right) CAS338 cultures at 28 days p.e. to Mov culture medium; blue, 4′,6-diamidino-2-phenylindole. (E and F) Same cultures as in D at higher magnification; green, GFAP. In infected cultures (B–D Right, and F), cells show a punctuate fluorescence assumed to reflect abnormal PrP accumulation, in contrast with the diffuse and homogenous PrP labeling in mock-infected cultures (A, D Left, and E). (Original magnification: A–C ×400, D ×200, E–F and Inset ×600; Bar = 10 μm.)
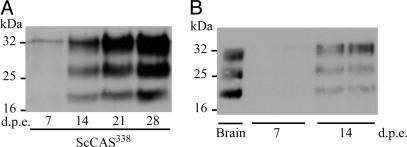
To establish whether astrocytes were permissive on their own to prion infection, we exposed CAS338 and CAS0/0 cultures highly enriched in astrocytes to ScMov culture medium. Labeling for PrP after GdnSCN treatment at 14 days p.e. revealed PrPSc-like staining in ≈80% of the cells in CAS338 cultures (Fig. 3 D–F). Moreover, Western blotting performed on lysates of such cultures revealed an accumulation of PrPres, with a 3- to 4-fold increase at 28 days p.e. (Fig. 4A). When assayed in tg338 mice (Table 1), scrapie-exposed CAS338 (ScCAS338) cell infectivity did not significantly increase between 14 and 28 days p.e., consistent with Western blotting data. However, ScCAS338 were found to contain 400- to 1,200-fold more infectivity than cell extracts from CAS0/0 exposed cultures. These data showed that cerebellar astrocytes are fully permissive to prion infection.
Fig. 4.
PrPres accumulation in purified astrocytes and neuron/astrocyte cocultures as assessed by Western blotting. (A) PrPres kinetics in ScCAS338 cultures exposed to ScMov culture medium. (B) Purified CGN338 neurons (99%) grown on top of a confluent nonpermissive CAS0/0 culture and exposed to 0.01% tg338 infectious brain homogenate. Duplicate experiments at 7 and 14 days p.e. show a de novo PrPres synthesis in neurons. PrPres was revealed by PK treatment of 40 μg of protein using mAb ICSM18.
Long-term culture of pure neurons is possible only through the presence of astrocytes, which provide a trophic support. Therefore, to further assess the contribution of the neurons to prion propagation in ScCGN cultures, we seeded a granular cell preparation highly enriched in neurons (>99%) on a preestablished CAS0/0 culture used as a feeder cell monolayer. Exposure of such cocultures to 0.01% infected brain homogenate again resulted in PrPres accumulation, which could readily be detected from 14 days p.e. onward (Fig. 4B; n = 2). Altogether, the above findings provided strong evidence that both neurons and astrocytes can be infected independently by sheep prion.
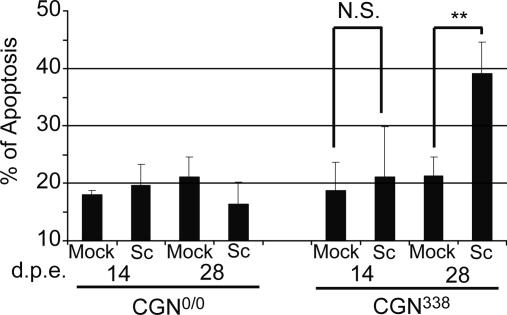
Infection-Induced Neuronal Apoptosis Occurs in ScCGN Cultures. Although substantial neuronal loss is commonly observed in TSE diseases, infection of the permissive neuronal cell lines currently available does not affect the cell viability, except for the GT1 cell line in which apoptosis was reported to occur in infected cultures, although inconsistently (16). As a first approach to document any prion-induced injury in ScCGN cells, we determined the proportion of cells showing signs of apoptosis in infected and mock-infected CGN338 and CGN0/0 cultures. Neuronal apoptosis was monitored by counting fragmented nuclei in GFAP-negative cells. Apoptosis was confirmed by colocalization of terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling-positive cells and fragmented nuclei. Between 7 and 14 days p.e., the apoptosis level increased from ≈7% to ≈20%, regardless of the PrP expression and infection status. It was thus assumed to correspond to the basal apoptotic rate in CGN cultures. At 28 days p.e., however, the apoptosis rate was ≈2-fold higher in ScCGN338 than in other cultures and approached 40% (Fig. 5). From these data, it was concluded that an apoptosis-like process was specifically triggered by prion infection in susceptible CGN neuronal cells.
Fig. 5.
Prion-induced neuronal apoptosis in infected CGN cultures. Quantification of neuronal apoptosis in CGN0/0 and CGN338 at 14 and 28 days p.e. to mock-infected (Mock) and infected (Sc) Mov culture medium. Apoptosis rate was determined by counting neurons with fragmented nuclei and expressed as a percentage of total number of neurons. Each quantification represents the mean of four independent experiments ± SEM. Apoptosis increased significantly at 28 days p.e. in ScCGN338 vs. CGN338 (Mann–Whitney U test; **, P < 0.01), whereas it did not differ significantly (N.S.) between ScCGN0/0 and CGN0/0.
Discussion
In the present study, we show that natural scrapie agent can actively propagate in de novo infected primarily grown differentiated neural cells. This TSE cell system consists of either granular neuron- or astrocyte-enriched cultures that are derived from the cerebellum of susceptible transgenic mice expressing ovine PrP and can be maintained for ≈1 month.
Several lines of evidence indicate that a bona fide prion propagation takes place in both kinds of culture, designated CGN338 and CAS338. First, abnormal PrP was shown to accumulate steadily in exposed cultures, as revealed by the detection of protease-resistant species and a change in the subcellular distribution of PrP. Second, the levels of infectivity associated with cultures at 2 and 4 weeks p.e. were several hundred-fold higher than in control cultures expressing no PrP, as assessed by mouse bioassay. One practical constraint inherent to the use of cells that cannot be subpassaged is to distinguish de novo generated infectivity from incoming prion. Indeed, infectivity was repeatedly found to persist at substantial levels in PrP0/0 cultures even at 4 weeks p.e. Interestingly, close examination of ScCGN0/0 and ScCGN338 cultures at this time point revealed PrPSc-like staining in small round CD11b-positive microglial cells (data not shown). This suggests that input PrPSc may be captured by and may persist in these resident CNS macrophages (representing <1% of cultured CGN cells), consistent with studies in mouse showing that microglial cells may carry infectivity at relatively high levels (10). Notwithstanding this persistence phenomenon, the net accumulation of infectivity was estimated to be 0.05 and 0.01 ID50/cell in CGN338 and CAS338 cultures, respectively, which compares to that reported for nonsubcloned neuroblastoma cells (12).
The PrPres signal observed in the permissive cultures essentially represented newly converted PrP because, due to the low dose of the inocula, no PrPres was detected in cultures at 1 week p.e. Remarkably, the PrPres patterns in CGN338 and CAS338 cultures closely resembled that in diseased mice brain. This is, to our knowledge, an unprecedented finding, because production of PrPres species of shorter size than in the donor tissue appears to be a common feature of the available cell systems permissive to sheep- or mouse-adapted prions (14, 31–33). Such N-terminally truncated species can be visualized before any PK digestion, indicating the involvement of cellular, presumably lysosomal, proteases (ref. 34; our unpublished data). Propagation of the sheep agent in CGN338 and CAS338 cells leads to the production of aglycosylated PrPres species ≈2 kDa larger than that in the Rov (31) and Mov (14) cell lines. It thus appears that distinct truncation events may occur depending on the cell system in which the prion propagates. It may be relevant in future studies to determine whether this differential truncation reflects a preferential intra- or extra-cellular accumulation of PrPSc, as recently proposed from in vivo studies (4). In any case, this finding suggests that these primarily grown neural cells recapitulate PrPSc processing in vivo more faithfully than the immortalized cell lines established so far, including neuronally derived cell lines.
Evidence has accumulated to indicate that both neurons and astrocytes are involved in prion propagation and pathogenesis, yet uncertainties remain about their respective roles. Transgenic mice expressing hamster PrP restricted to either neurons or astrocytes proved susceptible to infection by hamster prion (6, 11). Formally, however, this approach does not make it possible to rule out leaky PrP expression in nontargeted cell types, even though this issue was carefully addressed in the above studies. Here we provide direct evidence that both cell types can support active prion propagation, because astrocyte cultures essentially free of neurons on the one hand, and highly purified neurons cultivated in the presence of astrocytes lacking PrP on the other hand, can efficiently propagate the infectious agent. This finding further argues that each cell type is intrinsically permissive and could serve as primary target for replication of the scrapie agent in vivo. Recent data from immunohistochemical analyses performed on affected sheep-brain tissue have suggested that scrapie strains may substantially differ in their relative affinity for astrocytes or neurons (35). Such an issue could be addressed by exploiting the dual culture model described here to determine whether ability to infect and promote marked PrPSc accumulation in both CGN338 and CAS338 cells is a common trait of prions. Of interest in this regard, we found that CGN cultures were also capable of sustaining propagation of mouse-adapted prion.
How prions impair neuronal function and ultimately cause cell death remains largely unknown. Apoptosis is currently regarded as the principal mechanism of neuronal cell loss based on studies involving scrapie-infected mice and Creutzfeldt–Jakob disease-affected humans (36, 37). Cultivated primary neurons obviously provide a relevant context in which the precise mechanisms of prion-elicited cell death could be investigated. To date, such studies have focused only on the cytotoxic effects induced by purified PrPSc or by PrP-derived synthetic fragments, and among these the human PrP 106–126 peptide has been extensively used. Both PrPSc and the 106–126 peptide are reported to be toxic for mouse or rat primary neurons, leading to death through apoptosis within a few days p.e. (38–41). Such an effect was shown to depend on the expression of PrPc (42), yet its biological significance is still questioned. Here we present strong evidence that infected CGN cells entered an apoptotic pathway. In contrast to the acute toxic effect triggered by exogenous fibrillogenic PrP material, the apoptotic process observed in ScCGN338 cultures developed gradually and was detected within weeks p.e., after the accumulation of PrPSc became apparent. Investigation of the early primary events leading to neuronal cell death may thus benefit from studies performed on truly infected neurons. Cerebellar granule cells are ideally suited for such studies, because they form a very homogenous population of neurons. Moreover, through cocultivation of PrP0/0 or PrP+/+ neurons and ScCAS cells, our model should make it possible to further address the debated point of whether extraneuronally propagated prion is harmless for neurons that lack PrP (7, 11, 43), because it is conceivable that distinct pathogenic cascades could be activated depending on strain- or host cell-related factors. Finally, this experimental paradigm could also be harnessed to study the precise role of microglial cells, whose potential involvement in TSE neuropathogenesis has been emphasized both in vivo and ex vivo (10, 42, 44).
Conclusion
The propagation of prion in primary neurons and astrocytes has been achieved in this study, and the resulting findings support the view that this model may bring new opportunities to gain better insights into the mechanisms of neuronal injury in prion diseases.
Acknowledgments
We thank A. Ledur for help with the mouse bioassay; S. Hawke (Imperial College School of Medicine, London) and J. Grassi (Centre pour l'Energie Atomique, Saclay, France) for kindly providing the antibodies ICSM18, ICSM35, and SAF84.3, respectively; C. Weissmann (Imperial College School of Medicine, London) for the PrP0/0 and tga20 mice; and A. L. Haenni for carefully revising the manuscript. This project was supported by grants from the French government (GIS-Infections à Prion) and from the European Union (Neurodegeneration–QLG3CT2001). S.C. is a recipient of a French Ministry of Research fellowship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TSE, transmissible spongiform encephalopathy; PrP, prion protein; PrPc, cellular PrP; PK, proteinase K; PrPres, PK-resistant PrP; PrPSc, abnormal PrP; CGN, cerebellar granule neurons; CAS, cerebellar astrocytes; p.e., postexposure; moi, multiplicity of infection; GdnSCN, guanidine thiocyanate; GFAP, glial fibrillary acidic protein; ScMov, steady-state infected Mov cultures; ScCGN, scrapie-exposed CGN; ScCAS, scrapie-exposed CAS.
References
- 1.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weissmann, C. (1999) J. Biol. Chem. 274, 3–6. [DOI] [PubMed] [Google Scholar]
- 3.Collinge, J. (2001) Annu. Rev. Neurosci. 24, 519–550. [DOI] [PubMed] [Google Scholar]
- 4.Jeffrey, M., Martin, S. & Gonzalez, L. (2003) J. Gen. Virol. 84, 1033–1045. [DOI] [PubMed] [Google Scholar]
- 5.Budka, H. (2003) Br. Med. Bull. 66, 121–130. [DOI] [PubMed] [Google Scholar]
- 6.Race, R. E., Priola, S. A., Bessen, R. A., Ernst, D., Dockter, J., Rall, G. F., Mucke, L., Chesebro, B. & Oldstone, M. B. (1995) Neuron 15, 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallucci, G., Dickinson, A., Linehan, J., Klohn, P. C., Brandner, S. & Collinge, J. (2003) Science 302, 871–874. [DOI] [PubMed] [Google Scholar]
- 8.Diedrich, J. F., Bendheim, P. E., Kim, Y. S., Carp, R. I. & Haase, A. T. (1991) Proc. Natl. Acad. Sci. USA 88, 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye, X., Scallet, A. C., Kascsak, R. J. & Carp, R. I. (1998) Brain Res. 809, 277–287. [DOI] [PubMed] [Google Scholar]
- 10.Baker, C. A., Martin, D. & Manuelidis, L. (2002) J. Virol. 76, 10905–10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raeber, A. J., Race, R. E., Brandner, S., Priola, S. A., Sailer, A., Bessen, R. A., Mucke, L., Manson, J., Aguzzi, A., Oldstone, M. B., et al. (1997) EMBO J. 16, 6057–6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler, D. A., Scott, M. R., Bockman, J. M., Borchelt, D. R., Taraboulos, A., Hsiao, K. K., Kingsbury, D. T. & Prusiner, S. B. (1988) J. Virol. 62, 1558–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubenstein, R., Carp, R. I. & Callahan, S. M. (1984) J. Gen. Virol. 65, 2191–2198. [DOI] [PubMed] [Google Scholar]
- 14.Archer, F., Bachelin, C., Andreoletti, O., Besnard, N., Perrot, G., Langevin, C., Le Dur, A., Vilette, D., Baron-Van Evercooren, A., Vilotte, J. L., et al. (2004) J. Virol. 78, 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Follet, J., Lemaire-Vieille, C., Blanquet-Grossard, F., Podevin-Dimster, V., Lehmann, S., Chauvin, J. P., Decavel, J. P., Varea, R., Grassi, J., Fontes, M., et al. (2002) J. Virol. 76, 2434–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schatzl, H. M., Laszlo, L., Holtzman, D. M., Tatzelt, J., DeArmond, S. J., Weiner, R. I., Mobley, W. C. & Prusiner, S. B. (1997) J. Virol. 71, 8821–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milhavet, O., McMahon, H. E., Rachidi, W., Nishida, N., Katamine, S., Mange, A., Arlotto, M., Casanova, D., Riondel, J., Favier, A., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 13937–13942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaul, M., Garden, G. A. & Lipton, S. A. (2001) Nature 410, 988–994. [DOI] [PubMed] [Google Scholar]
- 19.Bajramovic, J. J., Munter, S., Syan, S., Nehrbass, U., Brahic, M. & Gonzalez-Dunia, D. (2003) J. Virol. 77, 12222–12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nargi-Aizenman, J. L. & Griffin, D. E. (2001) J. Virol. 75, 7114–7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilotte, J. L., Soulier, S., Essalmani, R., Stinnakre, M. G., Vaiman, D., Lepourry, L., Da Silva, J. C., Besnard, N., Dawson, M., Buschmann, A., et al. (2001) J. Virol. 75, 5977–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bueler, H., Fischer, M., Lang, Y., Bluethmann, H., Lipp, H. P., DeArmond, S. J., Prusiner, S. B., Aguet, M. & Weissmann, C. (1992) Nature 356, 577–582. [DOI] [PubMed] [Google Scholar]
- 23.Fischer, M., Rulicke, T., Raeber, A., Sailer, A., Moser, M., Oesch, B., Brandner, S., Aguzzi, A. & Weissmann, C. (1996) EMBO J. 15, 1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 24.Taraboulos, A., Serban, D. & Prusiner, S. B. (1990) J. Cell Biol. 110, 2117–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beringue, V., Mallinson, G., Kaisar, M., Tayebi, M., Sattar, Z., Jackson, G., Anstee, D., Collinge, J. & Hawke, S. (2003) Brain 126, 2065–2073. [DOI] [PubMed] [Google Scholar]
- 26.Hatten, M. E., Liem, R. K. & Mason, C. A. (1986) J. Neurosci. 6, 2676–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell, S. K., Rivas, R. J., Rodriguez-Boulan, E. & Hatten, M. E. (1997) J. Neurobiol. 32, 223–236. [DOI] [PubMed] [Google Scholar]
- 28.Bueler, H., Aguzzi, A., Sailer, A., Greiner, R. A., Autenried, P., Aguet, M. & Weissmann, C. (1993) Cell 73, 1339–1347. [DOI] [PubMed] [Google Scholar]
- 29.Chen, S. G., Teplow, D. B., Parchi, P., Teller, J. K., Gambetti, P. & Autilio-Gambetti, L. (1995) J. Biol. Chem. 270, 19173–19180. [DOI] [PubMed] [Google Scholar]
- 30.Laine, J., Marc, M. E., Sy, M. S. & Axelrad, H. (2001) Eur. J. Neurosci. 14, 47–56. [DOI] [PubMed] [Google Scholar]
- 31.Vilette, D., Andreoletti, O., Archer, F., Madelaine, M. F., Vilotte, J. L., Lehmann, S. & Laude, H. (2001) Proc. Natl. Acad. Sci. USA 98, 4055–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida, N., Harris, D. A., Vilette, D., Laude, H., Frobert, Y., Grassi, J., Casanova, D., Milhavet, O. & Lehmann, S. (2000) J. Virol. 74, 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanu, N., Imokawa, Y., Drechsel, D. N., Williamson, R. A., Birkett, C. R., Bostock, C. J. & Brockes, J. P. (2002) Curr. Biol. 12, 523–530. [DOI] [PubMed] [Google Scholar]
- 34.Caughey, B., Raymond, G. J., Ernst, D. & Race, R. E. (1991) J. Virol. 65, 6597–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez, L., Martin, S. & Jeffrey, M. (2003) J. Gen. Virol. 84, 1339–1350. [DOI] [PubMed] [Google Scholar]
- 36.Giese, A. & Kretzschmar, H. A. (2001) Curr. Top. Microbiol. Immunol. 253, 203–217. [DOI] [PubMed] [Google Scholar]
- 37.Hetz, C., Russelakis-Carneiro, M., Maundrell, K., Castilla, J. & Soto, C. (2003) EMBO J. 22, 5435–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forloni, G., Angeretti, N., Chiesa, R., Monzani, E., Salmona, M., Bugiani, O. & Tagliavini, F. (1993) Nature 362, 543–546. [DOI] [PubMed] [Google Scholar]
- 39.Brown, D. R., Herms, J. & Kretzschmar, H. A. (1994) Neuroreport 5, 2057–2060. [DOI] [PubMed] [Google Scholar]
- 40.Giese, A., Brown, D. R., Groschup, M. H., Feldmann, C., Haist, I. & Kretzschmar, H. A. (1998) Brain Pathol. 8, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller, W. E., Ushijima, H., Schroder, H. C., Forrest, J. M., Schatton, W. F., Rytik, P. G. & Heffner-Lauc, M. (1993) Eur. J. Pharmacol. 246, 261–267. [DOI] [PubMed] [Google Scholar]
- 42.Brown, D. R., Schmidt, B. & Kretzschmar, H. A. (1996) Nature 380, 345–347. [DOI] [PubMed] [Google Scholar]
- 43.Brandner, S., Isenmann, S., Raeber, A., Fischer, M., Sailer, A., Kobayashi, Y., Marino, S., Weissmann, C. & Aguzzi, A. (1996) Nature 379, 339–343. [DOI] [PubMed] [Google Scholar]
- 44.Marella, M. & Chabry, J. (2004) J. Neurosci. 24, 620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prusiner, S. B., Cochran, S. P., Groth, D. F., Downey, D. E., Bowman, K. A. & Martinez, H. M. (1982) Ann. Neurol. 11, 353–358. [DOI] [PubMed] [Google Scholar]