Abstract
Staphylococcus aureus is the leading cause of wound and hospital-acquired infections worldwide. The emergence of S. aureus strains with resistance to multiple antibiotics requires the identification of bacterial virulence genes and the development of novel therapeutic strategies. Herein, bursa aurealis, a mariner-based transposon, was used for random mutagenesis and for the isolation of 10,325 S. aureus variants with defined insertion sites. By screening for loss-of-function mutants in a Caenorhabditis elegans killing assay, 71 S. aureus virulence genes were identified. Some of these genes are also required for S. aureus abscess formation in a murine infection model.
Keywords: transposon, Caenorhabditis elegans
The Gram-positive bacterium Staphylococcus aureus is a physiological commensalism of the human skin, nares, and mucosal surfaces (1, 2). Staphylococci are also opportunistic and adaptable pathogens with the ability to infect, invade, persist, and replicate in any human tissue including skin, bone, visceral organs, or vasculature (1, 3). As a result of adaptation to specific tissues, staphylococci cause many diverse pathological syndromes such as abscess, bacteremia, osteomyelitis, toxic shock syndrome, food poisoning, and endocarditis (1, 4). Because they colonize the human skin continuously, S. aureus strains are exposed to all antibiotic therapies (5). Whenever drug-resistant microbes emerge, these strains can spread by direct contact very rapidly among human populations, as exemplified by the threat of methicillin-resistant S. aureus (MRSA) and vancomycin-resistant S. aureus (VRSA) worldwide (6–10).
Antibiotic therapy has focused on targets that are required by all bacteria, for example, cell wall synthesis, protein synthesis, and DNA replication (11). This therapeutic strategy cannot distinguish between resident nonpathogenic flora and invading pathogens and, in principle, does not require identification of etiologic disease agents or specific diagnosis to commence therapy (12). As bacteria distribute genetic information beyond species boundaries, the emergence of drug-resistant microbes profoundly impacts the selection of pathogenic microbes with resistance against all currently known antibiotics (12).
We analyzed the genome of S. aureus for genes that are uniquely required for the establishment of staphylococcal diseases. It is proposed that antiinfective therapy may provide a cure to otherwise fatal human disease in those cases where the etiologic agents of infectious diseases can be identified and matched with specific inhibitors. The 2.7–2.9 Mb genomes of several different S. aureus strains have been sequenced and encompass between 2,550 and 2,870 genes (13–16). Over the past several decades, S. aureus-secreted toxins, surface proteins, and regulatory factors were identified as virulence factors and characterized, often by using reverse genetic approaches (2). S. aureus strain Newman is a human clinical isolate (17) that stably maintains an agr+ phenotype, i.e., quorum-controlled toxin secretion and the ability to cause animal disease (18), whereas most other human clinical isolates rapidly lose the agr phenotype and with it the property of causing purulent infections in animals (19). We developed bursa aurealis, a mariner-based transposon (20, 21), which we used for the isolation of S. aureus strain Newman variants with defined insertion sites. By screening for loss-of-function mutants in a Caenorhabditis elegans killing assay (22), S. aureus virulence genes were identified.
Materials and Methods
Transposon Mutagenesis. S. aureus strain Newman was sequentially transformed with pFA545 and pBursa (Fig. 1). The resulting transformants were spread on tryptic soy agar (TSA) containing 10 μg·ml–1 erythromycin (TSAerm, tet) and incubated at 30°C overnight. Isolated single colonies were suspended in 200 μl of sterile water, spread on TSAerm, and then incubated at 43°C for 2 days to select against the plasmids. Isolated single colonies were inoculated in tryptic soy broth containing 10 μg·ml–1 erythromycin (TSBerm) and incubated at 43°C overnight. Cells were collected by centrifugation, suspended in 100 μl of TSM buffer (50 mM Tris·HCl, pH 7.5/0.5 M sucrose/10 mM MgCl2), and then treated with lysostaphin (0.1 mg·ml–1 final concentration) (23) at 37°C for 15 min. After collecting protoplasts by centrifugation, chromosomal DNA was purified with the Wizard Genomic DNA purification kit (Promega) according to the manufacturer's recommendation. AciI (NEB, Beverly, MA) was used to digest chromosomal DNA in 10 μl of reaction volumes for 1–3 h at 37°C. The digested DNA was ligated with T4 ligase in 100 μl of reaction volume for 5–16 h at room temperature, and the resulting DNA was purified with the QIAquick PCR purification kit (Qiagen, Valencia, CA). DNA fragments carrying transposon/chromosome junction sequences were amplified by PCR with the following primers: Martn-F (TTT ATG GTA CCA TTT CAT TTT CCT GCT TTT TC) and Martn-ermR (AAA CTG ATT TTT AGT AAA CAG TTG ACG ATA TTC). The annealing temperature was 63°C, and the DNA was amplified for 3 min with 40 cycles. After 0.8% agarose gel electrophoresis, PCR products were directly sequenced with the Martn-F primer at the sequencing facility of the University of Chicago Cancer Research Center.
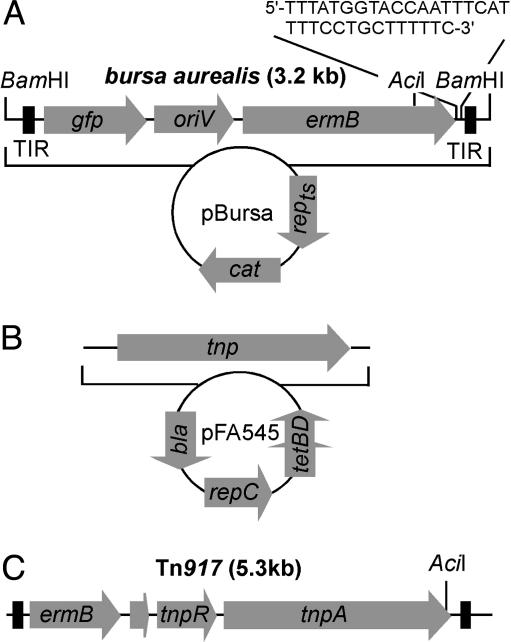
Fig. 1.
Plasmids used for transposon mutagenesis in S. aureus. (A) Bursa aurealis, a minimariner transposable element, was cloned into pTS2, with a temperature-sensitive plasmid replicon (repts) and chloramphenicol resistance gene (cat) to generate pBursa; the 3.2-kb bursa aurealis encompasses the mariner terminal inverted repeats (TIR), the green fluorescent protein gene (gfp), the R6K replication origin (oriV), and the erythromycin-resistance determinant (ermB). The position of restriction enzyme recognition sites (AciI and BamHI) is indicated. (B) Plasmid pFA545 encodes the mariner transposase (tnp), the origin of replication (repC), and the tetracycline- and ampicillin-resistance markers (tetBD and bla, respectively). (C) The structure of the 5.3-kb transposon Tn917 with the ermB erythromycin-resistance determinant, tnpR resolvase, and tnpA transposase is shown.
Nematode Killing Assay. The C. elegans infection assay was carried out as previously reported (22, 24) with the following modifications. To synchronize the growth of worms, eggs were collected via the hypochlorite method (25). Eggs were incubated in M9 buffer for 16–22 h at room temperature, and worms that had been arrested at the L1 larvae stage were plated on nematode growth medium agar that had been inoculated with Escherichia coli strain OP50. Agar plates were incubated at 15°C for 72 h to generate L4 larvae or for 84–88 h to generate adult worms. For initial screening procedures (screens 1–3 in Table 1, which is published as supporting information on the PNAS web site), 30–50 adult worms were used, whereas 50–100 L4 larvae or adult worms were infected for the final screening assay (screen 4 in Table 1) after transduction of transposon insertions into the wild-type strain background. Worms were transferred with a platinum wire and autoclaved Elmer's Glue to TSA plates that had been inoculated with staphylococci and incubated at 25°C for 48 h, and live worms were counted under a stereomicroscope (Nikon SMZ645). The nematode strain N2 and E. coli strain OP50 used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis).
Murine Intravenous Infection and Abscess Formation. S. aureus transposon mutants were grown at 37°C overnight in tryptic soy broth containing 10 μg·ml–1 erythromycin (TSBerm), diluted 100-fold in fresh media, and incubated at 37°C for 2.5 h until the cultures reached OD600 1.0. Bacteria were collected by centrifugation, washed, and suspended in PBS. Viable staphylococci were enumerated by colony formation on TSA plates to quantify the infectious dose. One hundred microliters of bacterial suspension (≈1.3 × 107 colony-forming units) was administered intravenously via retroorbital injection into each of 10 6- to 8-week-old BALB/c mice. Four days after the injection, the mice were killed by CO2 asphyxiation, and the kidneys and liver were removed. The organs were homogenized in 1 ml of PBS, and dilutions of the homogenates were plated on TSA. Student's t test was performed for statistical analysis by using the software analyze-it (Analyse-it Software, Leeds, U.K.).
Results
Bursa Aurealis. Bursa aurealis was derived from the Himar1 (mariner) element; it encompasses the short inverted repeats of the hornfly transposon (20, 21) as well as the ermB resistance marker (26) and a promotorless Aequorea victoria green fluorescent protein gene (Fig. 1). Insertion of bursa aurealis into genome sequences conferred resistance to erythromycin and expression of gfp when the insertion occurred in the vicinity of promoter sequences. Bursa aurealis was cloned into the pTS2 vector with a temperature-sensitive S. aureus replicon and chloramphenicol resistance gene (27), thereby generating pBursa (Fig. 1 A). The Himar1 transposase (21) was cloned along with the xylose repressor of pIK64 (28) into plasmid pSPT246ts (29), resulting in the transposase vector pFA545 (Fig. 1 A). Plasmids were transformed into S. aureus strain Newman by electroporation, and mutants were isolated by temperature shift to 43°C and by streaking for erythromycin-resistant phenotype. Isolated mutants were stored frozen, and the site of transposition for 10,325 isolates was examined by inverse PCR of bursa aurealis flanking sequences (Fig. 2). Analysis of insertion sequences revealed that bursa aurealis generates TA duplications at the insertion sites and does not exhibit sequence preference in the S. aureus genome (Fig. 3 and data not shown).
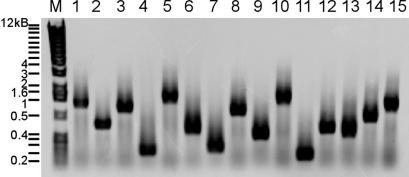
Fig. 2.
Mapping insertion sites by inverse PCR. Fifteen bursa aurealis transposon mutants of S. aureus strain Newman were subjected to DNA purification, AciI restriction, fragment ligation, inverse PCR, and agarose gel electrophoresis. M indicates the molecular weight marker (1-kb DNA ladder).
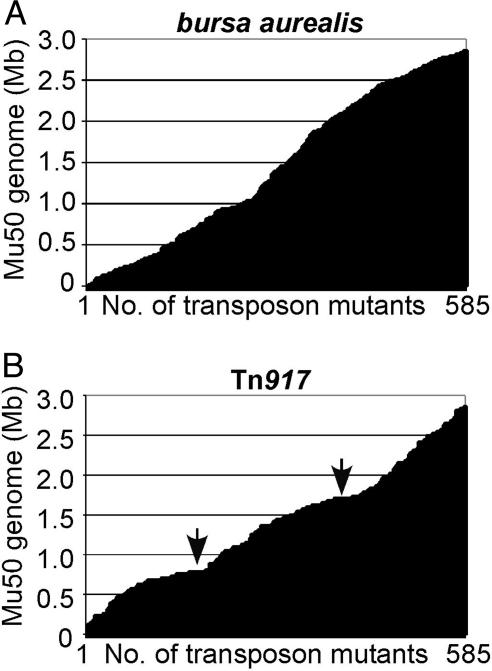
Fig. 3.
Comparison of the transposon insertion sites of bursa aurealis (A) and Tn917 (B). S. aureus strain Newman was subjected to mutagenesis, and 585 insertion mutants of each transposon were mapped by DNA sequencing and aligned according to their insertion site on the genome of S. aureus strain Mu50.
Analysis of Bursa Aurealis and Transposon 917 (Tn917) Transposition Events. Plasmid pTV1 carries Tn917 (30), a transposon that has been used for insertional mutagenesis in Gram-positive microbes (31, 32). We compared populations of transposon insertion mutants generated by Tn917 and bursa aurealis mutagenesis. After transformation of pTV1 and temperature-induced selection of transposon insertions, 960 S. aureus strain Newman mutants were generated with Tn917. Of 960 mutants, 585 mutants produced readable sequencing results, and the Tn917 insertion site was determined by using the same technique as described above for bursa aurealis. Insertion in noncoding DNA sequences occurred at a higher frequency for Tn917 (36% of all insertions as compared with 18% for bursa aurealis). Tn917 exhibited a strong bias for insertion in two regions on the staphylococcal chromosome centered at Mb 0.75 and 1.7, respectively (Fig. 3). Insertions of Tn917 (n = 585 insertions) were located in 271 different genes, whereas the same number of bursa aurealis insertions hit 350 different genes. In addition, bursa aurealis insertions did not display the same bias for insertion at Mb 0.75 and 1.7 as is reported here for Tn917 (Fig. 3).
Bursa Aurealis Mutagenesis of S. aureus Strain Newman. A total of 10,325 bursa aurealis isolates were generated and sequenced, and the ORF insertions were mapped (Fig. 4). Comparison of the data set with the genome sequence of S. aureus strain Mu50 indicated that 6,917 mutants carried bursa aurealis insertions within coding sequences, whereas 1,533 insertions occurred within intergenic or noncoding DNA sequences (ΦNΞ, Phoenix library; see Table 2, which is published as supporting information on the PNAS web site). The entire pBursa plasmid inserted at the S. aureus pre plasmid recombination site, yielding chloramphenicol/erythromycin double-resistant colonies that accounted for 7.6% of erythromycin-resistant colonies after mutagenesis. Together these experiments provided several insights. (i) The mutagenesis of the S. aureus chromosome with 10,325 bursa aurealis insertions was not saturated. When using the acquired data set for prediction, ≈20,000 random insertion mutants would be required to achieve saturating mutagenesis. (ii) Based on the comparison of raw sequence data with the published genome sequence of S. aureus strain Mu50, it appears that S. aureus strain Newman encodes ≈2,700 genes. Approximately 67% of all ORFs, 1,812 genes, have been disrupted by bursa aurealis insertion (Fig. 4).
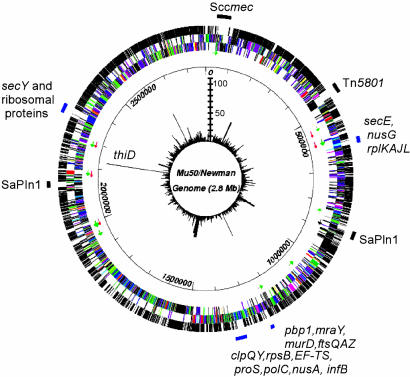
Fig. 4.
Bursa aurealis transposon insertions in the genome of S. aureus strain Newman. Four regions of the genome, in which very few transposon mutants were isolated, are indicated by blue bars with the names of the genes residing in each of the four regions. The first (outer) circle represents ORFs that were disrupted (black) or not disrupted (white) by bursa aurealis. The second and third circles represent ORFs in the genome of S. aureus strain Mu50, in which different colors were assigned according to their functions. Green and red arrows indicate tRNA and rRNA, respectively. The fourth circle shows the scale of the map (in bp). The fifth (inner) circle is a circular bar graph showing the number of transposon mutants per ORF (scale bar with each ladder step representing 10 mutants). Sccmec, staphylococcal cassette chromosome mec, and SaPIn1, staphylococcal pathogenicity island 1, are indicated. Bursa aurealis insertions (116) were isolated at the same position within thiD, but these insertions are likely to represent siblings, because the isolated mutants arose from the same batch of transposon insertion mutants.
Genes Required for S. aureus Growth. By generating a larger collection of bursa aurealis mutants, for example, a population of 20,000 mutants, all nonessential genes of S. aureus strain Newman may in theory be disrupted. It is unlikely, however, that one can isolate bursa aurealis insertions in nonessential genes whose transcription precedes that of essential genes in polycistronic operons. Based on the DNA sequence of the Mu50 strain, we assume that 700–800 genes were not hit by the transposon. Of these, 252 reside within operons in which upstream genes within the same predicted transcriptional unit have been disrupted by bursa aurealis, suggesting that these operons will likely not encode essential genes. The remaining 450–550 ORFs may encode genes that are essential for the growth of S. aureus strain Newman on laboratory media. Previous work compared the contents of S. aureus genome sequences and pointed to genes that occur in some, but not in all, staphylococcal isolates (13, 14). With the assumption that essential or housekeeping gene functions may be shared among staphylococci, some of the S. aureus strain Newman genes that are not found in other staphylococcal isolates may be uniquely responsible for animal virulence of strain Newman. For example, a comparison of the S. aureus MW2 genome sequence [a community-acquired methicillin-resistant S. aureus (MRSA) strain] with the sequences of S. aureus strains N315 and Mu50 (hospital-acquired MRSA) revealed 94.7% and 94.8% sequence identities, respectively (N315 and Mu50 display a 99.7% nucleotide match) (14). When analyzed with respect to the bursa aurealis pool of mutants, 234 insertions occurred in S. aureus strain Newman genes that did not display homology to other staphylococcal gene sequences and that may therefore represent mobile genetic elements or even virulence genes. Fig. 4 shows that several mobile elements of strain Mu50 display only limited or no homology to S. aureus strain Newman gene sequences.
S. aureus Virulence Genes Identified by Nematode Killing. Ausubel, Sifri, Calderwood and coworkers (22, 24) introduced the nematode C. elegans as a model host for the pathogenesis of S. aureus infections. When incubated on agar plates, staphylococci enter the worm oral cavity, pass the food grinder, and multiply in the C. elegans digestive tract. Colonization with S. aureus causes distension of the digestive tract and ultimately killing of infected worms over a period of 2–7 days (24). Using this assay, Sifri and coworkers (24) demonstrated that several known S. aureus virulence factors are also required for the killing of C. elegans. We therefore chose nematode killing as a first assay to identify virulence genes. S. aureus strain Newman mutants (n = 1,736) with a bursa aurealis insertion were screened on agar plates for the ability to kill between 30–50 worms. Adult C. elegans proved to be more susceptible to S. aureus infection than larvae and were used to screen the population of inserts. Both live and dead worms were enumerated under a dissecting microscope after a 48-h incubation at 25°C on agar plates. Statistical significance was examined by χ2 analysis, and mutants with a P value <0.1 were submitted to a second and third screening procedure with the same parameters. After three screening assays, 174 insertion mutants displayed reduced virulence and were analyzed further (P < 0.05). To determine whether unrelated mutations caused the observed reduced virulence phenotypes, each strain was lysed with phage φ 85 (33), and transposon insertions were transduced into S. aureus strain Newman wild type. The transduced bursa aurealis insertion sites were again mapped by inverse PCR and DNA sequencing. The transduced variants were then screened for defects in virulence by using agar plate infections of either adult worms or L4 larvae. Seventy-one mutants of the original 174 displayed reduced virulence in both assays (P < 0.05) (Table 3, which is published as supporting information on the PNAS web site), encompassing 41 genes with assigned function and 30 hypothetical ORFs (13). Genes with assigned functions could be classified into three categories: metabolism, regulation, and extracellular functions.
S. aureus Mutants with Insertions in Metabolic Genes. Approximately 70% of the bursa aurealis mutants with virulence defects carry insertions in genes responsible for energy metabolism or intermediary metabolism. This observation corroborates earlier work using in vivo expression technology (IVET) transposon screening techniques to define virulence defects in S. aureus, Salmonella species, or other enteric pathogens (34, 35). Further, enzymes of the S. aureus TCA cycle such as OdhB and CitB are required for S. aureus virulence in a murine model of bacteremia. CitB (aconitase) uses an iron–sulfur cluster to catalyze the conversion of citrate to isocitrate (36). Limited iron conditions affect the integrity of this iron–sulfur cluster, leading to an arrest of the TCA cycle, an accumulation of citrate, and a switch in the activity of CitB from hydrolyase to RNA binding. In E. coli and Bacillus subtilis, the regulatory RNA-binding protein CitB is involved in the oxidative stress response. In S. aureus, CitB inactivation may affect the level of other factors such as RNAIII (37), the regulatory molecule that controls the output of the Agr-dependent quorum-sensing system (38). Killing of C. elegans by S. aureus was markedly decreased in mutants interfering with other metabolic pathways such as urea cycle, pyrimidine biosynthesis, and sugar and amino acid metabolism (Table 3). Nematode killing was also reduced for two mutants impaired in DNA helicase activity (recG and recQ). Bursa aurealis insertions of recA have thus far not been isolated. However, signature-tag mutagenesis previously identified recA as required for virulence in a murine bacteremia model (31). Together these results suggest that replication, recombination, and, thus, functional maintenance of chromosomal DNA are important for virulence. Quality control of protein biogenesis may also be required for virulence. We observed that a ftsH mutation exhibited decreased virulence in the nematode killing assay. This observation is in agreement with ftsH mutants displaying attenuated virulence in a murine skin abscess model (39). ATP-dependent proteases such as Clp and FtsH are key factors in bacterial adaptation to environmental stress (39). It seems plausible that these bifunctional chaperones/proteases may also modulate the activity of S. aureus virulence factors.
S. aureus Mutants with Insertions in Regulatory Genes. The genome of S. aureus encodes five two-component regulatory systems and numerous transcription factors that are known to affect the production of virulence factors of S. aureus, albeit that important strain variations and differences in regulation occur (18). Of these, three two-component regulatory systems were hit by bursa aurealis (saeRS, srrAB, and agrAC), whereas insertions in arlRS were not identified (40). The two-component regulator encoded by the yycGF genes is known to be essential for S. aureus growth (41). Only insertions in two of the sensory systems, saeRS and srrAB (42, 43), caused defects in the worm killing assay. Previously, an agrA mutant was shown to display reduced virulence in the nematode killing assay (24). Interestingly, in our experiments, agrA, agrB, and agrC mutants did not display a consistent and significant reduction in virulence and were therefore not included in the list of virulence genes. Bursa aurealis insertions in 11 sarA homologs were isolated (44). Surprisingly, insertion in SarA did not cause a significant defect in virulence, and only insertions in the transcriptional regulator sarH2 diminished the ability of S. aureus strain Newman variants to kill worms. Mutations in msrR and tcaAB affect the susceptibility of S. aureus to methicillin and teicoplanin and are thought to affect cell wall metabolism (45, 46). Inactivation of msrR or tcaAB also affected the virulence of S. aureus during C. elegans infection.
S. aureus Mutants with Insertions in Genes Encoding Extracellular Factors. In S. aureus, synthesis and secretion of extracellular factors follow a quorum-controlled program, whereby surface adhesion genes are expressed before secreted hemolysins and host-tissue degradative or invasive enzymes (18). Only two extracellular factors, secreted nuclease and α-hemolysin, were required for the killing of C. elegans during S. aureus strain Newman infection. Further, attenuation of virulence was observed only for infections with L4 larvae, not with adult worms, suggesting that the screening assay conditions may be too stringent to identify secreted virulence factors that play redundant or at least partially overlapping functions during pathogenesis. Furthermore, the requirement for surface proteins in this model system was directly addressed by transducing and testing mutants with bursa aurealis insertions in 10 genes encoding surface proteins. None of the insertions reduced the C. elegans virulence properties of strain Newman, not even insertions in sortase (srtA), a gene required for the cell surface display of ≈20 different adhesins (33, 47). In Staphylococcus epidermidis, biofilm production has been shown to involve synthesis and secretion of polysaccharide intercellular adhesin (PIA), a β-1,6-linked N-acetylglucosamine polysaccharide. Several mechanisms contribute to biofilm formation, which requires the icaADBC genes for PIA biosynthesis and the atlE, ssp1, and clfA genes encoding adhesive surface proteins, as well as genes specifying other cofactors (dltA, aap, and bap) (48). Although most of these genes have been hit by bursa aurealis, the isolated mutants infected and killed worms in a manner that was indistinguishable from wild-type staphylococci. Thus, we conclude that sortase-anchored surface adhesins and the PIA biofilm of S. aureus are dispensable for staphylococcal replication in the C. elegans digestive track (Table 1). In contrast, insertions in genes involved in capsular polysaccharide biosynthesis, a known virulence factor in animals (49–51), caused reduced killing of adult worms and L4 larvae (Table 3).
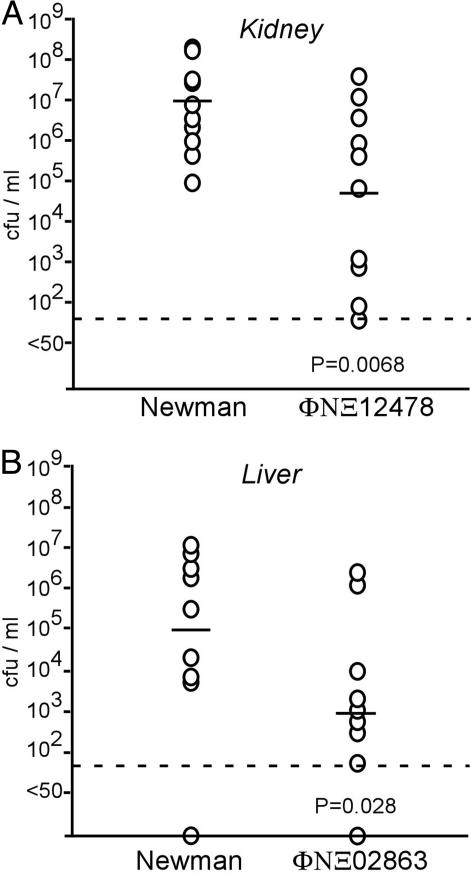
Murine Infection Experiments Identify S. aureus Virulence Genes. Our screen identified 30 hypothetical genes that are required for the pathogenesis of S. aureus infection in C. elegans. To gauge whether these genes represent virulence factors that are involved in general disease strategies, S. aureus strains were inoculated intravenously into mice, which were killed 4 days after infection (49). The kidney and liver were removed, inspected for abscess formation, and then homogenized and plated on agar medium to quantify staphylococcal replication in host tissues. Of the 11 strains that were analyzed in this manner (Table 3), nine mutant strains caused organ abscesses in the liver and kidney in a way that was indistinguishable from that of S. aureus wild-type strain Newman. Two strains, however, displayed significant defects in the ability to cause disease (Fig. 5). When compared with S. aureus strain Newman, strain ΦNΞ12478 with a bursa aurealis insertion in SAV0862, a hypothetical gene encoded by a lysogenic phage of S. aureus strain Newman, displayed a 2-log reduction in bacterial replication within kidney abscesses (Fig. 5). An association of virulence genes with lysogenic S. aureus phages has been described (13). ORF SAV2567 encodes a putative acyltransferase. Bursa aurealis insertion in SAV2567 caused a 2-log reduction in the ability of the mutant ΦNΞ02863 to establish liver abscesses in infected mice (Fig. 5). It should be noted that the observed defects in virulence for both strains were restricted to specific host organ tissues. ΦNΞ12478 did not show statistically significant defects in liver abscess formation, whereas the ΦNΞ02863 mutant caused kidney abscesses in a manner that was indistinguishable from that of wild-type strain Newman (data not shown).
Fig. 5.
Identification of S. aureus virulence genes in a mouse model of infection. S. aureus strain Newman or bursa aurealis insertion mutants ΦNΞ12478 (A) or ΦNΞ02863 (B) were used to infect 10 mice each via retroorbital injection [≈1.3 × 107 colony-forming units (cfu)]. After 4 days, mice were killed and organs (kidney and liver) were removed. Homogenized tissues were incubated on agar medium for S. aureus colony formation and enumeration. Each circle indicates an animal experiment. The dashed line represents the limit of detection, and horizontal bars indicate the mean.
Discussion
Previous work used Tn917 and signature-tagged mutagenesis to identify staphylococcal virulence factors (31, 32, 52). Libraries of 1,248 or 1,520 randomly chosen (nonsequenced) transposon insertion mutants of S. aureus were analyzed in animal infections with mixed populations to reveal a competitive disadvantage of individual variants. Based on the distribution of Tn917 knockout mutations that were analyzed here, we estimate that these studies examined ≈400–500 S. aureus genes. Even though only ≈20% of all S. aureus genes were analyzed, these studies provided important forays in defining the roles of specific S. aureus genes during the pathogenesis of specific infectious diseases (31, 32, 52). Here we report that bursa aurealis is significantly more efficient and more random than Tn917.
Although bursa aurealis insertion in many metabolic genes did not cause growth defects when staphylococci were grown on laboratory media (Table 3), host infection represents a unique ecological niche with nutritional challenges that may place mutants with specific blocks in metabolic pathways at a disadvantage. Thus, mutations that globally or specifically affect metabolic pathways may cause defects in pathogenesis.
Gene function of S. aureus has been previously examined by using antisense technology (53, 54). By cloning gene fragments in reversed orientation under control of an inducible promoter, the ability of antisense nucleotide sequences to interfere with S. aureus growth on agar plates was used to identify essential genes. In two independent studies, only 30% of all gene sequences (350 genes) in each study identified the same gene (75 genes total) (53, 54). Bursa aurealis insertions occurred in 110 of these presumed essential genes, suggesting that at least some of the assignments may not be correct (Table 1). Comparison of the data set described here with the recently reported characterization of essential genes in B. subtilis identified an overlap of ≈150–200 homologous genes (55).
Results presented here and elsewhere corroborate the usefulness of the nematode killing assay to identify virulence genes that are also required for staphylococcal infection of warm-blooded animals (22, 24). Nevertheless, there seem to be significant differences in the genetic requirements of S. aureus to cause disease in nematodes and mammals. One significant difference arises from the temperature during infection, because worms grow at 25°C but not at 37°C, whereas the temperature of mammalian tissues before infection is 37°C and during later stages of disease is between 32°C and 43°C (56). It is not yet clear how these temperature changes affect gene expression and virulence properties of staphylococci.
Our results and other studies show that genes required for the pathogenesis of one staphylococcal infection can be dispensable in a different disease model (52). In fact, very few genes seem to be required for the pathogenesis of all S. aureus infections in mammalian hosts, and even fewer genes play a role in both nematodes and warm-blooded animals. Thus, it appears that specific disease processes must be uniquely modeled during animal infection to obtain detailed views on the genetic requirements for disease. The collection of bursa aurealis mutants (ΦNΞ, Phoenix library) generated here will provide a useful resource in establishing the genetic requirements for the pathogenesis of staphylococcal disease entities (1, 22). This, in turn, will permit the rational design of antiinfective therapies that disrupt mechanistic processes of disease without implementing generalized antibiotic therapy.
Supplementary Material
Acknowledgments
We thank Costi Sifri, Barbara Page, and Karen Yook for their assistance in nematode killing assays; Kristin DeBord for help with the murine infection model; and members of our laboratories for critical reading of this manuscript. E.M.G. acknowledges support from the U.S. Department of Energy under Contract W-31-109-ENG-38. These studies were supported as University of Chicago Seed Project Awards (to D.M.M. and O.S.).
Abbreviations: TSA, tryptic soy agar; Tn917, transposon 917.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY672108 (pFA545) and AY672109 (pBursa)].
References
- 1.Lowy, F. D. (1998) New Engl. J. Med. 339, 520–532. [DOI] [PubMed] [Google Scholar]
- 2.Archer, G. L. (1998) Clin. Infect. Dis. 26, 1179–1181. [DOI] [PubMed] [Google Scholar]
- 3.Gillet, Y., Issartel, B., Vanhems, P., Fournet, J. C., Lina, G., Bes, M., Vandenesch, F., Piemont, Y., Brousse, N., Floret, D. & Etienne, J. (2002) Lancet 359, 753–759. [DOI] [PubMed] [Google Scholar]
- 4.Archer, G. L. & Climo, M. W. (2001) N. Engl. J. Med. 344, 55–56. [DOI] [PubMed] [Google Scholar]
- 5.Neu, H. C. (1992) Science 257, 1064–1073. [DOI] [PubMed] [Google Scholar]
- 6.Brumfitt, W. & Hamilton-Miller, J. (1989) N. Engl. J. Med. 320, 1188–1199. [DOI] [PubMed] [Google Scholar]
- 7.Hiramatsu, K., Hanaki, H., Ino, T., Yabuta, K., Oguri, T. & Tenover, F. C. (1997) J. Antimicrob. Chemother. 40, 135–136. [DOI] [PubMed] [Google Scholar]
- 8.Chang, S., Sievert, D. M., Hageman, J. C., Boulton, M. L., Tenover, F. C., Downes, F. P., Shah, S., Rudrik, J. T., Pupp, G. R., Brown, W. J., et al. (2003) N. Engl. J. Med. 348, 1342–1347. [DOI] [PubMed] [Google Scholar]
- 9.Weigel, L. M., Clewell, D. B., Gill, S. R., Clark, N. C., McDougal, L. K., Flannagan, S. E., Kolonay, J. F., Shetty, J., Killgore, G. E. & Tenover, F. C. (2003) Science 302, 1569–1571. [DOI] [PubMed] [Google Scholar]
- 10.Oliveira, D. C., Tomasz, A. & De Lencastre, H. (2002) Lancet Infect. Dis. 2, 180–189. [DOI] [PubMed] [Google Scholar]
- 11.Walsh, C. T. (2000) Nature 406, 775–781. [DOI] [PubMed] [Google Scholar]
- 12.Alksne, L. E. & Projan, S. J. (2000) Curr. Opin. Biotechnol. 11, 625–636. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda, M., Ohta, T., Uchiyama, I., Baba, T., Yuzawa, H., Kobayashi, I., Cui, L., Oguchi, A., Aoki, K., Nagai, Y., et al. (2001) Lancet 357, 1225–1240. [DOI] [PubMed] [Google Scholar]
- 14.Baba, T., Takeuchi, F., Kuroda, M., Yuzawa, H., Aoki, K., Oguchi, A., Nagai, Y., Iwama, N., Asano, K., Naimi, T., et al. (2002) Lancet 359, 1819–1827. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald, J. R., Sturdevant, D. E., Mackie, S. M., Gill, S. R. & Musser, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holden, M. T., Feil, E. J., Lindsay, J. A., Peacock, S. J., Day, N. P., Enright, M. C., Foster, T. J., Moore, C. E., Hurst, L., Atkin, R., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 9786–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duthie, E. S. & Lorenz, L. L. (1952) J. Gen. Microbiol. 6, 95–107. [DOI] [PubMed] [Google Scholar]
- 18.Novick, R. P. (2003) Mol. Microbiol. 48, 1429–1449. [DOI] [PubMed] [Google Scholar]
- 19.Somerville, G. A., Beres, S. B., Fitzgerald, J. R., DeLeo, F. R., Cole, R. L., Hoff, J. S. & Musser, J. M. (2002) J. Bacteriol. 184, 1430–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson, H. M. & Lampe, D. J. (1995) Mol. Biol. Evol. 12, 850–862. [DOI] [PubMed] [Google Scholar]
- 21.Lampe, D. J., Churchill, M. E. & Robertson, H. M. (1996) EMBO J. 15, 5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 22.Garsin, D. A., Sifri, C. D., Mylonakis, E., Qin, X., Singh, K. V., Murray, B. E., Calderwood, S. B. & Ausubel, F. M. (2001) Proc. Natl. Acad. Sci. USA 98, 10892–10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schindler, C. A. & Schuhardt, V. T. (1964) Proc. Natl. Acad. Sci. USA 51, 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sifri, C. D., Begun, J., Ausubel, F. M. & Calderwood, S. B. (2003) Infect. Immun. 71, 2208–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, J. A. & Fleming, J. T. (1995) Methods Cell Biol. 48, 4–27. [PubMed] [Google Scholar]
- 26.Trieu-Cuot, P., Carlier, C., Poyart-Salmeron, C. & Courvalin, P. (1990) Nucleic Acids Res. 18, 4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzgerald, S. N. & Foster, T. J. (2000) J. Bacteriol. 182, 1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kullik, I., Giachino, P. & Fuchs, T. (1998) J. Bacteriol. 180, 4814–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janzon, L. & Arvidson, S. (1990) EMBO J. 9, 1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartley, R. W. & Paddon, C. J. (1986) Plasmid 16, 45–51. [DOI] [PubMed] [Google Scholar]
- 31.Mei, J. M., Nourbakhsh, F., Ford, C. W. & Holden, D. W. (1997) Mol. Microbiol. 26, 399–407. [DOI] [PubMed] [Google Scholar]
- 32.Schwan, W. R., Coulter, S. N., Ng, E. Y., Langhorne, M. H., Ritchie, H. D., Brody, L. L., Westbrock-Wadman, S., Bayer, A. S., Folger, K. R. & Stover, C. K. (1998) Infect. Immun. 66, 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazmanian, S. K., Liu, G., Jensen, E. R., Lenoy, E. & Schneewind, O. (2000) Proc. Natl. Acad. Sci. USA 97, 5510–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowe, A. M., Beattie, D. T. & Deresiewicz, R. L. (1998) Mol. Microbiol. 27, 967–976. [DOI] [PubMed] [Google Scholar]
- 35.Mahan, M. J., Slauch, J. M. & Mekalanos, J. J. (1993) Science 259, 686–688. [DOI] [PubMed] [Google Scholar]
- 36.Kohler, C., von Eiff, C., Peters, G., Proctor, R. A., Hecker, M. & Engelmann, S. (2003) J. Bacteriol. 185, 6928–6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somerville, G. A., Said-Salim, B., Wickman, J. M., Raffel, S. J., Kreiswirth, B. N. & Musser, J. M. (2003) Infect. Immun. 71, 4724–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novick, R. P., Ross, H. F., Projan, S. J., Kornblum, J., Kreiswirth, B. & Moghazeh, S. (1993) EMBO J. 12, 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lithgow, J. K., Ingham, E. & Foster, S. J. (2004) Microbiology 150, 373–381. [DOI] [PubMed] [Google Scholar]
- 40.Fournier, B., Klier, A. & Rapoport, G. (2001) Mol. Microbiol. 41, 247–261. [DOI] [PubMed] [Google Scholar]
- 41.Dubrac, S. & Msadek, T. (2004) J. Bacteriol. 186, 1175–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giraudo, A. T., Calzolari, A., Cataldi, A. A., Bogni, C. & Nagel, R. (1999) FEMS Microbiol. Lett. 177, 15–22. [DOI] [PubMed] [Google Scholar]
- 43.Pragman, A. A., Yarwood, J. M., Tripp, T. J. & Schlievert, P. M. (2004) J. Bacteriol. 186, 2430–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheung, A. L., Projan, S. J. & Gresham, H. (2002) Curr. Infect. Dis. Rep. 4, 400–410. [DOI] [PubMed] [Google Scholar]
- 45.Rossi, J., Bischoff, M., Wada, A. & Berger-Bächi, B. (2003) Antimicrob. Agents Chemother. 47, 2558–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brandenberger, M., Tschierske, M., Giachino, P., Wada, A. & Berger-Bächi, B. (2000) Biochim. Biophys. Acta 1523, 135–139. [DOI] [PubMed] [Google Scholar]
- 47.Mazmanian, S. K. & Schneewind, O. (2002) in Bacillus subtilis and Its Closest Relatives, eds. Sonenshine, A., Losick, R. & Hoch, J. (Am. Soc. Microbiol., Washington, DC), pp. 57–70.
- 48.Götz, F. (2002) Mol. Microbiol. 43, 1367–1378. [DOI] [PubMed] [Google Scholar]
- 49.Albus, A., Arbeit, R. D. & Lee, J. C. (1991) Infect. Immun. 59, 1008–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee, J. C., Betley, M. J., Hopkins, C. A., Perez, N. E. & Pier, G. B. (1987) J. Infect. Dis. 156, 741–750. [DOI] [PubMed] [Google Scholar]
- 51.Sau, S., Bhasin, N., Wann, E. R., Lee, J. C., Foster, T. J. & Lee, C. Y. (1997) Microbiology 143, 2395–2405. [DOI] [PubMed] [Google Scholar]
- 52.Coulter, S. N., Schwan, W. R., Ng, R., Langhorne, E. Y., Ritchie, M. H., Westbrock-Wadman, S., Hufnagle, W. O., Folger, K. R., Bayer, A. S. & Stover, C. K. (1998) Mol. Microbiol. 30, 393–404. [DOI] [PubMed] [Google Scholar]
- 53.Ji, Y., Zhang, B., Van, S. F., Horn, Warren, P., Woodnutt, G., Burnham, M. K. & Rosenberg, M. (2001) Science 293, 2266–2269. [DOI] [PubMed] [Google Scholar]
- 54.Forsyth, R. A., Haselbeck, R. J., Ohlsen, K. L., Yamamoto, R. T., Xu, H., Trawick, J. D., Wall, D., Wang, L., Brown-Driver, V., Froelich, J. M., et al. (2002) Mol. Microbiol. 43, 1387–1400. [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi, K., Ehrlich, S. D., Albertini, A., Amati, G., Andersen, K. K., Arnaud, M., Asai, K., Ashikaga, S., Aymerich, S., Bessieres, P., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vlach, K. D., Boles, J. W. & Stiles, B. G. (2000) Comp. Med. 50, 160–166. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.