Abstract
The plant hormone auxin can regulate gene expression by destabilizing members of the Aux/IAA family of transcriptional repressors. Auxin-induced Aux/IAA degradation requires the protein-ubiquitin ligase SCFTIR1, with auxin acting to enhance the interaction between the Aux/IAAs and SCFTIR1. SKP1, Cullin, and an F-box-containing protein (SCF)-mediated degradation is an important component of many eukaryotic signaling pathways. In all known cases to date, the interaction between the targets and their cognate SCFs is regulated by signal-induced modification of the target. The mechanism by which auxin promotes the interaction between SCFTIR1 and Aux/IAAs is not understood, but current hypotheses propose auxin-induced phosphorylation, hydroxylation, or proline isomerization of the Aux/IAAs. We found no evidence to support these hypotheses or indeed that auxin induces any stable modification of Aux/IAAs to increase their affinity for SCFTIR1. Instead, we present data suggesting that auxin promotes the SCFTIR1–Aux/IAA interaction by affecting the SCF component, TIR1, or proteins tightly associated with it.
The hormone auxin influences nearly every aspect of plant growth, from patterning in the early embryo to the control of adult plant architecture (1). These diverse effects are mediated partly by the ability of auxin to regulate the expression of numerous genes by controlling the abundance of transcriptional repressor proteins of the Aux/IAA family (2–5).
Aux/IAA proteins, of which there are 29 in Arabidopsis, are characterized by four conserved domains named I–IV (6, 7). A very early biochemical manifestation of the auxin response (≤5 min) is the dose-dependent increase in the interaction between Aux/IAAs and the E3 ubiquitin-protein ligase SCFTIR1, as monitored by pull-down assays in cell-free extracts (2).
SKP1, Cullin, and F-box-containing proteins (SCFs) act to ubiquitinate their targets, marking them for degradation by the 26S proteasome (reviewed in ref. 8). They are multisubunit enzymes named after their first three characterized subunits; SKP1, Cullin, and an F-box-containing protein (8). The Cullin interacts with a fourth subunit, RBX1, to catalyze ubiquitination (8). The SKP1 protein acts as a scaffold to link the Cullin-RBX dimer to the F-box protein. F-box proteins are characterized by an N-terminal F-box motif through which they interact with SKP1. Their C termini include one of a diverse range of protein–protein-interaction domains, which confer substrate specificity to the SCF complex (8). In the case of SCFTIR1, the TIR1 F-box protein contains leucine-rich repeats (9), which are required to destabilize the Aux/IAAs (2).
Domain II of the Aux/IAA is the site of the destabilization signal required for interaction with SCFTIR1 (10–12). A 13-aa core region of domain II has been shown to be necessary and sufficient to confer auxin-regulated SCFTIR1 interaction and instability on translationally fused reporter proteins and hence constitutes a degron (11). Of the degron consensus sequence (QVVGWPPVRSYRK), the central GWPPV residues are absolutely essential to both fusion-protein instability and interaction with SCFTIR1 (11). Mutations in this core region result in increased and auxin-resistant Aux/IAA stability, reduced SCFTIR1 interaction, and, in the context of an otherwise intact Aux/IAA in planta, a plethora of auxin-response phenotypes (2, 7, 13). Thus, the ability of auxin to regulate plant development depends on its ability to regulate the interaction between Aux/IAA domain II and SCFTIR1. Unfortunately, the mechanism by which auxin influences this interaction is poorly understood.
Several models have been proposed to account for the effect of auxin. Foremost has been the idea that the auxin-enhanced Aux/IAA–SCFTIR1 interaction requires covalent modification of domain II. Most of the characterized canonical SCF–target protein interactions from yeast and mammals depend on the phosphorylation of the target protein (8). However, several lines of evidence suggest that phosphorylation does not play a role in regulating the SCFTIR1–Aux/IAA interaction: The 13-aa degron, necessary and sufficient for auxin-induced destabilization of Aux/IAAs, does not contain any essential phosphorylatable residues (11), and a broad spectrum of kinase and phosphatase inhibitors does not interfere with the interaction (13, 14). There are a few known exceptions to the phosphorylation paradigm. For example, the human SCFFbx2 has been shown to recognize N-glycosylated proteins rejected by the endoplasmic reticulum (15). A second exception is in the degradation of the transcription factor hypoxia-inducible factor α (HIFα) by von Hippel–Lindau tumor suppressor complex, an E3 ligase structurally and functionally analogous to the SCF (16, 17). Here interaction depends on proline hydroxylation. This is an attractive hypothesis for regulating the Aux/IAA–SCFTIR1 interaction, because the degron contains two adjacent central proline residues that are essential for the auxin-regulated Aux/IAA–SCFTIR1 interaction (2). However, a variety of inhibitors and enhancers of proline hydroxylation, known to affect the stability of HIFα, have no effect on SCFTIR1–Aux/IAA interaction or Aux/IAA stability in planta (13, 14). Furthermore, a synthetic proline-hydroxylated domain II peptide was shown to interact in an auxin-regulated manner with SCFTIR1 (14).
An alternative model is that auxin regulates the Aux/IAA–SCFTIR1 interaction by altering the isomerization state of the domain II prolines. This hypothesis is supported by the observation that juglone, an inhibitor of the parvulin class of peptidyl proline isomerases (PPIases) is able to inhibit the Aux/IAA–SCFTIR1 interaction (13, 14). An additional hypothetical link to a role for proline isomerization has come from the recent identification and characterization of the SIR1 gene, mutation in which results in resistance to sirtinol, a synthetic compound that was shown to destabilize Aux/IAA proteins (18). SIR1 encodes a protein with a C-terminal rhodanese-like domain that shares homology with the C-terminal domain of a single member of the Arabidopsis parvulin PPIase family. It has been suggested that these two proteins might cooperate to regulate the isomerization of domain II prolines and hence the ability of Aux/IAAs to interact with SCFTIR1 (18).
To date, these various hypotheses about the mode of auxin action in increasing the Aux/IAA–SCFTIR1 interaction are largely derived from predicting a specific domain II modification and then testing for the effects of this modification or its predicted inhibitors on the interaction. We have taken more generic approaches to look for domain II modifications. We are unable to find any evidence to support the idea that auxin causes the modification of Aux/IAAs to increase their affinity for SCFTIR1. Instead, we propose that auxin acts through modification of TIR1 or a tightly associated protein.
Methods
Plant Materials and Extracts. All transgenic and mutant lines have been described (19). Seedlings for protein extraction were grown at 22°C under continuous illumination for 7 days in liquid Arabidopsis thaliana salt medium under sterile conditions (19). Extracts were made by grinding in liquid nitrogen and solubilizing in extraction buffer (0.15 M NaCl/0.5% Nonidet P-40/0.1 M Tris·HCl, pH 7.5/β-glycerophosphate at 10 mM/PMSF/DTT/NaF at 1 mM/MG132 at 10 μM/pepstatin A at 2 μM). Extracts were cleared by centrifugation at 18,000 × g for 20 min. Protein levels were determined by using Coomassie Plus reagent (Pierce) according to manufacturer instructions.
Peptides and GST Fusion Proteins. The peptide biotinyl-NH-AKAQVVGWPPVRNYRKN-COOH was synthesized by Thermo Hybaid (Ulm, Germany). GST-AXR2 has been described (2). To construct GST-AXR2dII, the primers 5′-AAGGATCCCTTAAAGATCCTTCTAAGCCTCC-3′ and 5′-AAGAATTCTGCTGAGTCATCATGTTCTTC-3′ were used to amplify the domain II coding sequence from GST-AXR2. This fragment was cloned in frame into pGEX-6P-1 (Amersham Pharmacia) and transformed into BL21(DE3) Escherichia coli. GST-AXR2dII was purified on glutathione (GSH)-Sepharose (Amersham Pharmacia) according to manufacturer instructions. The mutant derivative GST-AXR2dIIPP-AA was generated by mutagenesis of the GST-AXR2dII plasmid with the QuikChange site-directed mutagenesis kit (Stratagene) and complimentary oligonucleotides with the sequence 5′-CACAAGTGGTGGGATGGGCAGCTGTGAGGAACTAC-3′ and expressed in BL21(DE3) E. coli as described above.
Electrophoresis, Staining, and Western Transfer. A 1D PAGE was performed on NuPAGE Novex 4–12% [bis(2-hydroxyethyl)amino]tris(hydroxymethyl) gels (Invitrogen) according to manufacturer instructions. Samples run for mass-spectrometric analysis were visualized by either Coomassie staining (SimplyBlue SafeStain, Invitrogen) or silver staining (SilverQuest silver staining kit, Invitrogen). For Western blotting of pull-down assay samples, 1D gels were electroblotted onto Invitrolon poly(vinylidene difluoride) by using NuPAGE transfer buffer (Invitrogen) according to manufacturer instructions.
Mass-Spectrometric Analysis. Twelve micrograms of GST-AXR2dII or GST-AXR2dIIPP-AA fusion protein, immobilized on GSH-Sepharose beads (Amersham Pharmacia), was incubated in 7.5 mg of crude Arabidopsis extract for 1 h at 4°C either with or without 10 μM 1-napthalene acetic acid (NAA) and then washed five times in extraction buffer. Over several experiments the treated fusion proteins were resolved by either 1D or 2D SDS/PAGE as described above. Protein bands or spots were excised and destained before tryptic digestion. Bands from 1D gels were also reduced and alkylated before digestion. These steps and the tryptic digests were performed according to protocols supplied with the SilverQuest silver staining kit (Invitrogen) except that 0.1% octyl-β-d-glucopyranoside was included in the digest reaction using Promega sequencing-grade modified trypsin. After overnight digestion, 1 μl of the digest was spotted onto the matrix-assisted laser desorption ionization (MALDI) plate and overlaid with ≈5 mg·ml–1 α-cyano-4-hydroxycinnamic acid in 0.1% trifluoroacetic acid/50% acetonitrile. For MS analysis of domain II peptides, the peptides were first immobilized on monomeric avidin agarose (SoftLink avidin agarose, Promega) according to manufacturer instructions. After treatment in plant extracts as described above, the peptides were washed and then eluted in 2 mM biotin/20 mM ammonium acetate, pH 7.0, dried, and resuspended in 10 μl of 0.1% trifluoroacetic acid. The peptides then were concentrated on μC18 ZipTips (Millipore) and eluted in matrix solution directly onto the MALDI plate according to manufacturer instructions. MALDI/time-of-flight (TOF) and MALDI-MS/MS analyses were performed on an Applied Biosystems 4700 proteomics analyzer.
Pulldowns, Immunoprecipitation, Conditioning Assays, Western Blotting, and Densitometry. Pulldowns with GST-AXR2dII were performed as described (2). For pulldowns using the biotinylated domain II peptide, 6.5 μg of peptide was used, recovered on streptavidin agarose (Novagen), and processed as described for GST-based pulldowns. Immunoprecipitation of TIR1myc was performed with anti-Myc 9E10 antibody (Covance, Berkeley, CA) crosslinked to protein G plus agarose (Novagen) (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
For all experiments except sirtinol treatment of intact plants, auxin (NAA) and inhibitors were added directly to plant extracts at the concentrations indicated. Juglone and sirtinol were purchased from Calbiochem and dissolved in DMSO. The DTT suppression of juglone was achieved by adding 2.5 mM DTT to extracts before juglone treatment. Sirtinol treatment of intact plants was 25 or 100 μM for 3 h, both added to the growing medium. Conditioning and partitioned inhibitor experiments were simply elaborations of the basic pull-down assay using similar quantities of GST fusions and domain II peptide. Thus, for domain II conditioning experiments (see Fig. 2b), 6.5 μg of immobilized domain II peptide or 4 μg of GST-AXR2dII protein were first incubated in 2.5 mg of crude tir1-1 extract (TIR1myc–) for 2 h at 4°C, then washed eight times (228 bed volumes) in a high-salt/detergent buffer (0.5 M NaCl/2% Nonidet P-40/0.1 M Tris·HCl, pH 7.5), and finally incubated in 2.5 mg of crude tir1-1[TIR1myc] extract (TIR1myc+) for 2 h at 4°C to assess recovery of TIR1myc. For the partitioned juglone/N-ethylmaleimide (NEM) experiments (see Fig. 3d), 2.5-mg aliquots of tir1-1 and tir1-1[TIR1myc] extract were treated with 10 μM NAA and either 100 μM juglone or 2.5 mM NEM. Paired aliquots then were mixed in the presence of 2.5 mM DTT and used for standard pull-down assays with 6.5 μg of domain II peptide. For TIR1 conditioning experiments (see Fig. 4 a and b), TIR1myc was immunoprecipitated by using 125 μl of crosslinked anti-Myc beads per 12.5 mg of tir1-1[TIR1myc] (TIR1myc+) extract treated with or without 10 μM NAA. The immunoprecipitates were washed eight times (640 bed volumes) in extraction buffer and eluted by vigorous agitation in 0.5 M NaCl/2% Nonidet P-40/10% dioxane/0.1 M Tris·HCl, pH 7.0. The eluates were diluted 1:10 with identical aliquots of tir1-1 (TIR1myc–) extract and used in standard pull-down assays with 6.5 μg of domain II peptide.
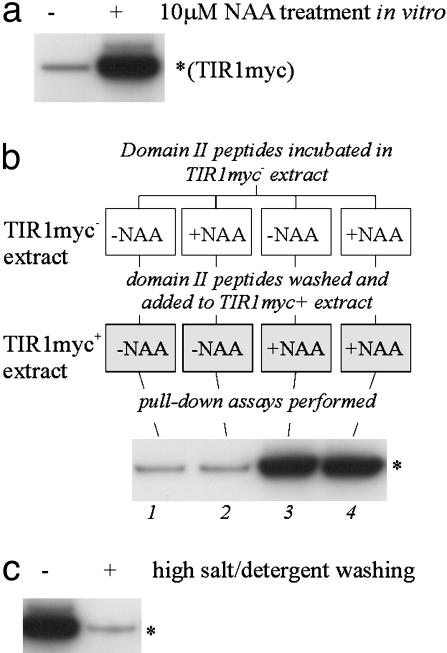
Fig. 2.
Auxin-enhanced SCFTIR1–Aux/IAA interaction is not mediated by stable modification of the Aux/IAA. (a) A synthetic Aux/IAA domain II peptide was used in pull-down assays from extracts of tir1-1[TIR1myc] plants without and with auxin (10 μM NAA) added in vitro. The recovery of TIR1myc on domain II peptide beads was assessed by immunoblotting with anti-c-Myc antibody (n = 40). (b) Immobilized domain II peptides were mock-treated or auxin-treated in TIR1myc– extract before being washed and used in pull-down assays from TIR1myc+ extract either mock- or auxin-treated as indicated in the schematic. The products were immunoblotted with anti-c-Myc antibody (n = 5). (c) The effect of high-salt/detergent washing of otherwise identical pulldowns (n = 1). n, the number of independent replicate, *, the position of the TIR1myc band.
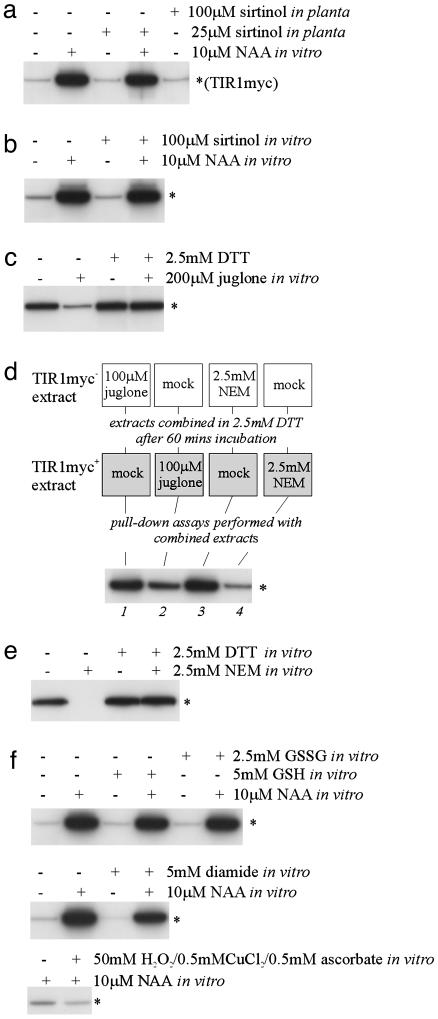
Fig. 3.
The inhibitors juglone and NEM but not sirtinol affect SCFTIR1–Aux/IAA interaction. (a) Standard domain II peptide pulldowns from extracts of tir1-1[TIR1myc] plants mock-treated or treated with sirtinol for 3 h before extraction (n = 2). (b) As described for a but with 100 μM sirtinol added directly to the pull-down assay (n = 2). (c) Standard domain II peptide pulldowns treated with 200 μM juglone and/or 2.5 mM DTT (n = 4). (d) Partitioned juglone/NEM experiment. Each lane represents a pulldown from an aliquot of TIR1myc– extract combined with an aliquot of TIR1myc+ extract. Before being mixed in the presence of DTT, the aliquots were pretreated with juglone or NEM or were mock-treated as indicated in the schematic (n = 3). (e) Standard domain II peptide pulldowns treated with 2.5 mM NEM and/or 2.5 mM DTT as indicated (n = 2). (c–e) All extracts were treated with 10 μM NAA in vitro.(f) Standard domain II peptide pulldowns treated with GSH/oxidized GSH (GSSG), diamide, and H2O2 as indicated. n, the number of independent replicates; *, the position of the TIR1myc band.
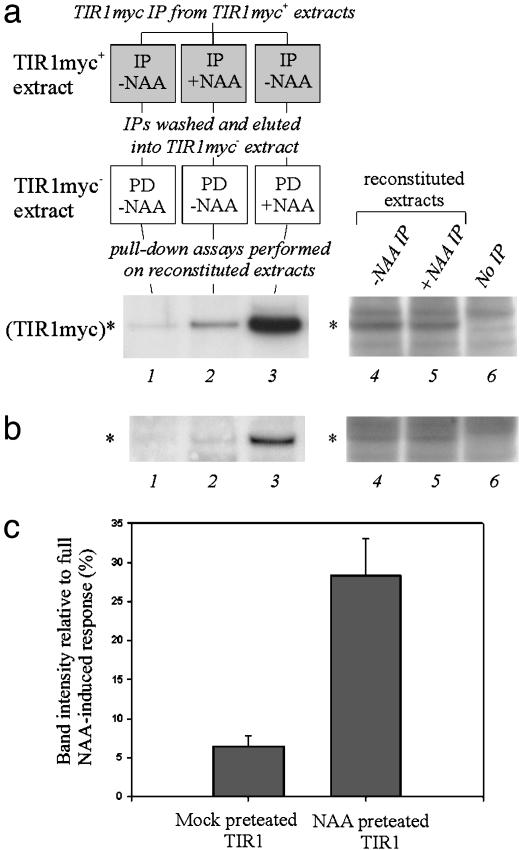
Fig. 4.
Auxin-enhanced SCFTIR1–Aux/IAA interaction may require modification of TIR1 or tightly associated proteins. (a) TIR1myc was immunoprecipitated (IP) from either mock- or auxin-treated tir1-1[TIR1myc] extract, washed, and eluted into tir1-1 extract. Pulldowns with domain II peptide were performed in these reconstituted extracts as indicated in the schematic. (b) As described for a but performed with additional controls for auxin carry through: in parallel with the immunoprecipitation stages, control beads (no antibody) were treated with and without auxin and combined with immunoprecipitated beads just before elution (–NAA IP with +NAA control, +NAA IP with –NAA control). (a and b) Lane 3 shows the full NAA-induced response where 10 μM NAA was added to the reconstituted extracts before pulldown. Lanes 4–6 in show the levels of TIR1myc in reconstituted extracts made with tir1-1 extract and –NAA IP, +NAA IP, and no immunoprecipitation, respectively. (c) Mean densitometric measurements of mock- and NAA-pretreated TIR1myc in pulled-down with domain II peptide as a percentage of the full NAA-induced response (n = 4). n, the number of independent replicates; *, the position of the TIR1myc band. (Error bars, SEM.)
The final processing of all pull-down assays described above consisted of washing three times in extraction buffer and elution in 1× lithium dodecyl sulfate loading buffer (Invitrogen), with the eluate being collected by microcentrifugation in minispin columns (Promega). The samples were electrophoresed and blotted as described above. Immunodetection of TIR1myc was performed by using a 1:1,000 dilution of monoclonal anti-c-Myc 9E10 antibody (Covance), followed by a 1:5,000 dilution of goat anti-mouse IgG γ-chain-specific peroxidase conjugate (Sigma) and chemiluminescent detection using ECL Plus reagents (Amersham Pharmacia). Densitometric measurements were made by using a genesnap/genetools documentation system (SynGene, Cambridge, U.K.).
Results
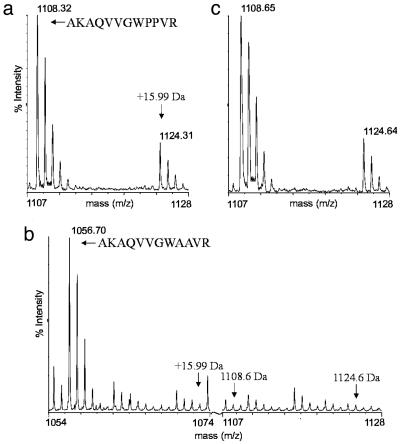
Auxin Does Not Induce Mass-Shifting Modifications to Domain II Peptides. To identify any auxin-induced posttranslational modifications to domain II that might regulate the interaction with SCFTIR1, we took a mass-spectrometric approach. A translational fusion of GST and domain II of AXR2 (GST-AXR2dII), capable of auxin-enhanced interaction with SCFTIR1, was immobilized on GSH-Sepharose beads and exposed to plant extract (initially without added auxin), washed, and resolved by SDS/PAGE. The protein then was excised and tryptically digested before MALDI/TOF MS. This analysis revealed that accompanying the 1,108.6-Da tryptic fragment (AQVVGWPPVR), corresponding to the major part of the 13-aa degron, was an additional fragment at +15.99 Da that could not be attributed to any other part of the digested protein (Fig. 1a). Such mass shifts are indicative of an oxidation event and, within this sequence, are most likely the result of hydroxylation of one of the prolines. This was confirmed by comparison of the 1,108- and 1,124-Da MALDI MS/MS spectra (data not shown) and changes in the MALDI MS spectrum of a trypsinized GST-AXR2dII derivative in which both prolines were substituted with alanines (GST-AXR2dIIPP-AA), in which the 1,108- and 1,124-Da fragments were replaced with just one of 1,056 Da (Fig. 1b). Given the established regulation of HIFα stability by proline hydroxylation (16, 17), this was a potentially interesting result. However, the relevance of this modification as a prerequisite for Aux/IAA–SCFTIR1 interaction is highly dubious, because the apparent hydroxylation is not affected by auxin (Fig. 1c) and indeed is observed, albeit at a lower level, in preparations of the bacterially expressed GST-AXR2dII that have not been exposed to plant extracts (data not shown). Additionally, as mentioned above, a synthetic domain II peptide with hydroxylated prolines has been shown to interact in an auxin-regulated manner with SCFTIR1, and a variety of inhibitors and enhancers of proline hydroxylation, known to affect the stability of HIFα in mammalian cells, have no effect on SCFTIR1–Aux/IAA interaction or Aux/IAA stability in planta (data not shown; refs. 13 and 14).
Fig. 1.
Auxin does not induce mass-shifting modifications within domain II. (a and c) MALDI/TOF MS spectra of trypsinized GST-AXR2dII exposed to plant extract without (a) and with (c) added auxin (10 μM NAA) (n = 5). The 1,108-Da peak corresponding to the major part of the domain II degron and the derivative peak at +15.99 Da are indicated. (b) MALDI/TOF MS spectrum of trypsinized GST-AXR2dIIPP-AA (both prolines replaced by alanines). The new 1,056-Da peak corresponding to the major part of the mutant domain II degron is marked. For comparison, the positions at 1,056 + 15.99 Da, 1,108 Da, and 1,124 Da are indicated (n = 2). n, the number of independent replicates.
This apparent hydroxylation event was the only mass shift we detected in these experiments. However, a region toward the C-terminal end of the degron might be underrepresented in our analysis because of the proximity of a number of tryptically susceptible arginine and lysine residues. Therefore, we performed similar experiments with a synthetic biotin-labeled domain II peptide (see below) immobilized on monomeric avidin agarose and eluted competitively with biotin. These eluates were prepared for MALDI/TOF spectrometry without electrophoresis or tryptic digestion. We found no evidence of any modification within the domain II degron other than proline hydroxylation.
Auxin-Enhanced SCFTIR1–Aux/IAA Interaction Is Not Mediated by Stable Modification of Domain II. To monitor the Aux/IAA–SCFTIR1 interaction, we previously used GST-tagged versions of the Aux/IAA proteins AXR2 and AXR3 in pull-down assays with extracts from tir1-1 mutant plants expressing a Myc-tagged TIR1 transgene (TIR1myc) (2). In addition, we synthesized a biotinylated 17-aa peptide corresponding to the core residues of domain II and encompassing the 13-aa degron. As with GST-Aux/IAA fusions, this peptide robustly supports the auxin-enhanced recovery of TIR1myc in pull-down assays (Fig. 2a).
To explore further whether auxin stably modifies this peptide to enhance its interaction with SCFTIR1, we conducted a series of conditioning experiments in which the auxin treatment of domain II was temporally separated from its exposure to TIR1myc. Biotinylated domain II peptide was incubated in TIR1myc– extract (tir1-1) either without or with 10 μM NAA (auxin pretreatment) for 2 h at 4°C. The peptides then were recovered on streptavidin beads and washed extensively in a high-salt/detergent buffer to remove auxin and dislodge bound proteins. The domain II peptide beads were then equilibrated in 0.1 M Tris·HCl, pH 7.5, and added to TIR1myc+ extract (tir1-1[TIR1myc]) either without or with 10 μM NAA, and the recovery of TIR1myc in the four assays was compared. Fig. 2b (lanes 1 and 2) shows that auxin pretreatment of the peptide had no effect on its ability to interact with TIR1myc compared with the mock-treated control. Additionally, Fig. 2b (lanes 3 and 4) demonstrates that the washing regime does not affect the ability of the peptide to interact with TIR1myc in an auxin-dependent manner. The effectiveness of the washing after pretreatment was judged by its ability to dislodge TIR1myc from domain II peptide beads. Fig. 2c shows that this washing is effective, removing the vast majority of TIR1myc. These conditioning experiments were repeated five times with domain II peptide and three times with GST-AXR2dII (data not shown) with identical results. Densitometric quantification of five domain II peptide conditioning experiments confirmed that there was no difference in the recovery of TIR1myc by auxin-pretreated and mock-pretreated peptides (t test: P = 0.77). Together, these data indicate that the domain II degron cannot be preconditioned with auxin to interact better with TIR1. Therefore, if any auxin-induced modification of domain II occurs, it must not only be non-mass-changing but also not stable or not sufficient to promote the SCFTIR1–Aux/IAA interaction.
Sirtinol Does Not Affect the SCFTIR1–Aux/IAA Interaction. Because no stable domain II modifications were induced by auxin, we next investigated in more detail the extant evidence for a role for proline isomerization, which could be unstable. This evidence comes from two sources. First, the ability of plants to respond to sirtinol, a drug that destabilizes Aux/IAAs, has been shown to require the Arabidopsis protein SIR1, which shares homology with an Arabidopsis PPIase (18). We therefore tested whether sirtinol could enhance the SCFTIR1–Aux/IAA interaction. Treatment with sirtinol, both of intact plants before extraction and in the pulldown, had no effect on the interaction (Fig. 3 a and b).
Juglone Inhibits SCFTIR1–Aux/IAA Interaction Through TIR1 or Associated Proteins. The second line of evidence to support a role for proline isomerization comes from the use of juglone, an inhibitor of the parvulin class of PPIases, which has been shown to reduce the SCFTIR1–Aux/IAA interaction (13, 14). Juglone inhibits parvulin-type PPIases by its covalent attachment to their cysteine sulfhydryl groups (20). This inhibitory effect is abolished in the presence of modest concentrations of DTT (20). We found this also to be true for the inhibition of SCFTIR1–Aux/IAA interaction by juglone (Fig. 3c).
The nullifying effect of DTT enabled us to perform the following experiment to assess on which of the interacting components juglone is acting. Aliquots of TIR1myc– seedling extract (tir1-1) and TIR1myc+ extract (tir1-1[TIR1myc]), equal in terms of volume and protein concentration, were used. In the first treatment (Fig. 3d, lane 1), one aliquot of TIR1myc– extract was treated with 10 μM NAA and 100 μM juglone, and one aliquot of TIR1myc+ extract was treated with 10 μM NAA alone. Both aliquots were incubated for 1 h at 4°C. The two aliquots then were combined in the presence of 2.5 mM DTT and used in a pull-down assay with biotinylated domain II peptide. The second treatment (Fig. 3d, lane 2) was the reciprocal of the first such that both TIR1myc– and TIR1myc+ extracts received 10 μM NAA, whereas juglone was administered only to the TIR1myc+ aliquot. If parvulin-type PPIases, which would be distributed similarly in the two extract types, were acting to change the conformation of proline(s) within the domain II degron, then there should be little difference in the recovery of TIR1myc from the two treatments; comparison of lanes 1 and 2 in Fig. 2d shows that this is not the case. The reduced interaction with TIR1myc from the treatment in which juglone was added to the TIR1myc+ aliquot is similar to that observed when juglone is added in the standard pull-down assay (compare with Fig. 3c), which indicates that juglone does not affect a PPIase operating on the domain II prolines and additionally that if proline isomerization is involved, it is of prolines in TIR1 or TIR1-associated proteins.
An alternative explanation is that the juglone adducts on the sulfhydryls of some or all of the 23 cysteines of TIR1 simply reduce its ability to interact with Aux/IAAs. Some support for this idea comes from our observation that the general sulfhydryl alkylating agent, NEM, also reduces SCFTIR1–Aux/IAA interaction (Fig. 3e). As with juglone, the inhibitory effect of NEM is abolished in the presence of DTT (Fig. 3e). An experiment identical to the partitioned juglone experiment but with juglone substituted by 2.5 mM NEM shows that NEM also affects TIR1 to reduce the SCFTIR1–Aux/IAA interaction (Fig. 3d, lanes 3 and 4). Although these effects of NEM and juglone may be quite general, we also considered the possibility that cysteine sulfhydryl chemistry might play a more specific role in regulating SCFTIR1–Aux/IAA interaction through redox sensing. We tested this theory by performing pulldowns, both with and without auxin, in the presence of 5 mM diamide (to promote disulfide bond formation) or 50 mM H2O2/0.5 mM CuCl2/0.5 mM ascorbate (to generate highly reactive oxygen species) and in extracts with artificially skewed ratios of reduced GSH/oxidized GSH. The manipulation of GSH/oxidized GSH ratios in either direction had no effect, whereas the imposition of relatively severe disulfide or reactive oxygen species stress resulted in only a modest reduction in TIR1myc recovery in both auxin-treated and untreated assays (Fig. 3f). These data are not consistent with redox regulation of SCFTIR1–Aux/IAA interaction.
Auxin-Enhanced SCFTIR1–Aux/IAA Interaction May Require Modification of TIR1 or Associated Proteins. Because auxin does not seem to affect domain II, we asked whether TIR1 could be predisposed to better interaction with Aux/IAAs by auxin treatment. This theory was tested by comparing the interaction between domain II peptides and TIR1myc recovered from auxin-treated and untreated extracts: anti-c-Myc antibody was used to immunoprecipitate TIR1myc from tir1-1[TIR1myc] extracts either mock-treated or treated with 10 μM NAA. These immunocomplexes were washed extensively (eight times = 640 bed volumes) and eluted by vigorous agitation in 0.5 M NaCl/2% Nonidet P-40/10% dioxane/0.1 M Tris·HCl, pH 7.0.
The eluates were diluted 1:10 with identical aliquots of tir1-1 extract (i.e., lacking TIR1myc) and used in pull-down assays with biotinylated domain II peptide. The reconstituted TIR1myc extracts contained equivalent levels of TIR1myc (Fig. 4a, lanes 4 and 5). Comparison of lanes 1 (mock-pretreated) and 2 (auxin-pretreated) of Fig. 4a shows that auxin pretreatment of TIR1myc subsequently enhanced its recovery on domain II peptide beads.
To control absolutely for possible auxin carry-through from the pretreatment, the experiment was repeated by using the following modification. Agarose beads without antibody were exposed to tir1-1 extract either mock-treated or treated with 10 μM NAA and then processed as described above. These eluates then were combined with the immunoprecipitated eluates such that –NAA IP eluate was added to +NAA control eluate, and +NAA IP eluate was added to –NAA control eluate. Each combined eluate then was diluted 1:10 with identical aliquots of tir1-1 extract and used in pull-down assays with biotinylated domain II peptide (Fig. 4b). Here again, the reconstituted extracts contained equivalent levels of TIR1myc (Fig. 4b, lanes 4 and 5), and auxin pretreatment of TIR1myc enhanced its recovery on domain II peptide beads (Fig. 4b, lanes 1 and 2).
The recovery of TIR1myc in these experiments is much lower than that in the standard pull-down assay, and thus the experiments are on the limits of Myc detection. This is because the reconstituted extracts contain considerably lower levels of TIR1myc than extracts made directly from tir1-1[TIR1myc] seedlings. Nonetheless, the increase in interaction caused by auxin pretreatment of TIR1myc is completely reproducible. Densitometric measurements over four experiments identical to that shown in Fig. 4b show that TIR1myc recovery is increased 4.41-fold (±0.13 SE) by auxin preconditioning of TIR1myc complexes (Fig. 4c), which suggests that there is an auxin-induced modification of TIR1 or TIR1-associated proteins, which facilitates enhanced interaction with Aux/IAAs. Addition of 10 μM NAA to these reconstituted TIR1myc extracts further enhanced the interaction with domain II peptide (Fig. 4 a and b, lane 3).
Discussion
The regulation of the interaction between SCFTIR1 and Aux/IAAs is unorthodox, conforming to none of the known modes of SCF–target interactions. A key question is whether auxin promotes the modification of target Aux/IAAs, increasing their affinity for SCFTIR1. We suggest that this is not the case and that instead the auxin-induced modification of TIR1 or tightly associated proteins is the regulatory step determining SCFTIR1–Aux/IAA interaction.
We have taken a generic approach to identify any auxin-regulated modifications to Aux/IAA domain II that might enhance interaction with SCFTIR1. Using MS, we could not detect any auxin-regulated modification of tagged Aux/IAA proteins and peptides. Our search was confined to the 13-aa degron of domain II, previously shown to be necessary and sufficient to confer auxin-enhanced instability on a translationally fused reporter protein (11). We did detect the apparent auxin-independent hydroxylation of one of the domain II prolines. Although seemingly irrelevant to the regulation of SCFTIR1–Aux/IAA interaction, this modification is puzzling, because it seems to be specific to the degron prolines. Several other tryptic fragments of GST-AXR2dII-containing prolines and even tandem prolines showed no signs of hydroxylation.
Similarly, it was not possible to predispose tagged domain II-containing proteins and peptides to interact better with TIR1 by pretreating them with auxin in plant extracts. In the first stage of these experiments, the domain II peptides/proteins were treated with auxin such that they could accumulate any auxin-induced modifications to which they might be susceptible before, and separate from, the second stage in which their ability to interact with TIR1myc was tested. To counter the possibility that the domain II targets might associate with endogenous TIR1-like proteins in the first stage, preventing later interaction with TIR1myc, a wash was performed before the second stage. As well as efficiently removing bound proteins, this wash also prevented the carry-over of exogenous auxin. Over the course of several experiments, there was never the slightest hint of a conditioning effect. Although it is possible that there are modifications to domain II peptides/proteins that are not revealed in these experiments, because they are not stable, there is a simpler explanation that accounts for all the data described here: that the auxin-induced interaction between Aux/IAAs and SCFTIR1 does not depend on auxin-regulated modification of domain II. In this model, domain II simply defines a structural module required for the interaction of Aux/IAA with SCFTIR1, with auxin acting elsewhere to promote the association.
This model is supported further by our finding that TIR1myc immunologically recovered from auxin-treated extracts interacts better with domain II peptides than that recovered from extracts not treated with auxin. This enhanced interaction with auxin pretreatment is less than the full auxin-enhanced interaction observed when auxin is added to extracts containing all the components together, and indeed, addition of auxin directly to the reconstituted extracts containing immunologically recovered TIR1myc further increases its interaction with domain II peptides (compare lanes 2 and 3 in Fig. 4 a and b). This result suggests that the modification to TIR1myc immunocomplexes does not fully survive the washing regime or does not persist at basal auxin levels in the second-stage extracts or does not represent the entire means by which auxin affects the enhancement of SCFTIR1–Aux/IAA interaction. In this context, it is interesting to note that the data do not distinguish between TIR1 or a protein tightly associated with it being modified in response to auxin or indeed a third-party protein becoming associated with or dissociated from TIR1 as a result of the auxin treatment. In any event, something happens to the TIR1 immunocomplexes to make their interaction with Aux/IAAs more likely and is thus the first indication of where auxin might act to promote the ubiquitination and destruction of Aux/IAAs. The obvious question is: What might this modification be?
Some information here can be derived from the pharmacological studies used to test for Aux/IAA modification. For example, there is now considerable literature describing how the pharmacological inhibition of phosphorylation and dephosphorylation do not affect the interaction between Aux/IAAs and SCFTIR1 (13, 14). To this we add our own finding that the kinase inhibitors staurosporine, U0126, and genistein and the phosphatase inhibitors calyculin A, okadaic acid, and NaF have no effect on the SCFTIR1–Aux/IAA interaction (data not shown). Although these studies do not definitively exclude a role for phosphorylation, they certainly give no indication of its involvement. Similarly, inhibitors of proline hydroxylation have no effect on the interaction (13, 14). In contrast, treatment of TIR1 with juglone and NEM, both of which form cysteine adducts, reduces auxin-enhanced SCFTIR1–Aux/IAA interaction. Because TIR1 contains 23 cysteine residues and the interaction seems not to involve redox regulation of cysteine sulfhydryl chemistry, this is likely a nonspecific effect.
The destruction of proteins by signal-induced SCF-mediated proteolysis is an important component of numerous and diverse eukaryotic signaling pathways. It will be interesting to determine what proportion of SCF–target interactions are regulated by modification of the target, the SCF complex, or both.
Supplementary Material
Acknowledgments
We thank R. Curwen and P. Ashton for assistance with MS and S. Day for critical reading of the manuscript. This work was supported by Biotechnology and Biological Science Research Council Grant 87/G14634.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SCF, SKP1, Cullin, and an F-box-containing protein; HIFα, hypoxia-inducible factor α; PPIase, peptidyl proline isomerase; GSH, glutathione; NAA, 1-napthalene acetic acid; MALDI, matrix-assisted laser desorption ionization; TOF, time of flight; NEM, N-ethylmaleimide.
References
- 1.Berleth, T. & Sachs, T. (2001) Curr. Opin. Plant Biol. 4, 57–62. [DOI] [PubMed] [Google Scholar]
- 2.Gray, W. M., Kepinski, S., Rouse, D., Leyser, O. & Estelle, M. (2001) Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- 3.Zenser, N., Ellsmore, A., Leasure, C. & Callis, J. (2001) Proc. Natl. Acad. Sci. USA 98, 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulmasov, T., Hagen, G. & Guilfoyle, T. J. (1999) Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiwari, S. B., Wang, X.-J., Hagen, G. & Guilfoyle, T. J. (2001) Plant Cell 13, 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abel, S. & Theologis, A. (1996) Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kepinski, S. & Leyser, O. (2002) Plant Cell 14, S81–S95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshaies, R. J. (1999) Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- 9.Ruegger, M., Dewey, E., Gray, W. M., Hobbie, L., Turner, J. & Estelle, M. (1998) Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rouse, D., Mackay, P., Stirnberg, P., Estelle, M. & Leyser, O. (1998) Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- 11.Ramos, J. A., Zenser, N., Leyser, O. & Callis, J. (2001) Plant Cell 13, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouellet, F., Overvoorde, P. J. & Theologis, A. (2001) Plant Cell 13, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian, Q., Nagpal, P. & Reed, J. W. (2003) Plant J. 36, 643–651. [DOI] [PubMed] [Google Scholar]
- 14.Dharmasiri, N., Dharmasiri, S., Jones, A. M. & Estelle, M. (2003) Curr. Biol. 13, 1418–1422. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida, Y., Tokunaga, F., Chiba, T., Iwai, K., Tanaka, K. & Tai, T. (2002) Nature 418, 438–442. [DOI] [PubMed] [Google Scholar]
- 16.Ivan, M., Kondo, K., Yang, H. F., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J., Lane, W. & Kaelin, W., Jr. (2001) Science 292, 464–448. [DOI] [PubMed] [Google Scholar]
- 17.Jaakkola, P., Mole, D. R., Tian, Y. M., Wilson, M. I., Gielbert, J., Gaskell, S., von Kriegsheim, A., Hebestreit, H., Mukherji, M., Schofield, C., et al. (2001) Science 292, 468–472. [DOI] [PubMed] [Google Scholar]
- 18.Zhao, Y., Dai, X., Blackwell, H. E., Schreiber, S. L. & Chory, J. (2003) Science 301, 1107–1110. [DOI] [PubMed] [Google Scholar]
- 19.Gray, W. M., del Pozo, J. C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W., Yang, M., Ma, H. & Estelle, M. (1999) Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennig, L., Christner, C., Kipping, M., Schelbert, B., Rücknagel, K. P., Grabley, S., Küllertz, G. & Fischer, G. (1998) Biochemistry 37, 5953–5960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.